Genetic Databases and Gene Editing Tools for Enhancing Crop Resistance against Abiotic Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Genome Databases of Abiotic Stress Gene
2.1. PlantStress
2.2. Plant Stress Gene Database
2.3. Plant Stress Proteome Database (PlantPReS)
2.4. Plant miRNA ENcyclopedia (PmiREN)
2.5. Network-Based Rice Expression Analysis (NetREx)
2.6. PncStress
2.7. Pearl Millet Drought Transcriptome Database (PMDTDb)
3. Functional Genomic Approaches and Abiotic Stress Tolerance
3.1. Sequencing-Based Approaches
3.2. Genome-Wide Association Studies (GWAS)
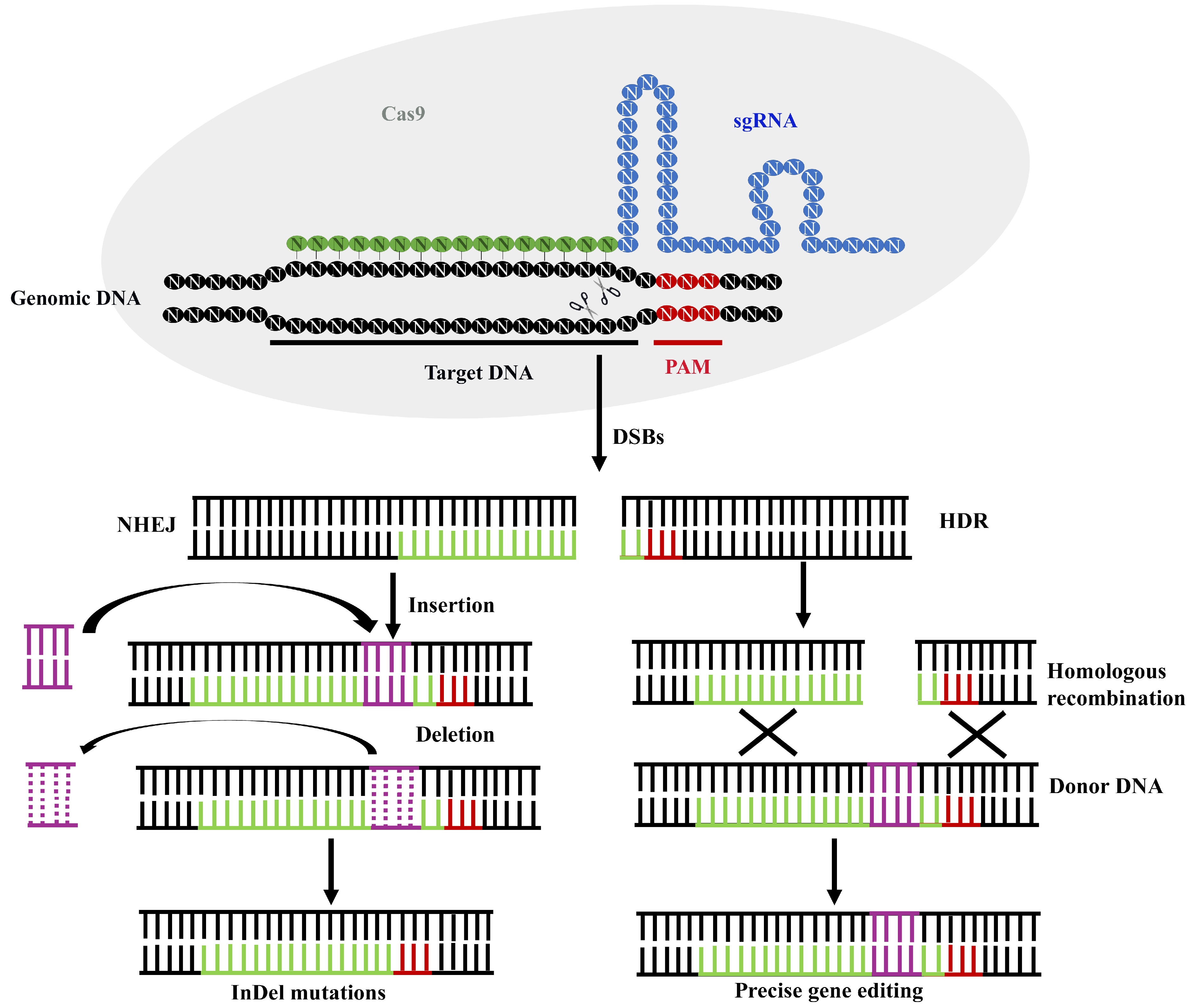
4. Mechanisms of CRISPR/CAS9 Genome Editing
5. Impact of CRISPR/Cas9-Based Genome Editing on Abiotic Stress Tolerance
5.1. Improvement in Drought Stress Tolerance using CRISPR/Cas System
5.2. Improvement in Salinity Stress Tolerance Using CRISPR/Cas System
5.3. Improvement in Heat Stress Tolerance Using CRISPR/Cas System
5.4. Improvement in Cold Stress Tolerance Using CRISPR/Cas System
5.5. Improvement in Metal and Herbicide Stress Tolerance Using CRISPR/Cas System
| Crops | Targeted Gene | Trait | References |
|---|---|---|---|
| Arabidopsis thaliana | OST2 | Drought tolerance | [117] |
| Arabidopsis thaliana | AVP1 | Drought tolerance | [3] |
| Arabidopsis thaliana | MIR169a and MIR827a | Drought tolerance | [118] |
| Arabidopsis thaliana | HAT | Drought tolerance | [119] |
| Arabidopsis thaliana | TRE1 | Drought tolerance | [120] |
| Arabidopsis thaliana | NAC07, NAC019, NAC055 | Drought tolerance | [121] |
| Arabidopsis thaliana | AITR3 and AITR4 | Drought and salinity tolerance | [7] |
| Arabidopsis thaliana | ACQO | Salinity tolerance | [141] |
| Arabidopsis thaliana | Oxp1 | Metal Stress tolerance | [13] |
| Brassica napus | BnaA6.RGA | Drought tolerance | [40] |
| Glycine max | AITR | Salinity tolerance | [41] |
| Glycine max | SOS1 | Salinity tolerance | [139] |
| Glycine max | ALS1 | Resistance to chlorsulfuron herbicide | [207] |
| Hordeum vulgare | ITPK1 | Salinity tolerance | [134] |
| Lactuca sativa | NCED4 | Heat tolerance | [179] |
| Lycopersicon esculentum | SlLBD40 | Drought tolerance | [127] |
| Lycopersicon esculentum | SlMAPK3 | Drought tolerance | [128,129] |
| Lycopersicon esculentum | SlHyPRP1 | Salinity tolerance | [37] |
| Lycopersicon esculentum | SlARF4 | Drought and salinity tolerance | [172] |
| Lycopersicon esculentum | SlCBF1 | Cold tolerance | [186] |
| Lycopersicon esculentum | SIAGL6 | Heat tolerance | [174] |
| Lycopersicon esculentum | CPK28, APX2 | Heat tolerance | [175] |
| Lycopersicon esculentum | BZR1 | Heat tolerance | [176] |
| Lycopersicon esculentum | ALS | Resistance to chlorsulfuron herbicide | [210] |
| Oryza sativa | SRL1, SRL2 | Drought tolerance | [133] |
| Oryza sativa | OsDST | Drought and salinity tolerance | [130] |
| Oryza sativa | OsERA1 | Drought tolerance | [132] |
| Oryza sativa | SAPK2 | Drought and salinity tolerance | [131] |
| Oryza sativa | RR22 | Salinity tolerance | [159] |
| Oryza sativa | miR535 | Drought and salinity tolerance | [166] |
| Oryza sativa | RAV2 | Salinity tolerance | [163] |
| Oryza sativa | RR9, RR10 | Salinity tolerance | [171] |
| Oryza sativa | NAC006 | Drought and heat tolerance | [180] |
| Oryza sativa | OTS1 | Salinity tolerance | [168] |
| Oryza sativa | HSP | Heat tolerance | [173] |
| Oryza sativa | HSA1 | Heat tolerance | [177] |
| Oryza sativa | MYB30 | Cold tolerance | [190] |
| Oryza sativa | Ann3 | Cold tolerance | [188] |
| Oryza sativa | PRP1 | Cold tolerance | [192] |
| Oryza sativa | WSL5 | Cold tolerance | [193,194] |
| Oryza sativa | HAK1 | Low cesium accumulation | [199] |
| Oryza sativa | LCT1,Nramp5 | Reduced cadmium accumulation | [198] |
| Oryza sativa | NRAMP1 | Reduced levels of heavy metals (Cd and Pb) | [14] |
| Oryza sativa | PRX2 | Potassium deficiency tolerance | [200] |
| Oryza sativa | ARM1 | Increase tolerance to high Arsenic | [201] |
| Oryza sativa | ALS | Resistance to Imazethapyr and imazapic herbicides | [209] |
| Oryza sativa | ALS | Herbicide resistance | [204] |
| Oryza sativa | ALS1 | Resistance to bispyribac-sodium herbicide | [208] |
| Oryza sativa | ALS | Resistance to Sulfonylurea, imidazolinone, triazolopyrimidine, pyr-imidinyl-thiobenzoates and sulfonyl-aminocarbonyl-triazolinone herbicides | [206] |
| Oryza sativa | EPSPS | Resistance to glyphosate resistance | [212] |
| Oryza sativa | C287T | Resistance to imazamox herbicide | [211] |
| Oryza sativa | ALS, EPSPS | Herbicide resistance | [215] |
| Oryza sativa | BEL | Resistance to bentazon herbicide | [216] |
| Oryza sativa | OsTubA2 | Resistance to dinitroaniline herbicide | [217] |
| Oryza sativa | Osbhlh024 | Salinity tolerance | [144] |
| Oryza sativa | OsDERF1 | Drought tolerance | [116] |
| Triticum aestivum | DREB1A/CBF3 | Drought tolerance | [122] |
| Triticum aestivum | DREB2, ERF3 | Drought tolerance | [123] |
| Triticum aestivum | HAG1 | Salinity tolerance | [143] |
| Zea mays | ARGOS8 | Drought tolerance | [135] |
| Zea mays | HKTI | Salinity tolerance | [150] |
| Zea mays | TMS5 | Heat tolerance | [178] |
| Zea mays | ALS2 | Resistance to chlorsulfuron herbicide | [203] |
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asati, R.; Tripathi, M.K.; Tiwari, S.; Yadav, R.K.; Tripathi, N. Molecular Breeding and Drought Tolerance in Cicer arietinum. Life 2022, 12, 1846. [Google Scholar] [CrossRef] [PubMed]
- Karunarathne, S.; Walker, E.; Sharma, D.; Li, C.; Han, Y. Genetic resources and precise gene editing for targeted improvement of barley abiotic stress tolerance. J. Zhejiang Univ. Sci. B 2023, 28, 1–24. [Google Scholar] [CrossRef]
- Park, J.J.; Dempewolf, E.; Zhang, W.; Wang, Z.Y. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional genomics in plant abiotic stress responses and tolerance: From gene discovery to complex regulatory networks and their application in breeding. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2022, 98, 470–492. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Tripathi, M.K.; Tiwari, S.; Tripathi, N.; Asati, R.; Chauhan, S.; Tiwari, P.N.; Payasi, D.K. Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants. Life 2023, 13, 1456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ Balance and Transport Regulatory Mechanisms in Weedy and Cultivated Rice (Oryza sativa L.) Under Salt Stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021, 21, 137. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Erdoğan, İ.; Cevher-Keskin, B.; Bilir, Ö.; Hong, Y.; Tör, M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology 2023, 12, 1037. [Google Scholar] [CrossRef]
- Shkryl, Y.; Yugay, Y.; Avramenko, T.; Grigorchuk, V.; Gorpenchenko, T.; Grischenko, O.; Bulgakov, V. CRISPR/Cas9-Mediated Knockout of HOS1 Reveals Its Role in the Regulation of Secondary Metabolism in Arabidopsis thaliana. Plants 2021, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Nahakpam, S.; Chaturvedi, V.; Singh, P. Cadmium-Induced Anatomical Abnormalities in Plants. In Cadmium Toxicity and Tolerance in Plants; Hasanuzzaman, M., Prasad, M.N.V., Fujita, M., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 111–139. [Google Scholar]
- Baeg, G.J.; Kim, S.H.; Choi, D.M.; Tripathi, S.; Han, Y.J.; Kim, J. CRISPR/Cas9-Mediated Mutation of 5-Oxoprolinase Gene Confers Resistance to Sulfonamide Compounds in Arabidopsis. Plant Biotechnol. Rep. 2021, 15, 753–764. [Google Scholar] [CrossRef]
- Chu, C.; Huang, R.; Liu, L.; Tang, G.; Xiao, J.; Yoo, H.; Yuan, M. The rice heavy-metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homoeostasis. Plant Cell Environ. 2022, 45, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for Improving HDR Efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef]
- Lu, G.; Wang, C.; Wang, G.; Mao, G.; Habben, J.E.; Chen, G.; Liu, M.; Shi, Y.; Wang, W.; Wang, X.; et al. Knockouts of Drought Sensitive Genes Improve Rice Grain Yield Under Both Drought and Well-Watered Field Conditions. Adv. Crop. Sci. Technol. 2020, 8, 444. [Google Scholar]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics 2021, 15, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zhao, Y.; Wang, B.; Hao, Y.; Wang, Y.; Li, Y.; Luo, W.; Zong, W.; Li, G.; Chen, S.; et al. Efficient CRISPR/Cas9-based plant genomic fragment deletions by microhomology-mediated end joining. Plant Biotechnol. J. 2020, 11, 2161–2163. [Google Scholar] [CrossRef]
- Wiles, M.V.; Qin, W.; Cheng, A.W.; Wang, H. CRISPR-Cas9-mediated genome editing and guide RNA design. Mamm Genome 2015, 26, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants. 2017, 3, 17018. [Google Scholar] [CrossRef] [PubMed]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Nájera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome editing in cereal crops: An overview. Transgenic Res. 2021, 30, 461–498. [Google Scholar] [CrossRef]
- Ming, M.; Ren, Q.; Pan, C.; He, Y.; Zhang, Y.; Liu, S.; Zhong, Z.; Wang, J.; Malzahn, A.A.; Wu, J.; et al. CRISPR-Cas12b enables efficient plant genome engineering. Nat. Plants 2020, 6, 202–208. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Kumar, M.; Ayzenshtat, D.; Marko, A.; Bocobza, S. Optimization of T-DNA configuration with UBIQUITIN10 promoters and tRNA-sgRNA complexes promotes highly efficient genome editing in allotetraploid tobacco. Plant Cell Rep. 2022, 41, 175–194. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K.; et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2017, 15, 257–268. [Google Scholar] [CrossRef]
- Zafar, K.; Sedeek, K.E.M.; Rao, G.S.; Khan, M.Z.; Amin, I.; Kamel, R.; Mukhtar, Z.; Zafar, M.; Mansoor, S.; Mahfouz, M.M. Genome Editing Technologies for Rice Improvement: Progress, Prospects, and Safety Concerns. Front. Genome Ed. 2020, 2, 5. [Google Scholar] [CrossRef]
- Smedley, M.A.; Hayta, S.; Clarke, M.; Harwood, W.A. CRISPR-Cas9 Based Genome Editing in Wheat. Curr Protoc. 2021, 1, e65. [Google Scholar] [CrossRef]
- Garcia-Gimenez, G.; Jobling, S.A. Gene editing for barley grain quality improvement. J. Cereal Sci. 2022, 103, 103394. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, Y.; Ma, L.; Guo, Y.; Ran, Y. Efficient Genome Editing in Setaria italica Using CRISPR/Cas9 and Base Editors. Front. Plant Sci. 2022, 12, 815946. [Google Scholar] [CrossRef]
- Tran, M.T.; Son, G.H.; Song, Y.J.; Nguyen, N.T.; Park, S.; Thach, T.V.; Kim, J.; Sung, Y.W.; Das, S.; Pramanik, D.; et al. CRISPR-Cas9-based precise engineering of SlHyPRP1 protein towards multi-stress tolerance in Tomato. Front. Plant Sci. 2023, 14, 1186932. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, S.; Wang, W.; Xiong, X.; Meng, F.; Cui, X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015, 34, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Park, S., II; Kim, H.B.; Jeon, H.J.; Kim, H. Agrobacterium-mediated capsicum annuum gene editing in two cultivars, hot pepper CM334 and bell pepper dempsey. Int. J. Mol. Sci. 2021, 22, 3921. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA Protein BnaA6.RGA, in Modulating Drought Tolerance by Interacting with the ABA Signaling Component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Dong, Q.; Li, Y.; Cheng, Y.; Wang, C.; et al. Mutation of GmAITR Genes by CRISPR/Cas9 Genome Editing Results in Enhanced Salinity Stress Tolerance in Soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef] [PubMed]
- Oz, M.T.; Altpeter, A.; Karan, R.; Merotto, A.; Altpeter, F. CRISPR/Cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance. Front. Genome Ed. 2021, 3, 673566. [Google Scholar] [CrossRef]
- Ambrosino, L.; Colantuono, C.; Diretto, G.; Fiore, A.; Chiusano, M.L. Bioinformatics Resources for Plant Abiotic Stress Responses: State of the Art and Opportunities in the Fast Evolving-Omics Era. Plants 2020, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Prabha, R.; Ghosh, I.; Singh, D.P. Plant Stress Gene Database: A collection of plant genes responding to stress condition. ARPN J. Sci. Technol. 2011, 1, 28–31. [Google Scholar]
- Mousavi, S.A.; Pouya, F.M.; Ghaffari, M.R.; Mirzaei, M.; Ghaffari, A.; Alikhani, M.; Ghareyazie, M.; Komatsu, S.; Haynes, P.A.; Salekdeh, G.H. PlantPReS: A database for plant proteome response to stress. J. Proteom. 2016, 143, 69–72. [Google Scholar] [CrossRef]
- Naika, M.; Shameer, K.; Mathew, O.K.; Gowda, R.; Sowdhamini, R. STIFDB2: An Updated Version of Plant Stress-Responsive TranscrIption Factor DataBase with Additional Stress Signals, Stress-Responsive Transcription Factor Binding Sites and Stress-Responsive Genes in Arabidopsis and Rice. Plant Cell Physiol. 2013, 54, e8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From data annotation to functional exploration of plant microRNAs. Nucleic Acids Res. 2022, 50, D1475–D1482. [Google Scholar] [CrossRef]
- Sircar, S.; Musaddi, M.; Parekh, N. NetREx: Network-based Rice Expression Analysis Server for abiotic stress conditions. Database 2022, 2022, baac060. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Y.; Hu, D.; Zhou, Y.; Hu, Y.; Chen, Y.; Chen, M. PncStress: A manually curated database of experimentally validated stress-responsive non-coding RNAs in plants. Database 2020, 2020, baaa001. [Google Scholar] [CrossRef]
- Jaiswal, S.; Antala, T.J.; Mandavia, M.K.; Chopra, M.; Jasrotia, R.S.; Tomar, R.S.; Kheni, J.; Angadi, U.B.; Iquebal, M.A.; Golakia, B.A.; et al. Transcriptomic signature of drought response in pearl millet (Pennisetum glaucum (L.) and development of web-genomic resources. Sci. Rep. 2018, 8, 3382. [Google Scholar] [CrossRef] [PubMed]
- Akpınar, B.A.; Lucas, S.J.; Budak, H. Genomics approaches for crop improvement against abiotic stress. Sci. World J. 2013, 2013, 361921. [Google Scholar] [CrossRef]
- Kumar, R.; Mustafiz, A.; Sahoo, K.K.; Sharma, V.; Samanta, S.; Sopory, S.K.; Pareek, A.; Singla-Pareek, S.L. Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 2012, 79, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.A.; Kondawar, V.; Jain, P.K.; Karuppayil, S.M.; Raju, N.L.; Vadez, V.; Varshney, R.K.; Srinivasan, R. Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and-susceptible genotypes of Cicer arietinum under terminal drought stress. BMC Plant Biol. 2011, 11, 70. [Google Scholar] [CrossRef]
- Sun, M.; Zhou, G.; Lee, S.; Chen, J.; Shi, R.Z.; Wang, S.M. SAGE is far more sensitive than EST for detecting low-abundance transcripts. BMC Genom. 2004, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Molina, C.; Rotter, B.; Horres, R.; Udupa, S.M.; Besser, B.; Bellarmino, L.; Baum, M.; Matsumura, H.; Terauchi, R.; Kahl, G.; et al. SuperSAGE: The drought stress-responsive transcriptome of chickpea roots. BMC Genom. 2008, 9, 553. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.; Johnson, M.; Bridgham, J.; Golda, G.; Lloyd, D.H.; Johnson, D.; Luo, S.; McCurdy, S.; Foy, M.; Ewan, M.; et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000, 18, 630–634. [Google Scholar] [CrossRef]
- Reinartz, J.; Bruyns, E.; Lin, J.Z.; Burcham, T.; Brenner, S.; Bowen, B.; Kramer, M.; Woychik, R. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief Funct. Genom. Proteomic 2002, 1, 95–104. [Google Scholar] [CrossRef]
- Nakano, M.; Nobuta, K.; Vemaraju, K.; Tej, S.S.; Skogen, J.W.; Meyers, B.C. Plant MPSS databases: Signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006, 34, D731–D735. [Google Scholar] [CrossRef]
- Nobuta, K.; Vemaraju, K.; Meyers, B.C. Methods for analysis of gene expression in plants using MPSS. Methods Mol. Biol. 2007, 406, 387–408. [Google Scholar] [CrossRef]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Kawaura, K.; Mochida, K.; Ogihara, Y. Genome-wide analysis for identification of salt-responsive genes in common wheat. Funct. Integr. Genom. 2008, 8, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Pervaiz, T.; Amjid, M.W.; El-kereamy, A.; Niu, S.-H.; Wu, H.X. MicroRNA and cDNA-Microarray as Potential Targets against Abiotic Stress Response in Plants: Advances and Prospects. Agronomy 2022, 12, 11. [Google Scholar] [CrossRef]
- Wong, C.E.; Li, Y.; Labbe, A.; Guevara, D.; Nuin, P.; Whitty, B.; Diaz, C.; Golding, G.B.; Gray, G.R.; Weretilnyk, E.A. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray Analysis of Rice d1 (RGA1) Mutant Reveals the Potential Role of G-Protein Alpha Subunit in Regulating Multiple Abiotic Stresses Such as Drought, Salinity, Heat, and Cold. Front. Plant Sci. 2016, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop. J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Zhang, L.; Niyitanga, S.; Afzal, M.Z.; Xu, Y.; Zhang, L.; Qi, J. Principles and approaches of association mapping in plant breeding. Trop. Plant Biol. 2020, 13, 212–224. [Google Scholar] [CrossRef]
- Lv, Y.; Ma, J.; Wei, H.; Xiao, F.; Wang, Y.; Jahan, N.; Hazman, M.; Qian, Q.; Shang, L.; Guo, L. Combining GWAS, Genome-Wide Domestication and a Transcriptomic Analysis Reveals the Loci and Natural Alleles of Salt Tolerance in Rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 912637. [Google Scholar] [CrossRef] [PubMed]
- Bilgrami, S.S.; Ramandi, H.D.; Shariati, V.; Razavi, K.; Tavakol, E.; Fakheri, B.A.; Mahdi Nezhad, N.; Ghaderian, M. Detection of genomic regions associated with tiller number in Iranian bread wheat under different water regimes using genome-wide association study. Sci. Rep. 2020, 10, 14034. [Google Scholar] [CrossRef] [PubMed]
- Saini, D.K.; Srivastava, P.; Pal, N.; Gupta, P.K. Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Theoret. Appl. Genet. 2022, 135, 1049–1081. [Google Scholar] [CrossRef] [PubMed]
- Tanin, M.J.; Saini, D.K.; Sandhu, K.S.; Pal, N.; Gudi, S.; Chaudhary, J.; Sharma, A. Consensus genomic regions associated with multiple abiotic stress tolerance in wheat and implications for wheat breeding. Sci. Rep. 2022, 12, 13680. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhu, Y.; Liu, H.; Liang, Z.; Zhang, M.; Zou, C.; Yuan, G.; Gao, S.; Pan, G.; Shen, Y.; et al. A Combination of a Genome-Wide Association Study and a Transcriptome Analysis Reveals circRNAs as New Regulators Involved in the Response to Salt Stress in Maize. Int. J. Mol. Sci. 2022, 23, 9755. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ning, L.; Yu, W.; Zhao, W.; Huang, F.; Yu, D.; Wang, H.; Cheng, H. Detection of Candidate Loci and Genes Related to Phosphorus Efficiency at Maturity through a Genome-Wide Association Study in Soybean. Agronomy 2022, 12, 2031. [Google Scholar] [CrossRef]
- Wu, D.Z.; Sato, K.; Ma, J.F. Genome-wide association mapping of cadmium accumulation in different organs of barley. New Phytol. 2015, 208, 817–829. [Google Scholar] [CrossRef]
- Visioni, A.; Tondelli, A.; Francia, E.; Pswarayi, A.; Malosetti, M.; Russell, J.; Thomas, W.; Waugh, R.; Pecchioni, N.; Romagosa, I.; et al. Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.). BMC Genom. 2013, 14, 424. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.; Maurer, A.; Pillen, K.; Brien, C.; Dowling, K.; Berger, B.; Eglinton, J.K.; March, T.J. Genome-wide association of barley plant growth under drought stress using a nested association mapping population. BMC Plant Biol. 2019, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Tarawneh, R.A.; Alqudah, A.M.; Nagel, M.; Börner, A. Genome-wide association mapping reveals putative candidate genes for drought tolerance in barley. Environ. Exp. Bot. 2020, 180, 104237. [Google Scholar] [CrossRef]
- Cai, S.G.; Wu, D.H.; Jabeen, Z.; Huang, Y.Q.; Huang, Y.C.; Zhang, G.P. Genome-wide association analysis of aluminum tolerance in cultivated and Tibetan wild barley. PLoS ONE. 2013, 8, e69776. [Google Scholar] [CrossRef]
- Zhou, G.F.; Broughton, S.; Zhang, X.Q.; Zhou, M.X.; Li, C.D. Genome-wide association mapping of acid soil resistance in barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 406. [Google Scholar] [CrossRef] [PubMed]
- Long, N.V.; Dolstra, O.; Malosetti, M.; Kilian, B.; Graner, A.; Visser, R.G.; van der Linden, C.G. Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2013, 126, 2335–2351. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, G.; Shabala, S.; Chen, Z.; Cai, S.; Li, C.; Zhou, M. Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 94. [Google Scholar] [CrossRef]
- Samineni, S.; Mahendrakar, M.D.; Hotti, A.; Chand, U.; Rathore, A.; Gaur, P.M. Impact of heat and drought stresses on grain nutrient content in Cicer arietinum: Genome-wide marker-trait associations for protein, Fe and Zn. Environ. Exp. Bot. 2022, 194, 104688. [Google Scholar] [CrossRef]
- Choudhary, S.; Isobe, S.; Chahota, R.K. Elucidation of drought tolerance potential of horsegram (Macrotyloma uniflorum var.) germplasm using genome wide association studies. Gene 2022, 819, 146241. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Magadán, A.H.; Dupuis, M.È.; Villion, M.; Moineau, S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS ONE 2012, 7, e40913. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.M.; Wang, T.; Randolph, P.B.; Arbab, M.; Shen, M.W.; Huang, T.P.; Matuszek, Z.; Newby, G.A.; Rees, H.A.; Liu, D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020, 38, 471–481. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, Z.; Unver, T.; Zhang, B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2020, 29, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhang, Y.; Propson, N.E.; Howden, S.E.; Chu, L.F.; Sontheimer, E.J.; Thomson, J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 2013, 110, 15644–15649. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and Engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef]
- Abadi, S.; Yan, W.X.; Amar, D.; Mayrose, I. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput. Biol. 2017, 13, e1005807. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A Convenient Software Toolkit for CRISPR-Based Genome Editing. Mol. Plant. 2017, 10, 1246–1249. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant. 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Minkenberg, B.; Zhang, J.; Xie, K.; Yang, Y. CRISPR-PLANT v2: An online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol. J. 2019, 17, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, G.; Li, J.; Zhang, X.; Huang, S.; Xiang, S.; Hu, X.; Liu, C. CRISPRlnc: A manually curated database of validated sgRNAs for lncRNAs. Nucl. Acid. Res. 2019, 47, D63–D68. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Rodiger, J.; Chung, V.; Viswanatha, R.; Mohr, S.E.; Hu, Y.; Perrimon, N. SNP-CRISPR: A Web Tool for SNP-Specific Genome Editing. G3 2020, 10, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Siegner, S.M.; Karasu, M.E.; Schröder, M.S.; Kontarakis, Z.; Corn, J.E. PnB Designer: A web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinform. 2021, 22, 101. [Google Scholar] [CrossRef]
- Liu, C.T.; Mao, B.G.; Yuan, D.Y.; Chu, C.C.; Duan, M.J. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop. J. 2022, 10, 13. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Joshi, A.; Song, H.G.; Yang, S.Y.; Lee, J.H. Integrated Molecular and Bioinformatics Approaches for Disease-Related Genes in Plants. Plants 2023, 12, 2454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 System Produces Specific and Homozygous Targeted Gene Editing in Rice in One Generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.X.; Mao, L.; Chen, B.; Xu, Y.; et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 23890. [Google Scholar] [CrossRef]
- Roca Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in arabidopsis by CRISPR/dCas9 fusion with a histone Acetyl Transferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef]
- Nuñez-Muñoz, L.; Vargas-Hernández, B.; Hinojosa-Moya, J.; Ruiz-Medrano, R.; Xoconostle-Cázares, B. Plant Drought Tolerance Provided Through Genome Editing of the Trehalose Gene. Plant Signal. Behav. 2021, 16, 1877005. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Pellegrineschi, A.; Reynolds, M.; Pacheco, M.; Brito, R.M.; Almeraya, R.; Yamaguchi-Shinozaki, K.; Hoisington, D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2004, 47, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 Genome Editing in Wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced Drought Tolerance by CRISPR/Cas9-Mediated SlMAPK3 Mutagenesis in Tomato Plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wang, L.; Zhao, R.; Sheng, J.; Zhang, S.; Li, R.; Shen, L. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in Tomato plants. BMC Plant Biol. 2019, 19, 354. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef]
- Vlčko, T.; Ohnoutková, L. Allelic Variants of CRISPR/Cas9 Induced Mutation in an Inositol Trisphosphate 5/6 Kinase Gene Manifest Different Phenotypes in Barley. Plants 2020, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Yang, J.F.; Li, M.; Xu, Z.S.; Fu, J.D. The Maize WRKY Transcription Factor ZmWRKY40 Confers Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900–1916. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A Putative Plasma Membrane Na+/H+ Antiporter GmSOS1 Is Critical for Salt Stress Tolerance in Glycine max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef]
- Feki, K.; Quintero, F.J.; Khoudi, H.; Leidi, E.O.; Masmoudi, K.; Pardo, J.M.; Brini, F. A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep. 2014, 33, 277–288. [Google Scholar] [CrossRef]
- Kim, S.T.; Choi, M.; Bae, S.J.; Kim, J.S. The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis. Int. J. Mol. Sci. 2021, 22, 11389. [Google Scholar] [CrossRef]
- Lee, H.M.; Choi, J.W.; Choi, M.S. Role of Nitric Oxide and Protein S-Nitrosylation in Ischemia-Reperfusion Injury. Antioxidants 2022, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lin, J.; Liu, X.; Chu, W.; Li, J.; Gao, Y.; An, K.; Song, W.; Xin, M.; Yao, Y.; et al. Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 2021, 186, 1951–1969. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.I.; Qi, Y.; Satoshi, Y.; Izumi, S.; Deng, X.P.; Soo Sang, K.; Hironori, K.; Kiyoshi, T. Overexpression of a new rice vacuolar antiporter regulating protein OsARP improves salt tolerance in tobacco. Plant Cell Physiol. 2008, 49, 880–890. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, L.; Xue, Y.; Qian, Z.; Lu, W.; Shou, H. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. J. Plant Biol. 2010, 53, 444–452. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium Transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y.; et al. OsHKT1;4-mediated Na(+) transport in stems contributes to Na(+) exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016, 16, 22. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2017, 217, 1161–1176. [Google Scholar] [CrossRef]
- Vu, T.V.; Sivankalyani, V.; Kim, E.J.; Doan, D.T.H.; Tran, M.T.; Kim, J.; Sung, Y.W.; Park, M.; Kang, Y.J.; Kim, J.Y. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in Tomato. Plant Biotechnol. J. 2020, 18, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet. 2015, 11, e1005617. [Google Scholar] [CrossRef]
- Huang, X.Y.; Chao, D.Y.; Gao, J.P.; Zhu, M.Z.; Shi, M.; Lin, H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; Munns, R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot. 2008, 59, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Hasthanasombut, S.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance. Plant Cell Tissue Organ Cult. 2011, 104, 79–89. [Google Scholar] [CrossRef]
- Song, S.Y.; Chen, Y.; Chen, J.; Dai, X.Y.; Zhang, W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Liu, C.T.; Mao, B.G.; Ou, S.J.; Wang, W.; Liu, L.C.; Wu, Y.B.; Chu, C.C.; Wang, X.P. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Yuan, X.; Sun, H.; Tang, Z.; Tang, H.; Zhang, H.; Huang, J. A novel little membrane protein confers salt tolerance in rice (Oryza sativa L.). Plant Mol. Biol. Report. 2016, 34, 524–532. [Google Scholar] [CrossRef]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, C.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Liu, D.; Zhang, G.; Qian, Y.; Sun, Z.; et al. ARGONAUTE2 Enhances Grain Length and Salt Tolerance by Activating BIG GRAIN3 to Modulate Cytokinin Distribution in Rice. Plant Cell 2020, 32, 2292–2306. [Google Scholar] [CrossRef]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a Potential Genetic Editing Target for Drought and Salinity Stress Tolerance in Oryza sativa. Plants 2020, 9, 1337. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. OsNAC45 is Involved in ABA Response and Salt Tolerance in Rice. Rice 2020, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Zhang, C.; Caine, R.S.; Gray, J.; Sadanandom, A. Rice SUMO protease Overly Tolerant to Salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice. Plant J. 2017, 92, 1031–1043. [Google Scholar] [CrossRef]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 Interaction Controls Seedling Growth under Salt Stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef]
- Bo, C.; Chen, H.; Luo, G.; Li, W.; Zhang, X.; Ma, Q.; Cheng, B.; Cai, R. Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 2020, 39, 135–148. [Google Scholar] [CrossRef]
- Wang, W.C.; Lin, T.C.; Kieber, J.; Tsai, Y.C. Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Zouine, M. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct. 2019, 3, e00145. [Google Scholar] [CrossRef] [PubMed]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, J.; Ding, S. The protein kinase CPK28 phosphorylates ascorbate peroxidase and enhances thermotolerance in Tomato. Plant Physiol. 2021, 186, 1302–1317. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Qiu, Z.; Kang, S.; He, L.; Zhao, J.; Zhang, S.; Hu, J.; Zeng, D.; Zhang, G.; Dong, G.; Gao, Z.; et al. The newly identified heat-stress sensitive albino 1 gene affects chloroplast development in rice. Plant Sci. 2018, 267, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.; Si, X.; Tian, Y.; Chen, K.; Liu, J.; Chen, H.; Gao, C. Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genom. 2017, 44, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-Resolution Analysis of the Efficiency, Heritability, and Editing Outcomes of CRISPR/Cas9-Induced Modifications of NCED4 in Lettuce (Lactuca sativa). G3 2018, 8, 1513–1521. [Google Scholar] [CrossRef]
- Wang, B.; Zhong, Z.; Wang, X.; Han, X.; Yu, D.; Wang, C.; Song, W.; Zheng, X.; Chen, C.; Zhang, Y. Knockout of the OsNAC006 Transcription Factor Causes Drought and Heat Sensitivity in Rice. Int. J. Mol. Sci. 2020, 21, 2288. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 2004, 55, 225–236. [Google Scholar] [CrossRef]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004, 45, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, Y.; Liu, C.; Zheng, Y.; Xu, M.; Wu, N.; Sheng, J.; Shen, L. Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem. Biophys. Res. Commun. 2015, 464, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Han, J.; Li, X.; Li, W.; Yang, Q.; Li, Z.; Cheng, Z.; Lv, L.; Zhang, L.; Han, D. Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in Fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiol. Biochem. 2023, 196, 270–280. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPR-Cas9-Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Kakeshpour, T.; Tamang, T.M.; Motolai, G.; Fleming, Z.W.; Park, J.E.; Wu, Q.; Park, S. CGFS-type glutaredoxin mutations reduce tolerance to multiple abiotic stresses in Tomato. Physiol Plant. 2021, 173, 1263–1279. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYB30 Transcription Factor Suppresses Cold Tolerance by Interacting with a JAZ Protein and Suppressing β-Amylase Expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR-Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a Subfamily of Abscisic Acid Receptor Genes Promote Rice Growth and Productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Nawaz, G.; Han, Y.; Usman, B.; Liu, F.; Qin, B.; Li, R. Knockout of OsPRP1, a gene encoding proline-rich protein, confers enhanced cold sensitivity in rice (Oryza sativa L.) at the seedling stage. Biotech 2019, 9, 254. [Google Scholar] [CrossRef]
- Liu, X.; Lan, J.; Huang, Y.; Cao, P.; Zhou, C.; Ren, Y.; He, N.; Liu, S.; Tian, Y.; Nguyen, T.; et al. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 2018, 69, 3949–3961. [Google Scholar] [CrossRef]
- Molla, K.A.; Shih, J.; Yang, Y. Single-nucleotide editing for zebra3 and wsl5 phenotypes in rice using CRISPR/Cas9-mediated adenine base editors. aBiotech 2020, 1, 106–118. [Google Scholar] [CrossRef]
- Ramazan, A.K.N.; Najafi, F.; Rezaei, M. The influence of cadmium toxicity on some physiological parameters as affected by iron in rice (Oryza Sativa L.) plant. J. Plant Nutr. 2014, 37, 1202–1213. [Google Scholar] [CrossRef]
- Asgher, M.; Khan, M.I.; Anjum, N.A.; Khan, N.A. Minimising toxicity of cadmium in plants-role of plant growth regulators. Protoplasma 2015, 252, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zou, W.; Meng, L.; Fan, X.; Xu, G.; Ye, G. Advances in the Uptake and Transport Mechanisms and QTLs Mapping of Cadmium in Rice. Int. J. Mol. Sci. 2019, 20, 3417. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017, 7, 14438. [Google Scholar] [CrossRef] [PubMed]
- Nieves Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.A. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system. Plant J. 2017, 92, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zheng, Y.; Xiao, K.; Wei, Y.; Zhu, Y.; Cai, Q.; Chen, L.; Xie, H.; Zhang, J. OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 2018, 495, 461–467. [Google Scholar] [CrossRef]
- Wang, F.Z.; Chen, M.X.; Yu, L.J.; Xie, L.J.; Yuan, L.B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K.; et al. OsARM1, an R2R3 MYB Transcription Factor, Is Involved in Regulation of the Response to Arsenic Stress in Rice. Front. Plant Sci. 2017, 8, 1868. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Huang, Y.; Wang, K. The Development of Herbicide Resistance Crop Plants Using CRISPR/Cas9-Mediated Gene Editing. Genes 2021, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Mol. Plant. 2016, 9, 628–631. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, S.; Meng, X.; Chai, Z.; Wang, D.; Yuan, Y.; Chen, K.; Jiang, L.; Li, J.; Gao, C. Generating broad-spectrum tolerance to ALS-inhibiting herbicides in rice by base editing. Sci. China Life Sci. 2021, 64, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Z.B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Li, S.; Ren, B.; Yan, F.; Spetz, C.; Li, X.; Zhou, X.; Zhou, H. Base-Editing-Mediated Artificial Evolution of OsALS1 In Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant. 2020, 13, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, Y.; Li, W.; Chen, Z.; Wang, J.; Fan, F.; Tao, Y.; Jiang, Y.; Zhu, Q.H.; Yang, J. Creating a novel herbicide-tolerance OsALS allele using CRISPR/Cas9-mediated gene editing. Crop. J. 2021, 9, 305–312. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and Tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef]
- Sony, S.K.; Kaul, T.; Motelb, K.F.A.; Thangaraj, A.; Bharti, J.; Kaul, R.; Verma, R.; Nehra, M. CRISPR/Cas9-mediated homology donor repair base editing confers glyphosate resistance to rice (Oryza sativa L.). Front. Plant Sci. 2023, 14, 1122926. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, J.; Chen, B.; Mo, S.; Lian, L.; Luo, Y.; Ding, D.; Ding, Y.; Cao, Q.; Li, Y.; et al. A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants. 2021, 7, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, And EPSPS in Tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Xu, R.; Li, H.; Qin, R.; Wang, L.; Li, L.; Wei, P.; Yang, J. Gene targeting using the Agrobacterium tumefaciens mediated CRISPR-Cas system in rice. Rice 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Kuang, Y.; Yan, F.; Li, S.; Ren, B.; Gosavi, G.; Spetz, C.; Li, X.; Wang, X.; Zhou, X.; et al. Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol. J. 2021, 19, 5–7. [Google Scholar] [CrossRef]
| Name | Cas | Resources | PAM Sequence | PAM Location | Reference |
|---|---|---|---|---|---|
| SpCas9 | Cas9 | Streptococcus pyogenes | NGG | 3′ | [84] |
| St1Cas9 | Cas9 | Streptococcus thermophilus | NNAGAAW or | 3′ | [86] |
| NGGNG | |||||
| SaCas9 | Cas9 | Streptococcus aureus | NNGRRT | 3′ | [87] |
| NmCas9 | Cas9 | Neisseria meningitidis | NNNNGATT | 3′ | [97] |
| FnCas9 | Cas9 | Francisella novicida | NGG | 3′ | [98] |
| CjCas9 | Cas9 | Campylobacter jejuni | NNNNRYAC | 3′ | [99] |
| AsCas12a | Cas12a(cpf1) | Acidaminococcus sp. | TTTV | 5′ | [25] |
| LbCas12a | Cas12a(cpf1) | Lachnospiraceae bacterium | TTTV | 5′ | [25] |
| FnCas12a | Cas12a(cpf1) | Francisella novicida | TTTN or YTN | 5′ | [25] |
| LsCas13 | Cas13(C2c2) | Leptotrichia shahii | [100] | ||
| Cas14 | Cas14 | Archaea | [101] | ||
| FnCas9 variant | Cas9 | Modified FnCas9 | YG | 3′ | [98] |
| Modified SpCas9 | Cas9 | Engineered SpCas9 | NGA or NAG | 3′ | [102] |
| SaCas9-KKH | Cas9 | Engineered SaCas9 | NNNRRT | 3′ | [88] |
| SpCas9-HF | Cas9 | Engineered SpCas9 | NGG | 3′ | [89] |
| eSpCas9 | Cas9 | Engineered SpCas9 | NGG | 3′ | [90] |
| SpCas9-NG | Cas9 | Engineered SpCas9 | NG | 3′ | [85] |
| Sniper-Cas9 | Cas9 | Engineered SpCas9 | NGG | 3′ | [91] |
| evoCas9 | Cas9 | Mutated SpCas9 | NGG | 3′ | [92] |
| HypaCas9 | Cas9 | Mutated SpCas9-HF | NGG | 3′ | [93] |
| Cas9-NRNH | Cas9 | Engineered SpCas9 | NRNH | 3′ | [94] |
| SpG | Cas9 | Engineered SpCas9 | NGN | 3′ | [95] |
| SpRY | Cas9 | Engineered SpCas9 | NRN or NYN | 3′ | [95] |
| Tool | Organism | Major Feature | Weblink |
|---|---|---|---|
| CHOPCHOP | >100 species, including plants | Provides several predictive models and primers. Visualizing the genomic location of genes and targets [103]. | https://chopchop.cbu.uib.no/, accessed on 23 August 2023 |
| Cas-OFFinder | >100 species, including plants | Searches potential off-target sites [104]. | http://www.rgenome.net/cas-offinder/, accessed on 23 August 2023 |
| CCTop | >100 species | Predictes off-target impacts and sgRNA efficiency using CRISPRater with custom in vitro transcription. Searching for single and multiple queries [105]. | https://cctop.cos.uni-heidelberg.de/, accessed on 23 August 2023 |
| CRISTA | >100 species | Detectes off-target, providing machine learning framework, including DNA/RNA genomic information and RNA thermodynamics [106]. | https://crista.tau.ac.il/, accessed on 23 August 2023 |
| CRISPR-GE | >40 plant species | PCR sequencing result analysis. Provides software toolkits, primer design, and on-target amplification [107]. | http://skl.scau.edu.cn/, accessed on 23 August 2023 |
| CRISPR-P | 49 plant species | Providing on-target and off-target scoring and gRNA sequence analysis [108] | http://crispr.hzau.edu.cn/CRISPR2/, accessed on 23 August 2023 |
| CRISPR-PLANT V2 | 7 plant species | Allows selection of particular chromosomes and a resource for specific gRNA spacer sequences [109]. | http://omap.org/crispr2/, accessed on 23 August 2023 |
| CRISPRlnc | 10 species | Provides hundreds of lncRNAs and thousands of validated sgRNA [110]. | http://www.crisprlnc.org/, accessed on 23 August 2023 |
| SNP-CRISPR | 9 plants and animal species | Designing sgRNAs (NGG and NAG) for targeting SNPs or Indels [111]. | https://www.flyrnai.org/tools/snp_crispr/web/, accessed on 23 August 2023 |
| PnB Designer | O. sativa, V. vinifera | Designing sgRNAs for base editors and pegRNAs for prime editors [112]. | https://fgcz-shiny.uzh.ch/PnBDesigner/, accessed on 23 August 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, A.; Yang, S.-Y.; Song, H.-G.; Min, J.; Lee, J.-H. Genetic Databases and Gene Editing Tools for Enhancing Crop Resistance against Abiotic Stress. Biology 2023, 12, 1400. https://doi.org/10.3390/biology12111400
Joshi A, Yang S-Y, Song H-G, Min J, Lee J-H. Genetic Databases and Gene Editing Tools for Enhancing Crop Resistance against Abiotic Stress. Biology. 2023; 12(11):1400. https://doi.org/10.3390/biology12111400
Chicago/Turabian StyleJoshi, Alpana, Seo-Yeon Yang, Hyung-Geun Song, Jiho Min, and Ji-Hoon Lee. 2023. "Genetic Databases and Gene Editing Tools for Enhancing Crop Resistance against Abiotic Stress" Biology 12, no. 11: 1400. https://doi.org/10.3390/biology12111400
APA StyleJoshi, A., Yang, S.-Y., Song, H.-G., Min, J., & Lee, J.-H. (2023). Genetic Databases and Gene Editing Tools for Enhancing Crop Resistance against Abiotic Stress. Biology, 12(11), 1400. https://doi.org/10.3390/biology12111400







