Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Study Area and Samples
2.2. RNA Isolation
2.3. Transcriptome Analyses
2.4. Differential Gene Expression between Populations
2.5. Verification of Differentially Expressed Genes by Real-Time PCR
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, E.N. Relationships of the Palaearctic Lizards Assigned to the Genera Lacerta, Algyroides and Psammodromus (Reptilia: Lacertidae). Bull. Brit. Mus. Nat. Hist. Zool. 1973, 25, 291–366. [Google Scholar]
- Sacchi, R.; Scali, S.; Pupin, F.; Gentilli, A.; Galeotti, P.; Fasola, M. Microgeographic Variation of Colour Morph Frequency and Biometry of Common Wall Lizards. J. Zool. 2007, 273, 389–396. [Google Scholar] [CrossRef]
- Vercken, E.; Massot, M.; Sinervo, B.; Clobert, J. Colour Variation and Alternative Reproductive Strategies in Females of the Common Lizard Lacerta Vivipara. J. Evol. Biol. 2007, 20, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Arnold, N. Collins Field Guide to the Reptiles and Amphibians of Britain and Europe; Collins; New edition; Harper Collins: London, UK, 2002; ISBN 978-0-00-219964-3. [Google Scholar]
- Eimer, T. Untersuchungen über das Variiren der Mauereidechse: Ein Beitrag zur Theorie von der Entwicklung aus Constitutionellen Ursachen, Sowie zum Darwinismus; Nicolai (R. Stricker): Berlin, Germany, 1881. [Google Scholar]
- Podnar, M.; Mayer, W.; Tvrtković, N. Phylogeography of the Italian Wall Lizard, Podarcis Sicula, as Revealed by Mitochondrial DNA Sequences: Phylogeography of the Italian Wall Lizard. Mol. Ecol. 2005, 14, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Amori, G.; Angelici, F.; Frugis, S.; Gandol, G.; Groppali, R.; Lanza, B.; Relini, G.; Vicini, G. Vertebrata. In Checklist Delle Specie Della Fauna d’Italia; Calderini: Bologna, Italy, 1993; Volume 110, ISBN 88-7019-770-0. [Google Scholar]
- Corti, C.; Lo Cascio, P.; Böhme, W. The Lizards of Italy and Adjacent Areas; Frankfurt Contributions to Natural History/Frankfurter Beiträge zur Naturkunde; Edition Chimaira: Totnes, UK, 2002; Volume 14, ISBN 978-3-930612-68-0. [Google Scholar]
- Fulgione, D.; Lega, C.; Trapanese, M.; Buglione, M. Genetic Factors Implied in Melanin-based Coloration of the Italian Wall Lizard. J. Zool. 2015, 296, 278–285. [Google Scholar] [CrossRef]
- Raia, P.; Guarino, F.M.; Turano, M.; Polese, G.; Rippa, D.; Carotenuto, F.; Monti, D.M.; Cardi, M.; Fulgione, D. The Blue Lizard Spandrel and the Island Syndrome. BMC Evol. Biol. 2010, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, S.; Cipolla, R.; Corti, C.; Nappi, A. Notes on the Morphology of Podarcis sicula coerulea Compared to Other Nearby Islands and Mainland Populations of P. sicula. In Proceedings of the Atti del 6° Congresso Nazionale della Societas Herpetologica Italica; Stilgrafica: Rome, Italy, 2006. [Google Scholar]
- Monti, D.M.; Raia, P.; Vroonen, J.; Maselli, V.; Van Damme, R.; Fulgione, D. Physiological Change in an Insular Lizard Population Confirms the Reversed Island Syndrome: Physiology in Insular Lizards. Biol. J. Linn. Soc. Lond. 2013, 108, 144–150. [Google Scholar] [CrossRef]
- Buglione, M.; Ricca, E.; Petrelli, S.; Baccigalupi, L.; Troiano, C.; Saggese, A.; Rivieccio, E.; Fulgione, D. Gut Microbiota Plasticity in Insular Lizards under Reversed Island Syndrome. Sci. Rep. 2022, 12, 12682. [Google Scholar] [CrossRef]
- Baeckens, S.; Van Damme, R. Immunocompetence and Parasite Infestation in a Melanistic and Normally-Coloured Population of the Lacertid Lizard, Podarcis Siculus. Amphib.-Reptil. 2018, 39, 471–478. [Google Scholar] [CrossRef]
- Shuster, S.M.; Wade, M.J. Mating Systems and Strategies; Princeton University Press: Princeton, NJ, USA, 2003; ISBN 978-0-691-20688-2. [Google Scholar]
- Cooper, W.E.; Dimopoulos, I.; Pafilis, P. Sex, Age, and Population Density Affect Aggressive Behaviors in Island Lizards Promoting Cannibalism. Ethology 2015, 121, 260–269. [Google Scholar] [CrossRef]
- Lack, D. Competition for Food by Birds of Prey. J. Anim. Ecol. 1946, 15, 123. [Google Scholar] [CrossRef]
- Adler, G.H.; Levins, R. The Island Syndrome in Rodent Populations. Q. Rev. Biol. 1994, 69, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Blondel, J. Evolution and Ecology of Birds on Islands: Trends and Prospects. Vie Milieu 2000, 50, 205–220. [Google Scholar]
- McNab, B.K. Minimizing Energy Expenditure Facilitates Vertebrate Persistence on Oceanic Islands. Ecol. Lett. 2002, 5, 693–704. [Google Scholar] [CrossRef]
- Postma, E.; Van Noordwijk, A.J. Gene Flow Maintains a Large Genetic Difference in Clutch Size at a Small Spatial Scale. Nature 2005, 433, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Buglione, M.; Petrelli, S.; Maselli, V.; Trapanese, M.; Salvemini, M.; Aceto, S.; Di Cosmo, A.; Fulgione, D. Fixation of Genetic Variation and Optimization of Gene Expression: The Speed of Evolution in Isolated Lizard Populations Undergoing Reverse Island Syndrome. PLoS ONE 2019, 14, e0224607. [Google Scholar] [CrossRef] [PubMed]
- Rotger, A.; Tenan, S.; Igual, J.; Bonner, S.; Tavecchia, G. Life Span, Growth, Senescence and Island Syndrome: Accounting for Imperfect Detection and Continuous Growth. J. Anim. Ecol. 2023, 92, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.J.; Reed, R.D. Genome-Wide Regulatory Adaptation Shapes Population-Level Genomic Landscapes in Heliconius. Mol. Biol. Evol. 2019, 36, 159–173. [Google Scholar] [CrossRef]
- Verta, J.-P.; Jones, F.C. Predominance of Cis-Regulatory Changes in Parallel Expression Divergence of Sticklebacks. eLife 2019, 8, e43785. [Google Scholar] [CrossRef]
- Rosati, L.; Prisco, M.; Coraggio, F.; Valiante, S.; Scudiero, R.; Laforgia, V.; Andreuccetti, P.; Agnese, M. PACAP and PAC1 Receptor in the Reproductive Cycle of Male Lizard Podarcis Sicula. Gen. Comp. Endocrinol. 2014, 205, 102–108. [Google Scholar] [CrossRef]
- Agnese, M.; Rosati, L.; Prisco, M.; Coraggio, F.; Valiante, S.; Scudiero, R.; Laforgia, V.; Andreuccetti, P. The VIP/VPACR System in the Reproductive Cycle of Male Lizard Podarcis sicula. Gen. Comp. Endocrinol. 2014, 205, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ducrest, A.; Keller, L.; Roulin, A. Pleiotropy in the Melanocortin System, Coloration and Behavioural Syndromes. Trends Ecol. Evol. 2008, 23, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Meiri, S. Length–Weight Allometries in Lizards. J. Zool. 2010, 281, 218–226. [Google Scholar] [CrossRef]
- Trapanese, M.; Buglione, M.; Maselli, V.; Petrelli, S.; Aceto, S.; Fulgione, D. The First Transcriptome of Italian Wall Lizard, a New Tool to Infer about the Island Syndrome. PLoS ONE 2017, 12, e0185227. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Petrella, V.; Aceto, S.; Musacchia, F.; Colonna, V.; Robinson, M.; Benes, V.; Cicotti, G.; Bongiorno, G.; Gradoni, L.; Volf, P.; et al. De Novo Assembly and Sex-Specific Transcriptome Profiling in the Sand Fly Phlebotomus perniciosus (Diptera, Phlebotominae), a Major Old World Vector of Leishmania Infantum. BMC Genom. 2015, 16, 847. [Google Scholar] [CrossRef][Green Version]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Andrade, P.; Pinho, C.; Pérez i de Lanuza, G.; Afonso, S.; Brejcha, J.; Rubin, C.-J.; Wallerman, O.; Pereira, P.; Sabatino, S.J.; Bellati, A.; et al. Regulatory Changes in Pterin and Carotenoid Genes Underlie Balanced Color Polymorphisms in the Wall Lizard. Proc. Natl. Acad. Sci. USA 2019, 116, 5633–5642. [Google Scholar] [CrossRef]
- Musacchia, F.; Basu, S.; Petrosino, G.; Salvemini, M.; Sanges, R. Annocript: A Flexible Pipeline for the Annotation of Transcriptomes Able to Identify Putative Long Noncoding RNAs. Bioinformatics 2015, 31, 2199–2201. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Takahashi, K.; Taira, T.; Niki, T.; Seino, C.; Iguchi-Ariga, S.M.M.; Ariga, H. DJ-1 Positively Regulates the Androgen Receptor by Impairing the Binding of PIASxα to the Receptor. J. Biol. Chem. 2001, 276, 37556–37563. [Google Scholar] [CrossRef] [PubMed]
- Niki, T.; Takahashi-Niki, K.; Taira, T.; Iguchi-Ariga, S.M.M.; Ariga, H. DJBP: A Novel DJ-1-Binding Protein, Negatively Regulates the Androgen Receptor by Recruiting Histone Deacetylase Complex, and DJ-1 Antagonizes This Inhibition by Abrogation of This Complex. Mol. Cancer Res. 2003, 1, 247–261. [Google Scholar] [PubMed]
- Pigliucci, M.; Müller, G.B. Elements of an Extended Evolutionary Synthesis. In Evolution—The Extended Synthesis; Pigliucci, M., Müller, G.B., Eds.; The MIT Press: Cambridge, MA, USA, 2010; pp. 3–18. ISBN 978-0-262-51367-8. [Google Scholar]
- Laland, K.; Uller, T.; Feldman, M.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling-Smee, J.; Wray, G.A.; Hoekstra, H.E.; et al. Does Evolutionary Theory Need a Rethink? Nature 2014, 514, 161–164. [Google Scholar] [CrossRef]
- Hill, M.S.; Vande Zande, P.; Wittkopp, P.J. Molecular and Evolutionary Processes Generating Variation in Gene Expression. Nat. Rev. Genet. 2021, 22, 203–215. [Google Scholar] [CrossRef]
- Rosati, L.; Agnese, M.; Di Lorenzo, M.; Barra, T.; Valiante, S.; Prisco, M. Spermatogenesis and Regulatory Factors in the Wall Lizard Podarcis Sicula. Gen. Comp. Endocrinol. 2020, 298, 113579. [Google Scholar] [CrossRef]
- Ma, L.; Sun, B.; Cao, P.; Li, X.; Du, W. Phenotypic Plasticity May Help Lizards Cope with Increasingly Variable Temperatures. Oecologia 2018, 187, 37–45. [Google Scholar] [CrossRef]
- Losos, J.B.; Creer, D.A.; Glossip, D.; Goellner, R.; Hampton, A.; Roberts, G.; Haskell, N.; Taylor, P.; Ettling, J. Evolutionary Implications of Phenotypic Plasticity in the Hindlimb of the Lizard Anolis sagrei. Evolution 2000, 54, 301–305. [Google Scholar] [CrossRef]
- Price, T.D.; Qvarnström, A.; Irwin, D.E. The Role of Phenotypic Plasticity in Driving Genetic Evolution. Proc. R. Soc. Lond. B 2003, 270, 1433–1440. [Google Scholar] [CrossRef]
- Babu, M.M.; Luscombe, N.M.; Aravind, L.; Gerstein, M.; Teichmann, S.A. Structure and Evolution of Transcriptional Regulatory Networks. Curr. Opin. Struct. Biol. 2004, 14, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Gerstein, M. Genomic Analysis of the Hierarchical Structure of Regulatory Networks. Proc. Natl. Acad. Sci. USA 2006, 103, 14724–14731. [Google Scholar] [CrossRef] [PubMed]
- Moczek, A.P.; Sultan, S.; Foster, S.; Ledón-Rettig, C.; Dworkin, I.; Nijhout, H.F.; Abouheif, E.; Pfennig, D.W. The Role of Developmental Plasticity in Evolutionary Innovation. Proc. R. Soc. B 2011, 278, 2705–2713. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F.; Epel, D. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution; Sinauer Associates: Sunderland, MA, USA, 2009; Volume 4. [Google Scholar]
- Baldwin, J.M. A New Factor in Evolution (Continued). Am. Nat. 1896, 30, 536–553. [Google Scholar] [CrossRef]
- Simpson, G.G. The Baldwin Effect. Evolution 1953, 7, 110. [Google Scholar] [CrossRef]
- West-Eberhard, M.J. Developmental Plasticity and Evolution; Oxford University Press: Oxford, UK, 2003; ISBN 978-0-19-512234-3. [Google Scholar]


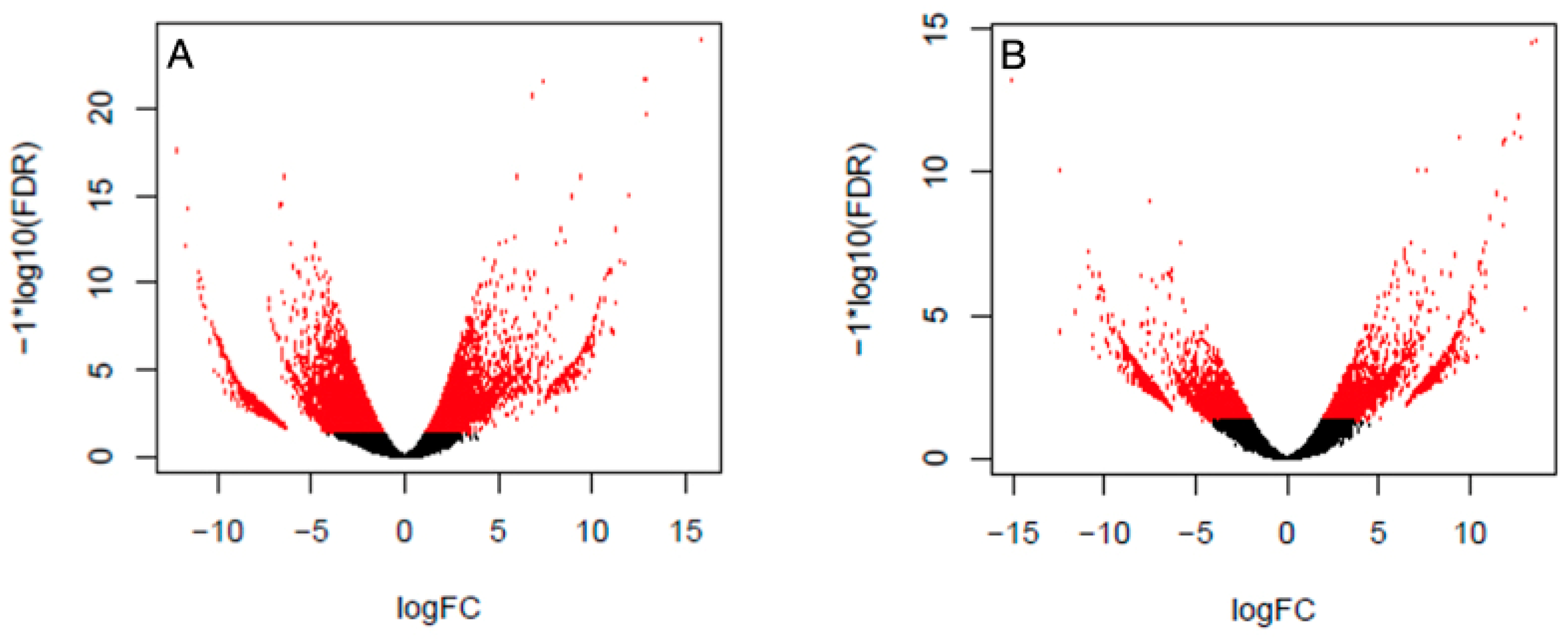
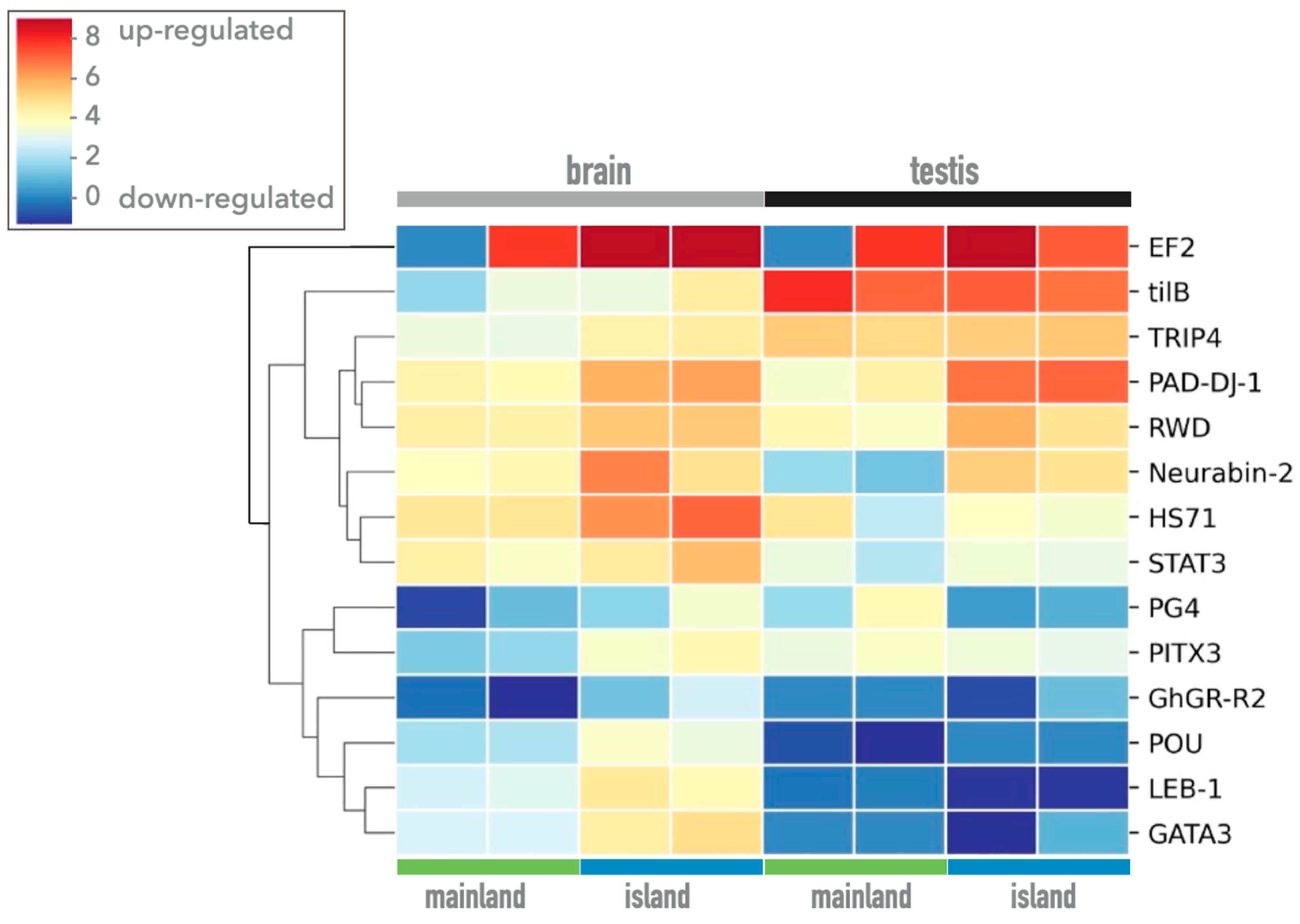

| Gene | Primer Name | Sequence Primer 5‘-3‘ | Amplicon Size (bp) |
|---|---|---|---|
| GnRH-R2 | GNRH_1524F | GTCAATGGAAACCCAACGGC | 166 |
| GNRH_1689R | ATGTTGAAGCAGGCGGAAGA | ||
| Actin | ACTINA_PS_R | GATCTGGCACCACACCTTCT | 104 |
| ACTINA_PSC_F | TCTTTTCTCTGTTGGCTTTGG |
| Transcripts FPKM > 1 | 373.072 |
| %GC | 45.04 |
| Average contig length | 821.89 |
| N50 (bp) | 1577 |
| Assembled bases (Mbp) | 307 |
| Gene | Description |
|---|---|
| GATA3 | Trans-acting T-cell-specific transcription factor GATA-3 |
| PAD-DJ-1 | Parkinsonism associated deglycase |
| PITX3 | Pituitary homeobox 2-like |
| HS71 | Heat shock cognate 71 kDa protein-like isoform X1 |
| tilB | Leucine rich repeat containing 6 |
| GnRH-R | Type 3/II gonadotropin-releasing hormone receptor |
| TRIP4 | Thyroid hormone receptor interactor 4 |
| STAT3 | Signal transducer and activator of transcription |
| EF2 | Eukaryotic elongation factor 2 |
| RWD | RWD domain containing 1 |
| PG4 | platelet glycoprotein 4 isoform X2 |
| POU | POU domain protein |
| LEB-1 | Isoform 6 of lymphoid enhancer-binding factor 1 |
| Neurabin-2 | Neurabin-2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fulgione, D.; Maselli, V.; Rivieccio, E.; Aceto, S.; Salvemini, M.; Buglione, M. Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation. Biology 2023, 12, 1478. https://doi.org/10.3390/biology12121478
Fulgione D, Maselli V, Rivieccio E, Aceto S, Salvemini M, Buglione M. Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation. Biology. 2023; 12(12):1478. https://doi.org/10.3390/biology12121478
Chicago/Turabian StyleFulgione, Domenico, Valeria Maselli, Eleonora Rivieccio, Serena Aceto, Marco Salvemini, and Maria Buglione. 2023. "Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation" Biology 12, no. 12: 1478. https://doi.org/10.3390/biology12121478
APA StyleFulgione, D., Maselli, V., Rivieccio, E., Aceto, S., Salvemini, M., & Buglione, M. (2023). Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation. Biology, 12(12), 1478. https://doi.org/10.3390/biology12121478








