Genetic Diversity and Signatures of Selection in the Roughskin Sculpin (Trachidermus fasciatus) Revealed by Whole Genome Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Sample Collection and Measurement of Growth Traits
- Total length: the vertical distance from the front end of the maxillary kiss to the end of the caudal fin;
- body length: the vertical distance from the front end of the maxillary kiss to the base of the caudal fin;
- body height: the vertical distance at the highest point of the fish body.
2.3. DNA Isolation and Sequencing
2.4. Variant Discovery and Quality Control
2.5. Population Genetic Diversity Analysis
2.6. Linkage Disequilibrium (LD) and Population Structure Analysis
2.7. Screening for Selective Sweeps
2.8. KEGG Enrichment Analysis of Candidate Genes
3. Results
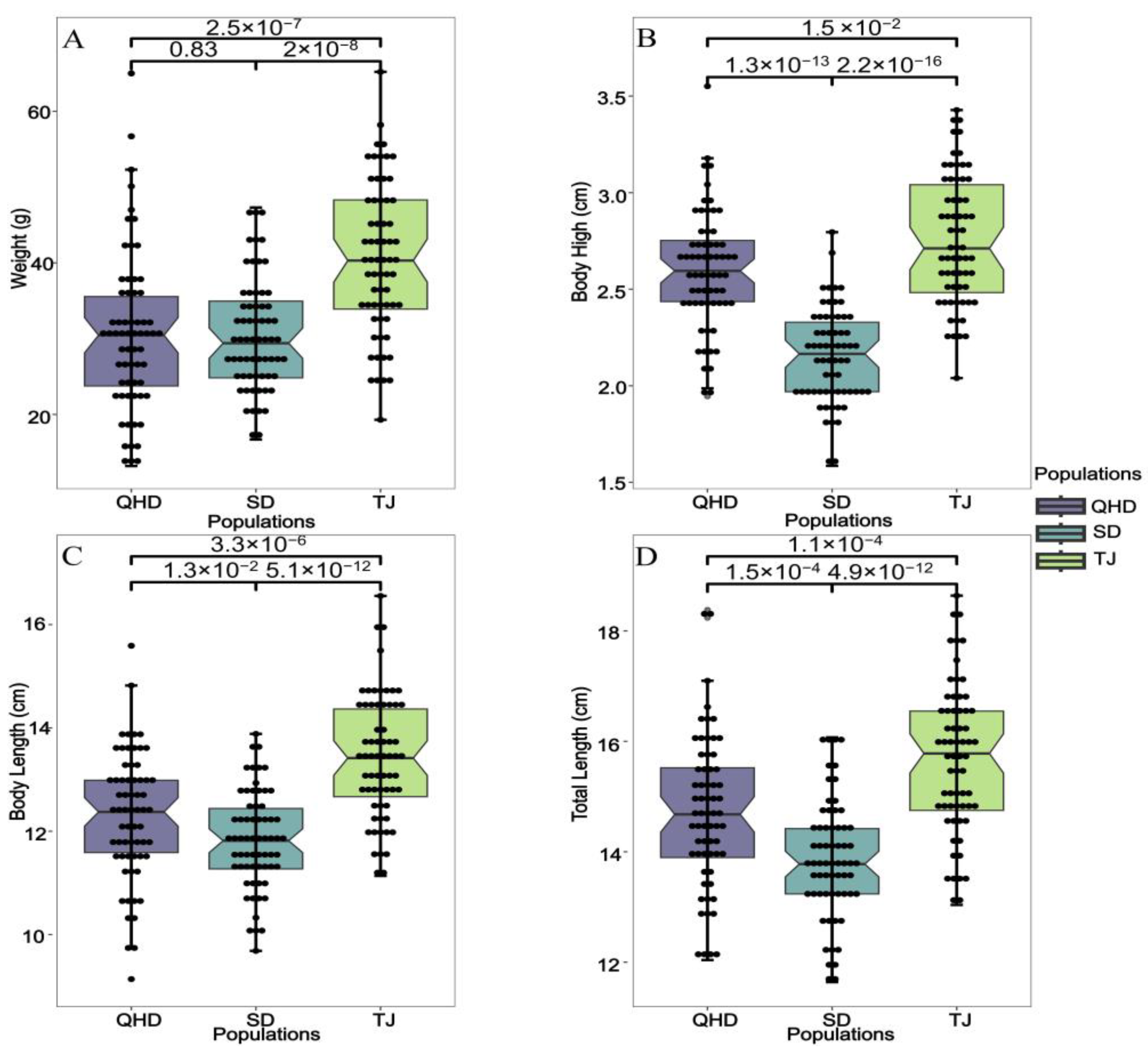
3.1. Comparison of Growth Traits in Different Geographic Groups
3.2. SNP Characteristics
3.3. Genetic Diversity
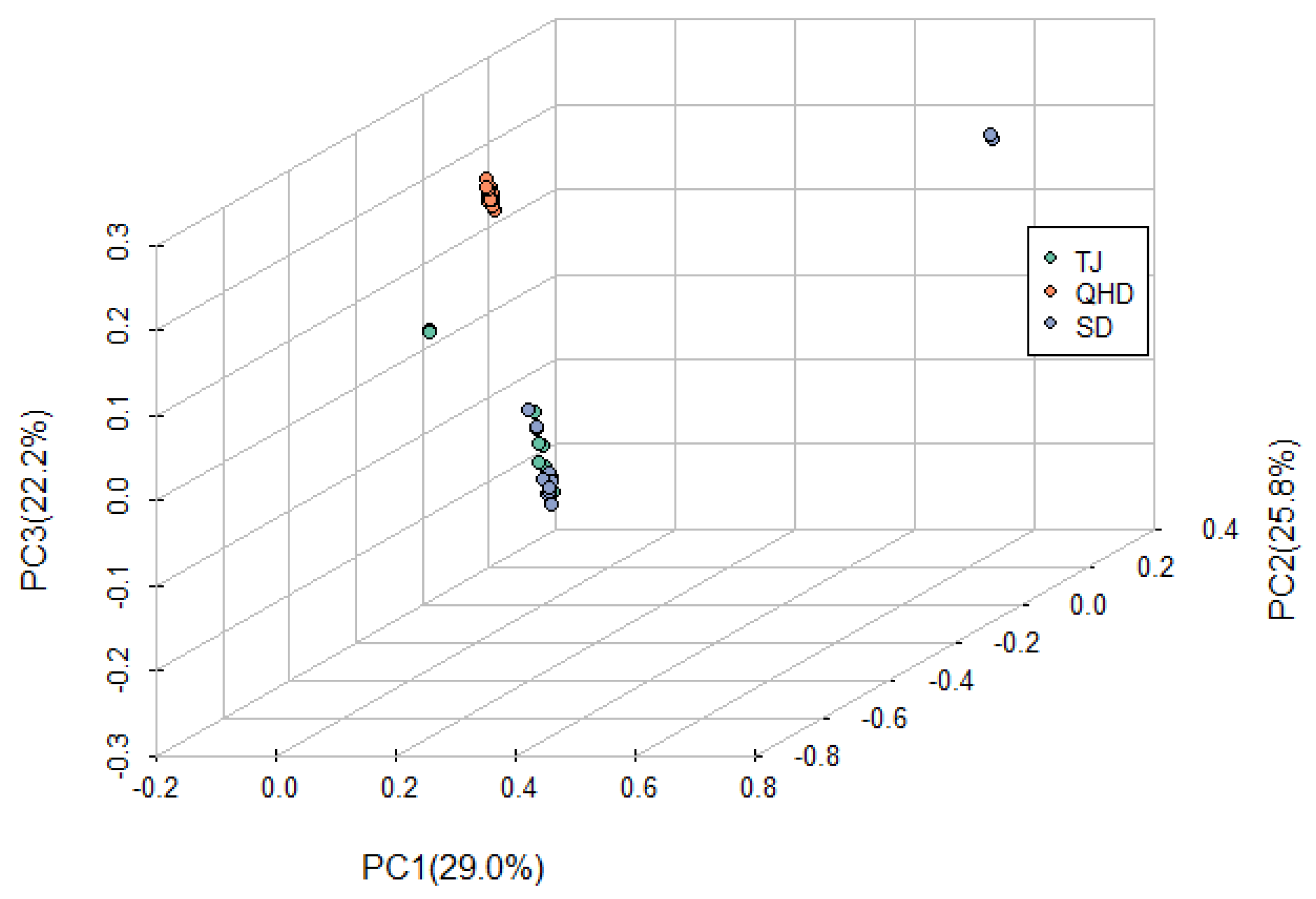
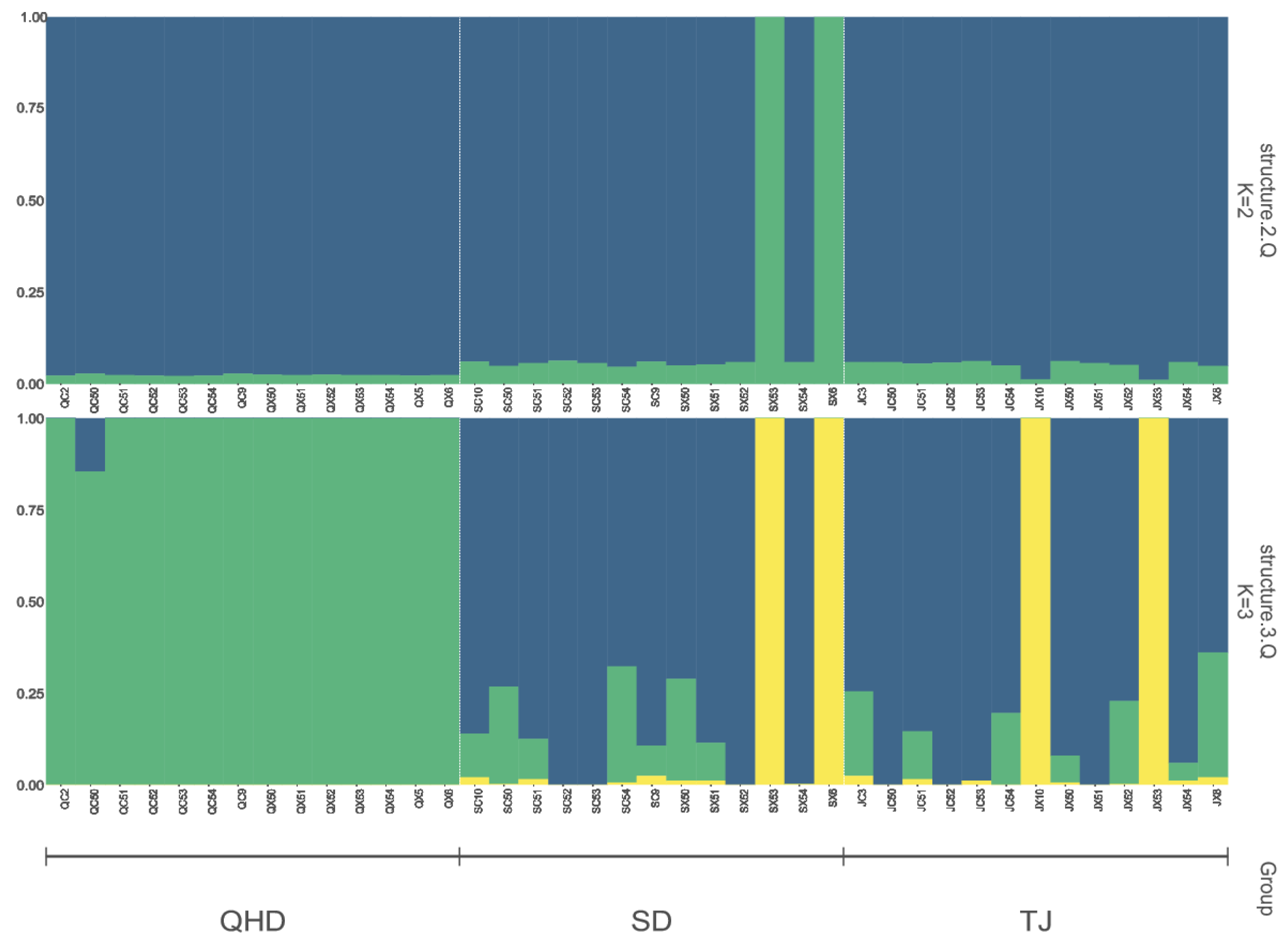
3.4. Population Differentiation and Linkage Disequilibrium
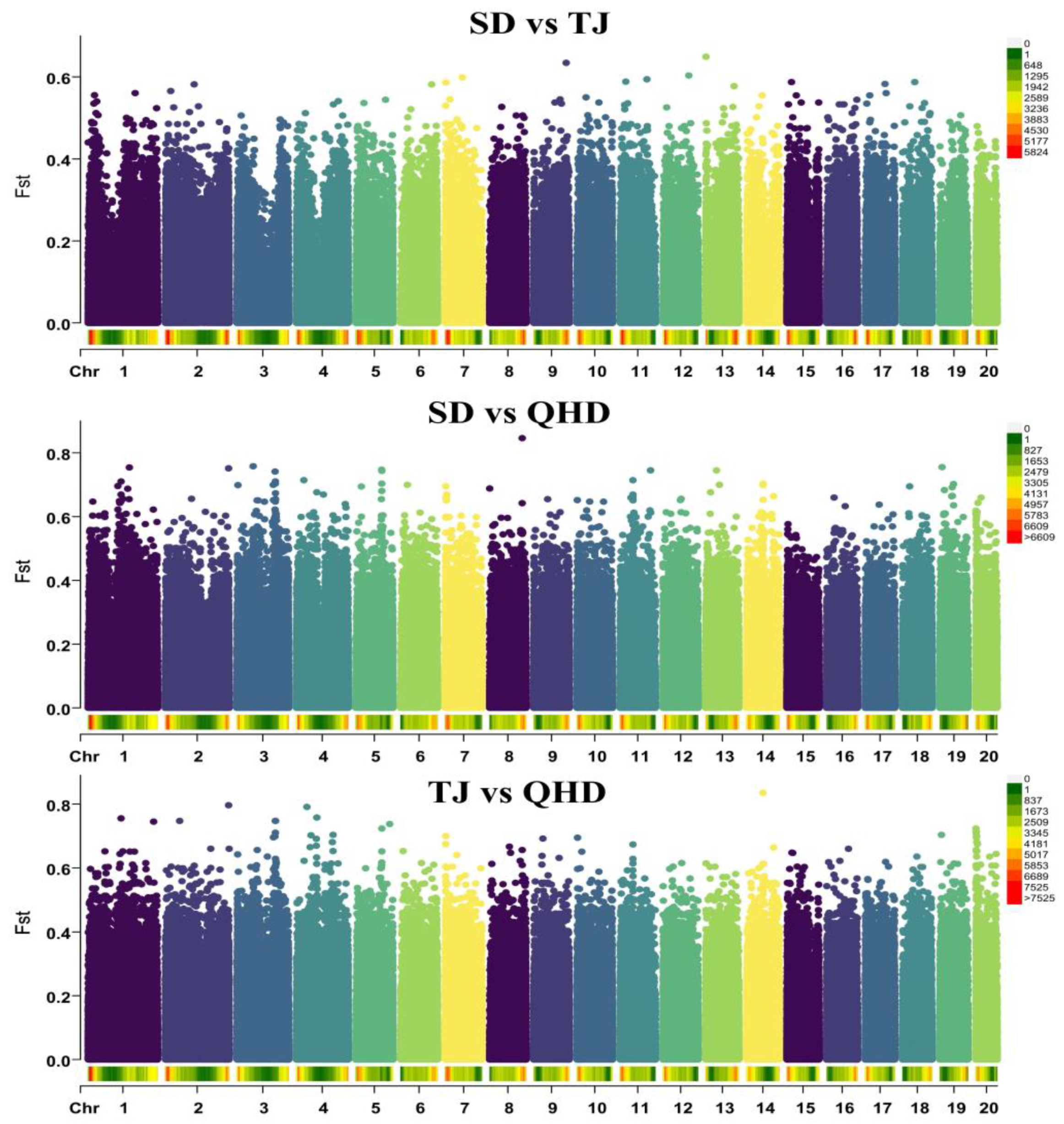
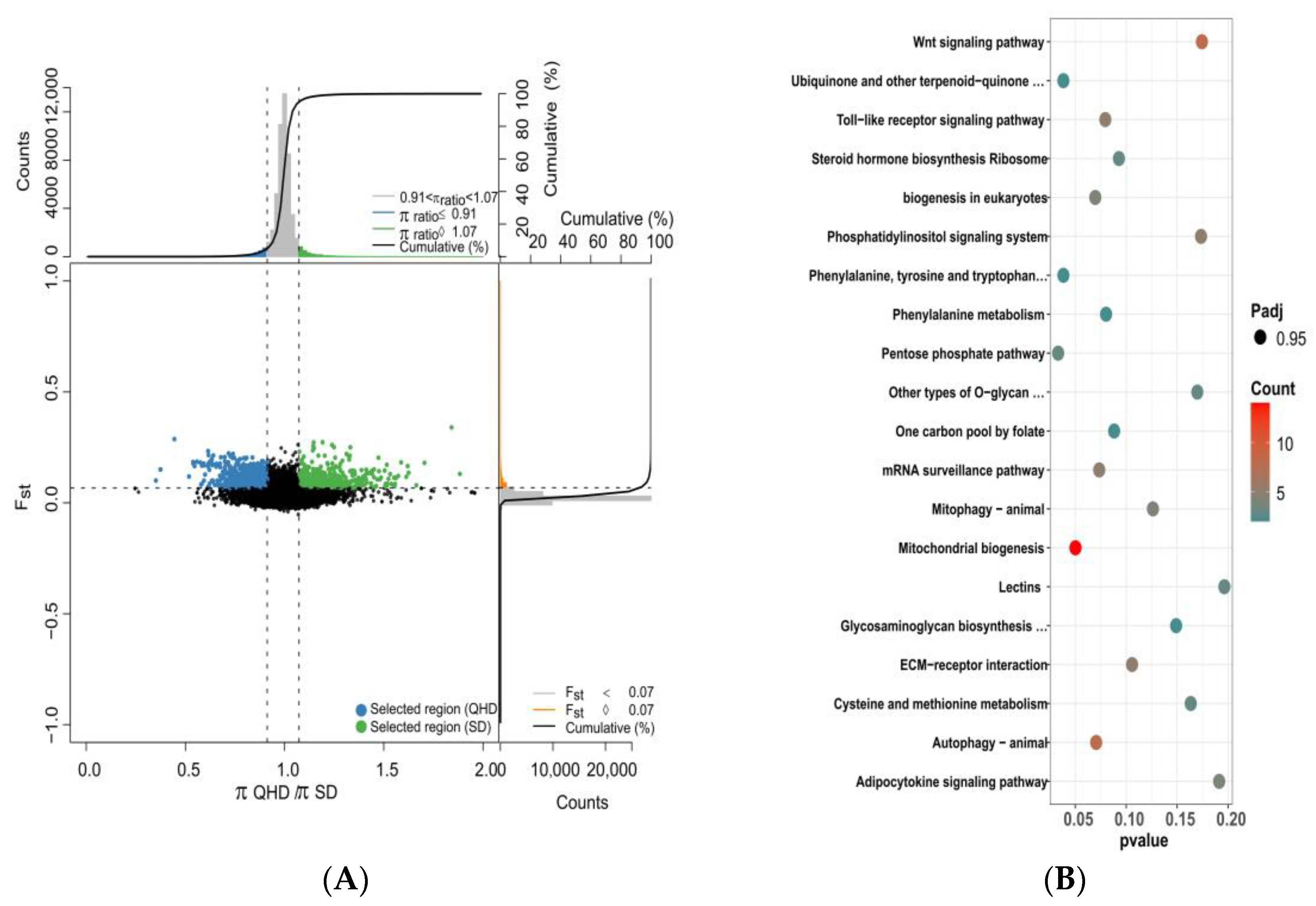
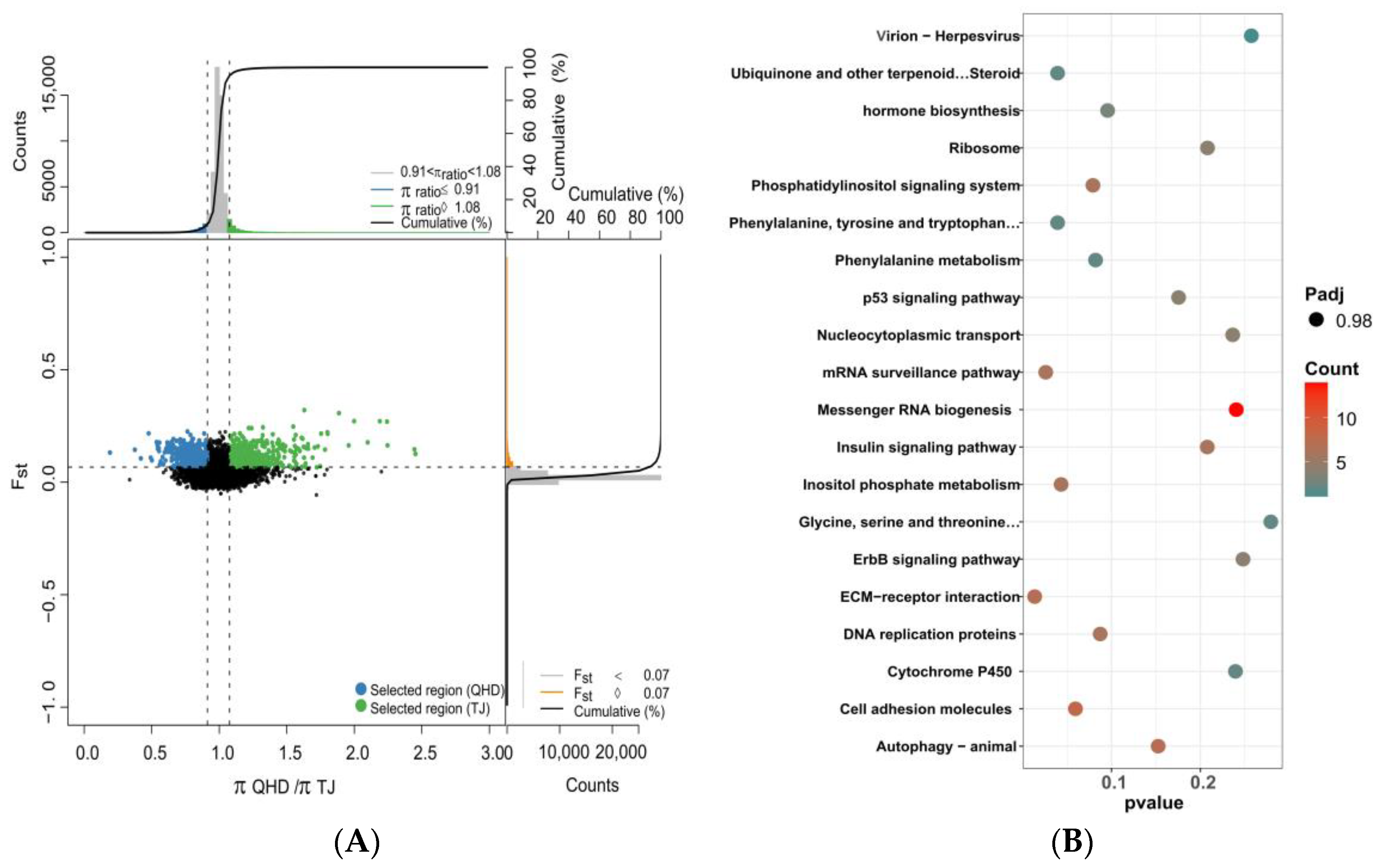
3.5. Selective Sweep and Selected Gene Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.Q.; Cheng, G.; Tang, Z.P. The Distribution of Roughskin Sculpin (Trachidermus fasciatus) in Yalu River Basin, China. J. Fudan Univ. 2001, 40, 471–476. (In Chinese) [Google Scholar]
- Wang, J.Q. Advances in studies on the ecology and reproductive biology of Trachidermus fasciatus Heckel. Acta Hydrobiol. Sin. 1999, 23, 729–734. (In Chinese) [Google Scholar]
- Yue, P.J.; Chen, Y.Y. China Red Data Book of Endangered Animal Species; Science Press: Beijing, China, 1988. (In Chinese) [Google Scholar]
- Fireston, J.L.; Jasieniuk, M. Small population size limits reproduction in an invasive grass through both demography and genetics. Oecologia 2013, 172, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Pan, L.D.; Liang, T.H.; Gan, H.C. Preliminary Study on Embryonic Development of the Roughskin Sculpin, Trachidermus fasciatus. J. Fudan Univ. 2004, 2, 250–254. (In Chinese) [Google Scholar]
- Zhang, W.; Xu, J.R.; Xu, X.M.; Yin, X.J. Comparative experiments on different factory culture modes of Trachidermus fasciatus. Fish. Mod. 2017, 44, 6–9. (In Chinese) [Google Scholar]
- Li, Y.L.; Xue, D.X.; Zhang, B.D.; Liu, J.X. Population genomic signatures of genetic structure and environmental selection in the catadromous roughskin sculpin Trachidermus fasciatus. Genome Biol. Evol. 2019, 11, 1751–1764. [Google Scholar] [CrossRef]
- Ren, G.; Hu, J.; Bao, Z.; Gao, T. Development and characterization of fourteen microsatellite loci in a threatened catadromous fish Trachidermus fasciatus. Conserv. Genet. Resour. 2011, 3, 685–687. [Google Scholar] [CrossRef]
- Bi, X.; Yang, Q.; Gao, T.; Li, C. The loss of genetic diversity during captive breeding of the endangered sculpin, Trachidermus fasciatus, based on ISSR markers: Implications for its conservation. Chin. J. Oceanol. Limnol. 2011, 29, 958–966. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, Z.Z.; Pan, L.D.; Tang, W.Q.; Wang, Q.; Geng, Y.H. Analysis of genetic diversity in wild populations of Trachidermus fasciatus by RAPD and the transformation of two SCAR markers. Zool. Res. 2012, 33, 203–210. (In Chinese) [Google Scholar] [CrossRef]
- Liu, Z.Z.; Zeng, Z.; Pan, L.D.; Tang, S.J.; Yang, J.Q.; Tang, W.Q. Isolation and characterization of polymorphic microsatellite markers for the endangered roughskin sculpin (Trachidermus fasciatus). Conserv. Genet. Resour. 2012, 4, 837–840. [Google Scholar] [CrossRef]
- Li, Y.L.; Xue, D.X.; Gao, T.X.; Liu, J.X. Genetic diversity and population structure of the roughskin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: Implications for its conservation and management. Conserv. Genet. 2016, 17, 921–930. [Google Scholar] [CrossRef]
- Ellegren, H. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 2014, 29, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A. Comparative genomics in ecological physiology: Toward a more nuanced understanding of acclimation and adaptation. J. Exp. Biol. 2012, 215, 884–891. [Google Scholar] [CrossRef]
- Nielsen, E.E.; Hemmer-Hansen, J.; Larsen, P.F.; Bekkevold, D. Population genomics of marine fishes: Identifying adaptive variation in space and time. Mol. Ecol. 2009, 18, 3128–3150. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, Z.; Zhang, X.; Zheng, X.; Li, J.; Jiang, Y.; Kuang, Y.; Zhang, Y.; Feng, J.; Li, C.; et al. Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio). BMC Genom. 2014, 15, 307. [Google Scholar] [CrossRef]
- Kjærner-Semb, E.; Ayllon, F.; Furmanek, T.; Wennevik, V.; Dahle, G.; Niemelä, E.; Ozerov, M.; Vähä, J.P.; Glover, K.A.; Rubin, C.J.; et al. Atlantic salmon populations reveal adaptive divergence of immune related genes—A duplicated genome under selection. BMC Genom. 2016, 17, 610. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.; Yang, L.; Gan, X.; Wu, B.; Gao, L.; Zeng, H.; Wang, X.; Liang, Z.; Wang, Y.; Fang, L.; et al. Whole genome sequencing of silver carp (Hypophthalmichthys molitrix) and bighead carp (Hypophthalmichthys nobilis) provide novel insights into their evolution and speciation. Mol. Ecol. Resour. 2021, 21, 912–923. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Q.; Tang, W.; Huang, Z.; Zhang, J. The evolutionary origin and domestication history of goldfish (Carassius auratus). Proc. Natl. Acad. Sci. USA 2020, 117, 29775–29785. [Google Scholar] [CrossRef]
- Xie, G.; Zhang, X.; Lv, F.; Sang, M.; Hu, H.; Wang, J.; Liu, D. Nanopore sequencing and Hi-C based de novo assembly of Trachidermus fasciatus genome. Genes 2021, 12, 692. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.; Danecek, P.; Scally, A.; Xue, Y.; Tyler-Smith, C.; Durbin, R. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 2016, 32, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Lorenzen, K.; Beveridge, M.C.M.; Mangel, M. Cultured fish: Integrative biology and management of domestication and interactions with wild fish. Biol. Rev. 2012, 87, 639–660. [Google Scholar] [CrossRef] [PubMed]
- González, S.; Flick, G.J.; O’Keefe, S.F.; Duncan, S.E.; McLean, E.; Craig, S.R. Composition of farmed and wild yellow perch (Perca flavescens). J. Food Compos. Anal. 2006, 19, 720–726. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Fernandez-Jover, D.; Black, K.D.; Ladoukakis, E.; Bayle-Sempere, J.T.; Sanchez-Jerez, P.; Dempster, T. Differentiating the wild or farmed origin of Mediterranean fish: A review of tools for sea bream and sea bass. Rev. Aquac. 2013, 5, 137–157. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Sanchez-Jerez, P.; Bayle-Sempere, J.T.; Sfakianakis, D.G.; Somarakis, S. Morphological differences between wild and farmed Mediterranean fish. Hydrobiologia 2012, 679, 217–231. [Google Scholar] [CrossRef]
- López, M.E.; Cádiz, M.I.; Rondeau, E.B.; Koop, B.F.; Yáñez, J.M. Detection of selection signatures in farmed coho salmon (Oncorhynchus kisutch) using dense genome-wide information. Sci. Rep. 2021, 11, 9685. [Google Scholar] [CrossRef]
- DeWoody, J.A.; Avise, J.C. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish. Biol. 2000, 56, 461–473. [Google Scholar] [CrossRef]
- Tseng, M.C.; Chen, C.A.; Kao, H.W.; Tzeng, W.N.; Lee, S.C. Polymorphisms of GA/GT microsateilite loci from Anguilla japonica. Mar. Biotechnol. 2001, 3, 275–280. [Google Scholar] [CrossRef]
- Maes, G.E.; Pujolar, J.M.; Raeymaekers, J.A.M.; Dannewitz, J.; Volckaert, F.A.M. Microsatellite conservation and Bayesian individual assignment in four Anguilla species. Mar. Ecol. Prog. Ser. 2006, 319, 251–261. [Google Scholar] [CrossRef][Green Version]
- Vu, N.T.; Ha, T.T.T.; Thuy, V.T.B.; Trang, V.T.; Nguyen, N.H. Population genomic analyses of wild and farmed striped catfish pangasianodon hypophthalmus in the lower Mekong river. J. Mar. Sci. Eng. 2020, 8, 471. [Google Scholar] [CrossRef]
- Fagbémi, M.N.A.; Pigneur, L.M.; André, A.; Smitz, N.; Gennotte, V.; Michaux, J.R.; Mélard, C.; Lalèyè, P.A.; Rougeot, C. Genetic structure of wild and farmed Nile tilapia (Oreochromis niloticus) populations in Benin based on genome wide SNP technology. Aquaculture 2021, 535, 736432. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Sremba, A.L.; Hancock-Hanser, B.; Branch, T.A.; LeDuc, R.L.; Baker, C.S. Circumpolar diversity and geographic differentiation of mtDNA in the critically endangered antarctic blue whale (Balaenoptera musculus intermedia). PLoS ONE 2012, 7, e32579. [Google Scholar] [CrossRef] [PubMed]
- Mbora, D.N.M.; McPeek, M.A. Endangered species in small habitat patches can possess high genetic diversity: The case of the Tana River red colobus and mangabey. Conserv. Genet. 2010, 11, 1725–1735. [Google Scholar] [CrossRef]
- Ellis, J.R.; Pashley, C.H.; Burke, J.M.; McCauley, D.E. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol. Ecol. 2006, 15, 2345–2355. [Google Scholar] [CrossRef]
- Pan, L.; Cai, F.; Ma, Z.; Jiang, X. Fisheries Science & Technology Information. Fish. Sci. Technol. Inf. 2010, 37, 211–214. (In Chinese) [Google Scholar]
- Yi, W.G.; Liang, P.; Liang, T.H.; Huang, Y.L. Roughskin Sculpin (Trachidermus fasciatus) in Yalu River Basin. Hebei Fish. 2006, 2, 53–54. (In Chinese) [Google Scholar]
- Oleksyk, T.K.; Smith, M.W.; O’Brien, S.J. Genome-wide scans for footprints of natural selection. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 185–205. [Google Scholar] [CrossRef]
- Bicskei, B.; Taggart, J.B.; Bron, J.E.; Glover, K.A. Transcriptomic comparison of communally reared wild, domesticated and hybrid Atlantic salmon fry under stress and control conditions. BMC Genet. 2020, 21, 57. [Google Scholar] [CrossRef]



| Category | Type | Count | Percent |
|---|---|---|---|
| Region | Intergenic | 3,331,449 | 47.0% |
| Intron | 2,985,075 | 41.4% | |
| Exon | 429,588 | 6.0% | |
| 5′ UTR | 125,011 | 3.9% | |
| 3′ UTR | 280,506 | 1.7% | |
| Mutation | Synonymous | 152,974 | 2.1% |
| Non-synonymous | 276,614 | 3.9% |
| Statistic | QHD | SD | TJ |
|---|---|---|---|
| Ho | 0.3524 | 0.3539 | 0.3549 |
| He | 0.3344 | 0.3343 | 0.3346 |
| MAF | 0.2437 | 0.2432 | 0.2434 |
| PN | 98.39% | 98.89% | 98.89% |
| D | 0.274212 | 0.278117 | 0.277829 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
San, L.; He, Z.; Liu, Y.; Zhang, Y.; Cao, W.; Ren, J.; Han, T.; Li, B.; Wang, G.; Wang, Y.; et al. Genetic Diversity and Signatures of Selection in the Roughskin Sculpin (Trachidermus fasciatus) Revealed by Whole Genome Sequencing. Biology 2023, 12, 1427. https://doi.org/10.3390/biology12111427
San L, He Z, Liu Y, Zhang Y, Cao W, Ren J, Han T, Li B, Wang G, Wang Y, et al. Genetic Diversity and Signatures of Selection in the Roughskin Sculpin (Trachidermus fasciatus) Revealed by Whole Genome Sequencing. Biology. 2023; 12(11):1427. https://doi.org/10.3390/biology12111427
Chicago/Turabian StyleSan, Lize, Zhongwei He, Yufeng Liu, Yitong Zhang, Wei Cao, Jiangong Ren, Tian Han, Bingbu Li, Guixing Wang, Yufen Wang, and et al. 2023. "Genetic Diversity and Signatures of Selection in the Roughskin Sculpin (Trachidermus fasciatus) Revealed by Whole Genome Sequencing" Biology 12, no. 11: 1427. https://doi.org/10.3390/biology12111427
APA StyleSan, L., He, Z., Liu, Y., Zhang, Y., Cao, W., Ren, J., Han, T., Li, B., Wang, G., Wang, Y., & Hou, J. (2023). Genetic Diversity and Signatures of Selection in the Roughskin Sculpin (Trachidermus fasciatus) Revealed by Whole Genome Sequencing. Biology, 12(11), 1427. https://doi.org/10.3390/biology12111427






