Bioinformatic Analysis Reveals the Role of Translation Elongation Efficiency Optimisation in the Evolution of Ralstonia Genus
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Ecological Diversity of Ralstonia
1.1.1. Soil Bacteria
1.1.2. Phytopathogens
1.1.3. Cupriavidus Necator: An Outgroup
1.2. The Genome Structure of Bacteria Belonging to Ralstonia Genus
2. Materials and Methods
2.1. Genome Assemblies Acquisition and Refinement of Their Taxonomical Identity
2.2. Elongation Efficiency Calculation
2.3. Potentially Highly Expressed Genes Analysis
2.4. The Functional Analysis of Potentially Highly Expressed Genes: Clusters of Orthologous Groups (COGs)
2.5. Analysis of Translation Elongation Characteristics of Orthologous Genes
2.6. Translation Elongation Efficiency and Divergence Index
3. Results
3.1. The Distribution of Ralstonia Genomes by Translation Elongation Optimization Types
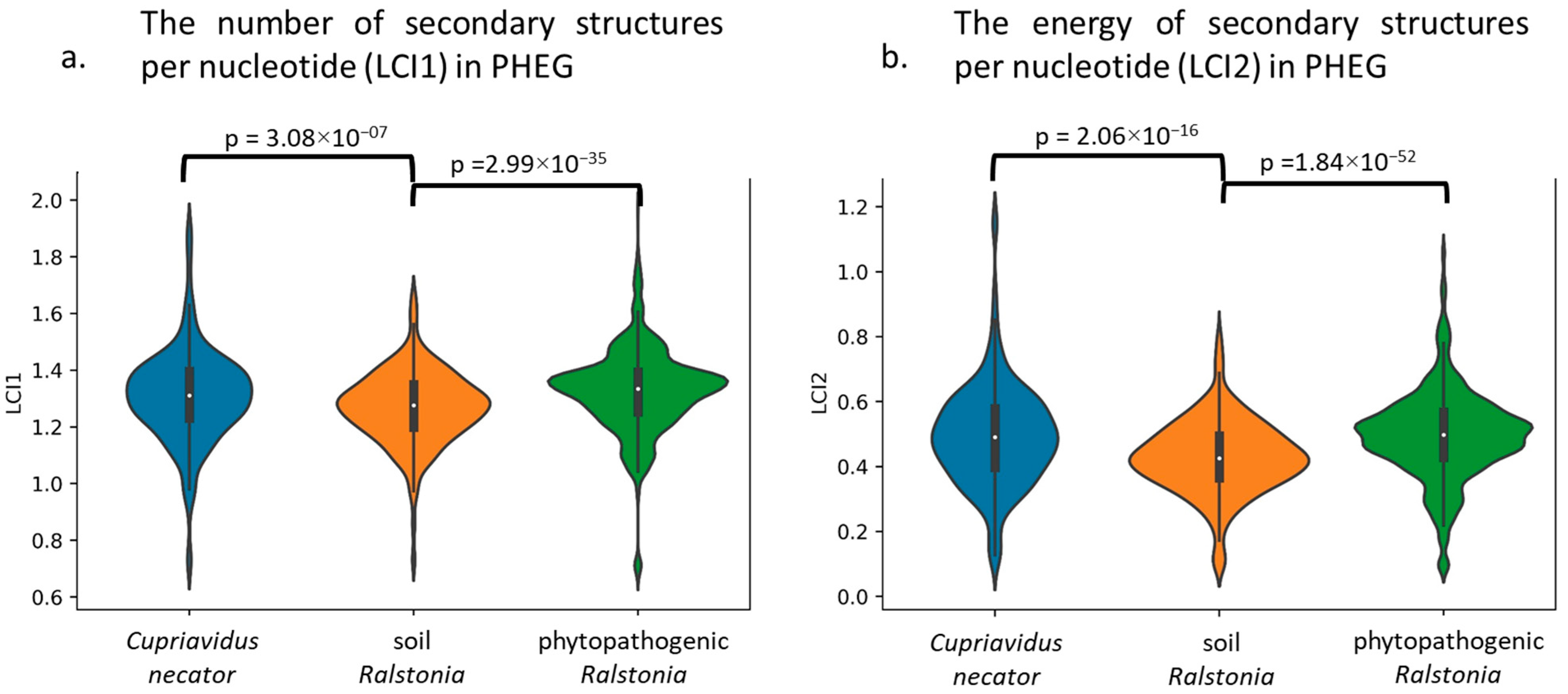
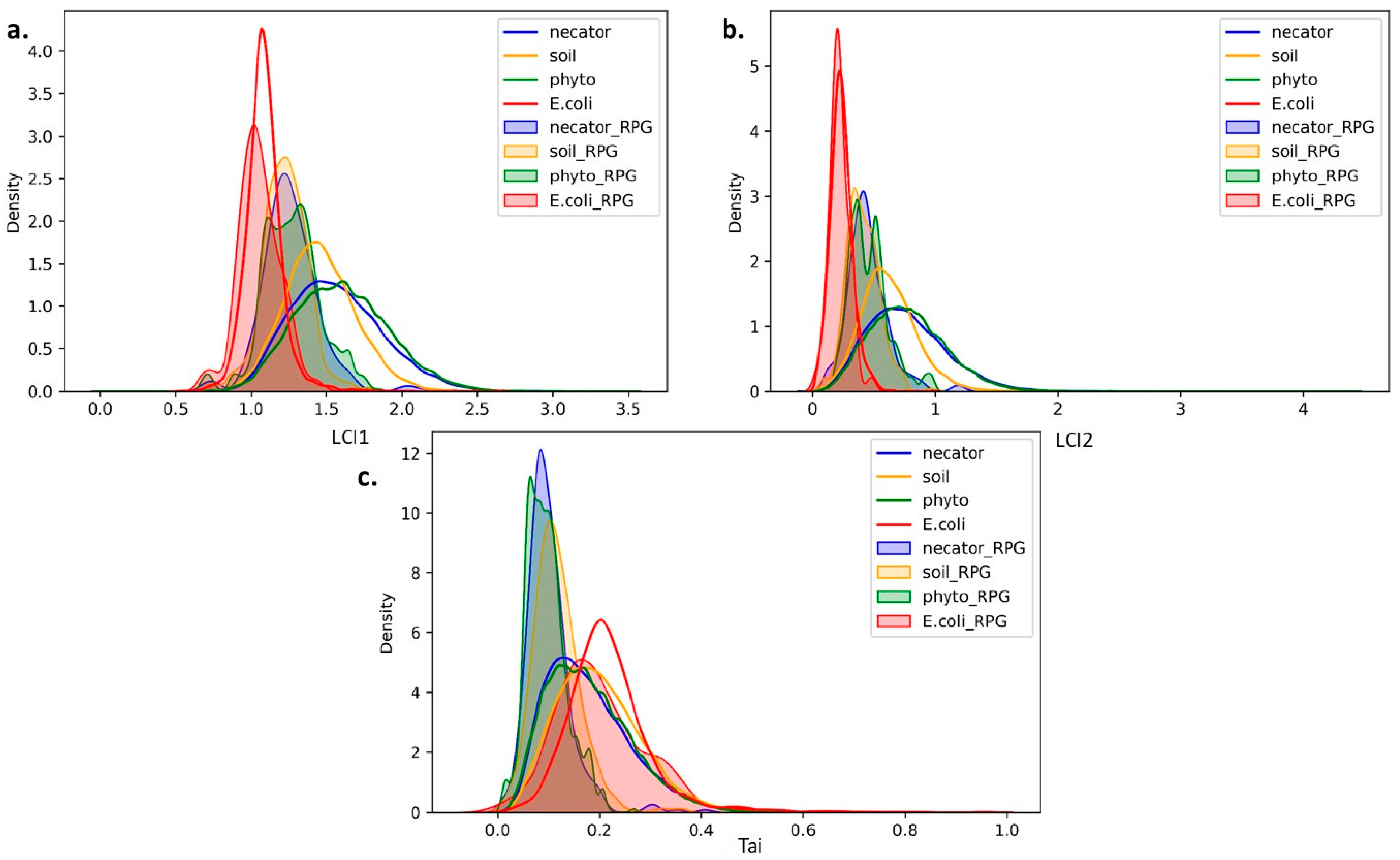
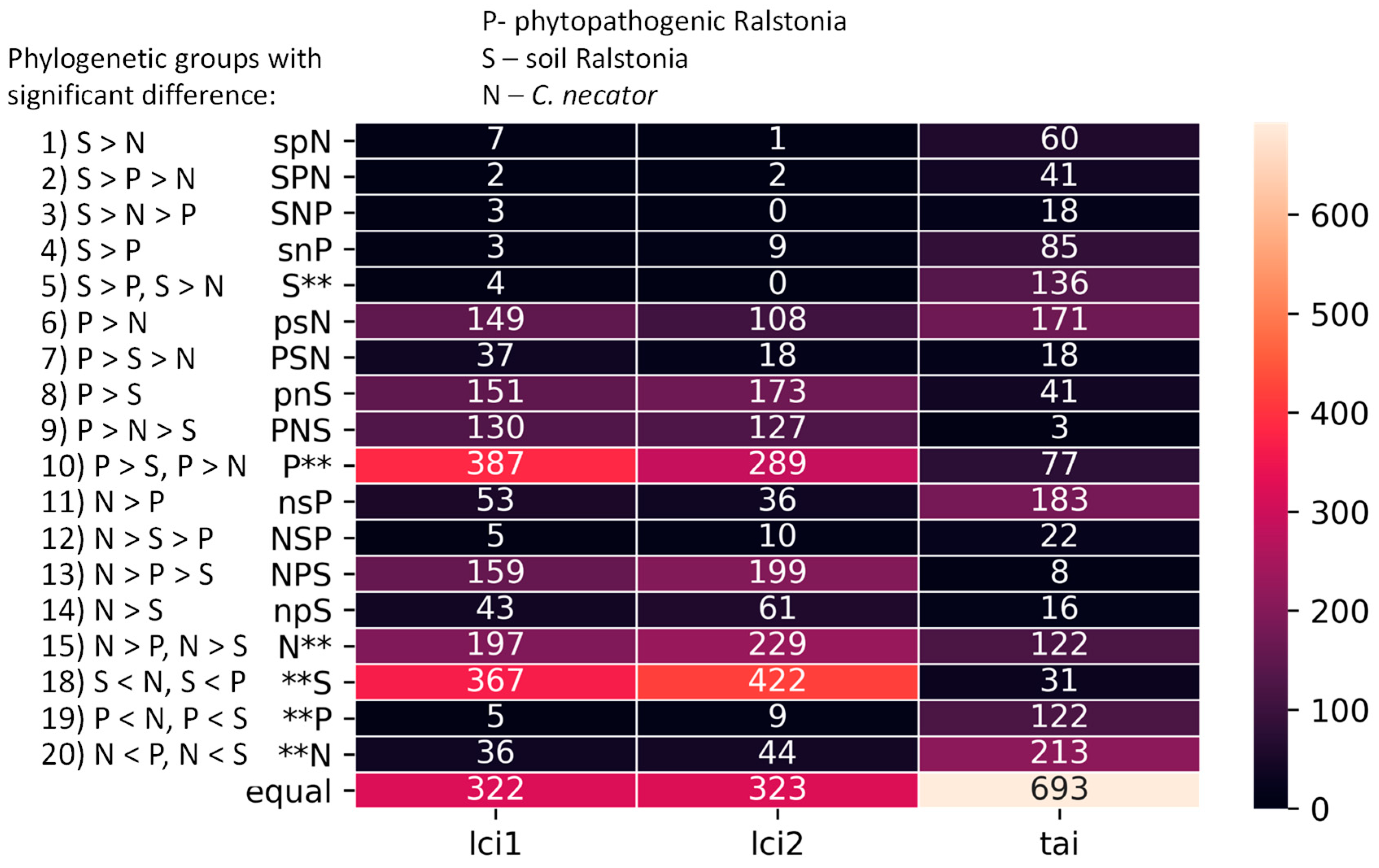
3.2. Comparative Analysis of Translation Elongation Efficiency of Genes in the Genomes of the Two Ralstonia branches and C. necator
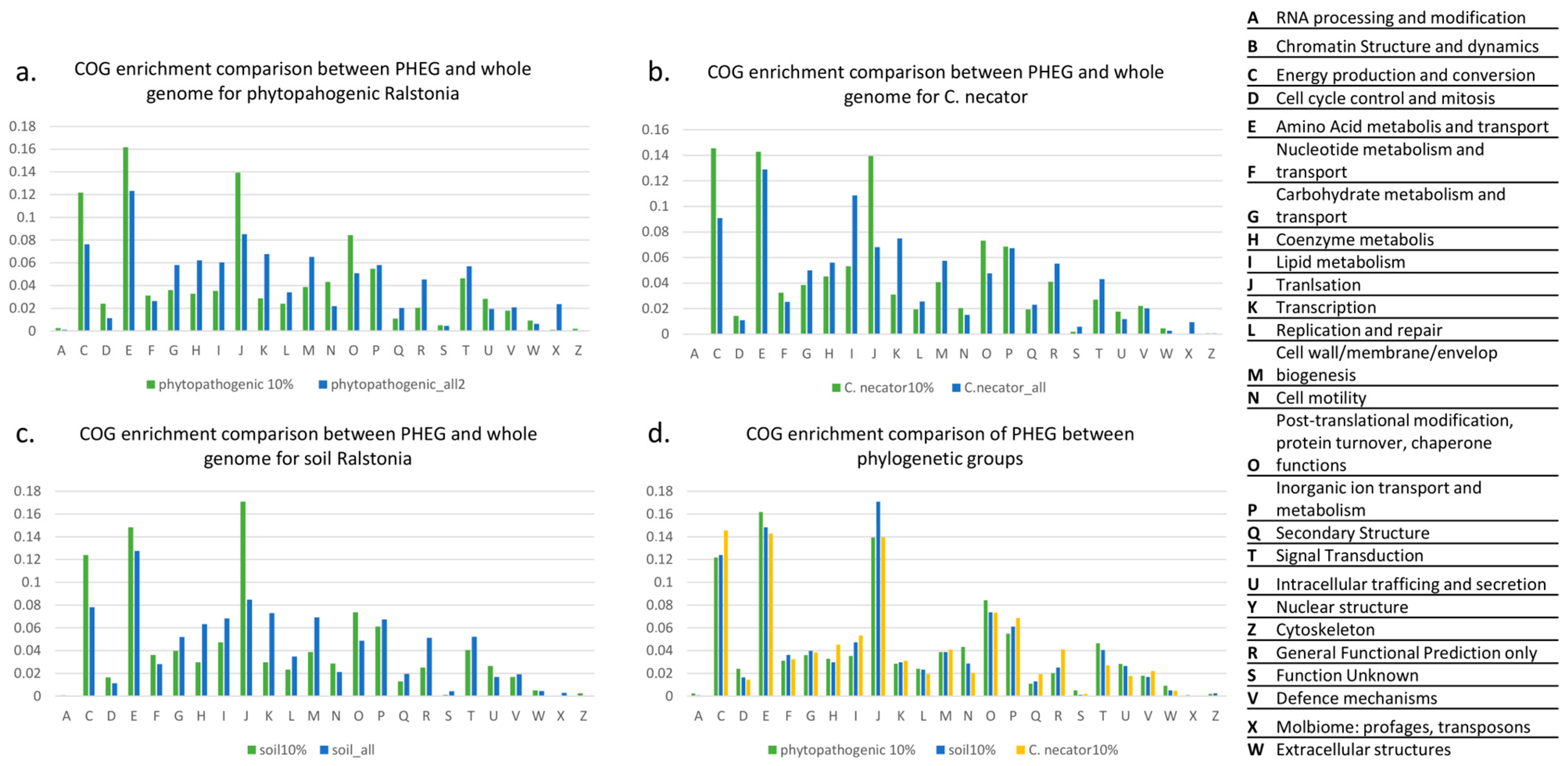
3.3. The Functional Analysis of Potentially Highly Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Organism | M1 | R1 | M2 | R2 | M3 | R3 | M4 | R4 | M5 | R5 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ralstonia pseudosolanacearum | −37 | 36 | 64 | 31 | 64 | 31 | 52 | 35 | 68 | 25 |
| Ralstonia solanacearum | −27 | 41 | 68 | 31 | 68 | 32 | 62 | 31 | 70 | 26 |
| Ralstonia syzygii | −23 | 40 | 67 | 30 | 68 | 27 | 59 | 35 | 68 | 27 |
| Ralstonia insidiosa | 74 | 31 | 57 | 36 | 55 | 36 | 87 | 19 | 86 | 20 |
| Ralstonia mannitolilytica | 39 | 42 | 70 | 25 | 69 | 27 | 81 | 22 | 80 | 20 |
| Ralstonia pickettii | 60 | 34 | 60 | 33 | 58 | 36 | 84 | 21 | 82 | 23 |
| Cupriavidus necator | 7 | 43 | 62 | 34 | 64 | 32 | 68 | 32 | 77 | 26 |
| COG | Comparison of COG Enrichment of PHEG Set between Groups | Comparison of COG Enrichment of Entire Gene Set between Groups | Comparison of COG Enrichment between PHEG and Entire Gene Set for Each Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ps_pval. | pn_pval. | sn_pval. | ps_pval_all | pn_pval_all | sn_pval_all | p_pval | n_pval | s_pval | |
| A | 1.52 × 10−1 | 8.40 × 10−3 | 3.81 × 10−1 | 6.38 × 10−2 | 1.05 × 10−2 | 7.23 × 10−1 | 1.04 × 10−9 | 6.78 × 10−1 | 6.36 × 10−1 |
| C | 8.55 × 10−1 | 3.19 × 10−3 | 6.42 × 10−2 | 5.82 × 10−1 | 2.59 × 10−10 | 1.02 × 10−4 | 1.43 × 10−153 | 6.82 × 10−14 | 5.47 × 10−18 |
| D | 3.71 × 10−2 | 3.68 × 10−3 | 6.75 × 10−1 | 9.33 × 10−1 | 7.62 × 10−1 | 7.62 × 10−1 | 4.27 × 10−66 | 3.88 × 10−2 | 1.30 × 10−1 |
| E | 1.82 × 10−1 | 3.49 × 10−2 | 7.33 × 10−1 | 1.82 × 10−1 | 6.24 × 10−2 | 7.62 × 10−1 | 1.76 × 10−78 | 5.21 × 10−3 | 4.82 × 10−2 |
| F | 2.63 × 10−1 | 8.44 × 10−1 | 6.09 × 10−1 | 2.58 × 10−1 | 5.82 × 10−1 | 1.81 × 10−1 | 1.70 × 10−6 | 3.18 × 10−2 | 3.88 × 10−2 |
| G | 4.64 × 10−1 | 6.50 × 10−1 | 8.73 × 10−1 | 3.61 × 10−3 | 4.95 × 10−5 | 5.82 × 10−1 | 1.71 × 10−65 | 9.51 × 10−3 | 9.51 × 10−3 |
| H | 6.09 × 10−1 | 4.21 × 10−3 | 1.02 × 10−2 | 7.23 × 10−1 | 3.70 × 10−3 | 1.33 × 10−2 | 4.70 × 10−114 | 2.43 × 10−13 | 2.13 × 10−2 |
| I | 8.13 × 10−3 | 6.29 × 10−5 | 5.12 × 10−1 | 2.20 × 10−4 | 3.74 × 10−111 | 1.11 × 10−36 | 1.25 × 10−82 | 4.44 × 10−5 | 1.33 × 10−22 |
| J | 2.51 × 10−4 | 9.93 × 10−1 | 8.13 × 10−3 | 8.61 × 10−1 | 4.27 × 10−14 | 7.72 × 10−8 | 6.13 × 10−195 | 8.06 × 10−39 | 2.17 × 10−34 |
| K | 8.55 × 10−1 | 6.09 × 10−1 | 8.94 × 10−1 | 2.64 × 10−2 | 1.25 × 10−3 | 6.36 × 10−1 | 2.81 × 10−196 | 1.35 × 10−19 | 5.65 × 10−21 |
| L | 9.51 × 10−1 | 2.25 × 10−1 | 4.89 × 10−1 | 7.23 × 10−1 | 1.67 × 10−8 | 7.48 × 10−6 | 4.38 × 10−22 | 2.66 × 10−3 | 4.90 × 10−2 |
| M | 9.88 × 10−1 | 7.04 × 10−1 | 8.55 × 10−1 | 8.57 × 10−2 | 2.51 × 10−4 | 4.95 × 10−5 | 9.39 × 10−86 | 4.74 × 10−10 | 3.40 × 10−4 |
| N | 1.84 × 10−3 | 4.25 × 10−9 | 1.31 × 10−1 | 7.23 × 10−1 | 5.75 × 10−9 | 1.12 × 10−4 | 1.13 × 10−100 | 2.08 × 10−2 | 4.78 × 10−2 |
| O | 1.48 × 10−1 | 1.14 × 10−1 | 9.88 × 10−1 | 3.43 × 10−1 | 1.03 × 10−1 | 7.34 × 10−1 | 2.77 × 10−118 | 4.92 × 10−7 | 5.17 × 10−8 |
| P | 3.03 × 10−1 | 1.02 × 10−2 | 4.32 × 10−1 | 9.35 × 10−6 | 6.90 × 10−6 | 1.00 | 2.09 × 10−2 | 2.44 × 10−1 | 8.24 × 10−1 |
| Q | 5.12 × 10−1 | 1.84 × 10−3 | 1.48 × 10−1 | 7.34 × 10−1 | 2.35 × 10−2 | 5.50 × 10−2 | 8.33 × 10−34 | 2.08 × 10−2 | 2.54 × 10−1 |
| R | 2.09 × 10−1 | 1.80 × 10−9 | 4.84 × 10−3 | 1.55 × 10−3 | 4.80 × 10−8 | 1.77 × 10−1 | 5.63 × 10−116 | 3.64 × 10−10 | 2.15 × 10−3 |
| S | 5.15 × 10−3 | 5.77 × 10−2 | 6.75 × 10−1 | 1.00 | 2.81 × 10−2 | 1.25 × 10−1 | 1.30 × 10−1 | 8.15 × 10−3 | 8.65 × 10−3 |
| T | 3.03 × 10−1 | 8.94 × 10−6 | 2.19 × 10−2 | 2.11 × 10−2 | 1.03 × 10−13 | 4.08 × 10−4 | 2.24 × 10−15 | 1.44 × 10−2 | 4.65 × 10−5 |
| U | 7.76 × 10−1 | 3.19 × 10−3 | 7.31 × 10−2 | 4.03 × 10−2 | 1.68 × 10−12 | 4.57 × 10−4 | 1.11 × 10−23 | 1.32 × 10−3 | 1.47 × 10−2 |
| V | 8.63 × 10−1 | 2.25 × 10−1 | 3.10 × 10−1 | 2.36 × 10−1 | 7.34 × 10−1 | 6.42 × 10−1 | 1.15 × 10−3 | 4.91 × 10−1 | 5.68 × 10−1 |
| W | 7.32 × 10−2 | 3.54 × 10−2 | 9.16 × 10−1 | 2.11 × 10−2 | 4.03 × 10−9 | 1.33 × 10−2 | 1.66 × 10−8 | 7.85 × 10−1 | 1.19 × 10−1 |
| X | 2.25 × 10−1 | 2.25 × 10−1 | 1.00 | 2.47 × 10−99 | 2.70 × 10−39 | 1.37 × 10−13 | 3.78 × 10−261 | 2.15 × 10−3 | 2.81 × 10−11 |
| Z | 6.75 × 10−1 | 2.31 × 10−1 | 2.09 × 10−1 | 7.62 × 10−1 | 4.12 × 10−1 | 7.49 × 10−1 | 1.45 × 10−15 | 8.25 × 10−3 | 1.00 |
| COG | Fraction of COG in the PHEG Set | Fraction of COG in the Whole Genome | ||||
|---|---|---|---|---|---|---|
| Phyto_10% | Soil_10% | Necator_10% | Phyto_All | Soil_All | Necator_All | |
| A | 0.002477 | 0.000718 | 0 | 0.001038 | 0.000499 | 0.000357 |
| C | 0.121803 | 0.123922 | 0.145534 | 0.076358 | 0.078066 | 0.090947 |
| D | 0.024021 | 0.016523 | 0.014386 | 0.011323 | 0.01142 | 0.010959 |
| E | 0.161698 | 0.148348 | 0.142857 | 0.123321 | 0.127551 | 0.128946 |
| F | 0.031242 | 0.036279 | 0.032452 | 0.026414 | 0.028206 | 0.025372 |
| G | 0.036016 | 0.039871 | 0.038474 | 0.057995 | 0.051919 | 0.04997 |
| H | 0.032763 | 0.029813 | 0.045166 | 0.062162 | 0.063276 | 0.056045 |
| I | 0.03533 | 0.047414 | 0.053195 | 0.060326 | 0.068268 | 0.108517 |
| J | 0.139438 | 0.170977 | 0.139512 | 0.085339 | 0.084805 | 0.068195 |
| K | 0.028616 | 0.029813 | 0.031114 | 0.067729 | 0.072886 | 0.074926 |
| L | 0.02408 | 0.023348 | 0.019404 | 0.033984 | 0.034758 | 0.02567 |
| M | 0.038701 | 0.038793 | 0.040816 | 0.065183 | 0.069204 | 0.057475 |
| N | 0.043267 | 0.028736 | 0.020408 | 0.021905 | 0.021217 | 0.015068 |
| O | 0.084296 | 0.073635 | 0.073269 | 0.050951 | 0.048799 | 0.047647 |
| P | 0.054785 | 0.061063 | 0.068585 | 0.058128 | 0.067457 | 0.067421 |
| Q | 0.011011 | 0.012931 | 0.019404 | 0.020233 | 0.019657 | 0.023169 |
| R | 0.02038 | 0.025144 | 0.041151 | 0.045348 | 0.051357 | 0.055331 |
| S | 0.005102 | 0.001078 | 0.002007 | 0.004468 | 0.004431 | 0.005837 |
| T | 0.04637 | 0.040589 | 0.027099 | 0.057055 | 0.052168 | 0.04318 |
| U | 0.028377 | 0.02658 | 0.017732 | 0.019431 | 0.016849 | 0.011852 |
| V | 0.017993 | 0.016882 | 0.022081 | 0.020836 | 0.019158 | 0.02025 |
| W | 0.009161 | 0.005029 | 0.004684 | 0.006252 | 0.004618 | 0.00274 |
| X | 0.001044 | 0 | 0 | 0.023704 | 0.002871 | 0.00941 |
| Z | 0.002029 | 0.002514 | 0.000669 | 0.000516 | 0.000562 | 0.000715 |
References
- Zhang, Y.; Qiu, S. Phylogenomic Analysis of the Genus Ralstonia Based on 686 Single-Copy Genes. Antonie Leeuwenhoek 2016, 109, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Moriuchi, R.; Dohra, H.; Kanesaki, Y.; Ogawa, N. Complete Genome Sequence of 3-Chlorobenzoate-Degrading Bacterium Cupriavidus Necator NH9 and Reclassification of the Strains of the Genera Cupriavidus and Ralstonia Based on Phylogenetic and Whole-Genome Sequence Analyses. Front. Microbiol. 2019, 10, 133. [Google Scholar] [CrossRef]
- Waugh, J.B.; Granger, W.M.; Gaggar, A. Incidence, Relevance and Response for Ralsfonia Respiratory Infections. Clin. Lab. Sci. 2010, 23, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Feng, Y.; Feng, P.; Wang, X.; Zong, Z. Nosocomial Bloodstream Infection and the Emerging Carbapenem-Resistant Pathogen Ralstonia Insidiosa. BMC Infect. Dis. 2019, 19, 334. [Google Scholar] [CrossRef] [PubMed]
- Peeters, N.; Guidot, A.; Vailleau, F.; Valls, M. Ralstonia Solanacearum, a Widespread Bacterial Plant Pathogen in the Post-Genomic Era. Mol. Plant Pathol. 2013, 14, 651–662. [Google Scholar] [CrossRef]
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Su, M.; Luff, J.; Valecha, M.; Everett, J.K.; Acton, T.B.; Xiao, R.; et al. Codon Influence on Protein Expression in E.Coli. Nature 2016, 529, 358–363. [Google Scholar] [CrossRef]
- Mohammad, F.; Green, R.; Buskirk, A.R. A Systematically-Revised Ribosome Profiling Method for Bacteria Reveals Pauses at Single-Codon Resolution. Elife 2019, 8, e42591. [Google Scholar] [CrossRef]
- Quax, T.E.F.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Guimaraes, J.C.; Rocha, M.; Arkin, A.P. Transcript Level and Sequence Determinants of Protein Abundance and Noise in Escherichia Coli. Nucleic Acids Res. 2014, 42, 4791–4799. [Google Scholar] [CrossRef]
- Zhang, D.; Li, S.H.J.; King, C.G.; Wingreen, N.S.; Gitai, Z.; Li, Z. Global and Gene-Specific Translational Regulation in Escherichia Coli across Different Conditions. PLoS Comput. Biol. 2022, 18, e1010641. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, Z.; Zou, Q.; Fang, J.; Wang, Q.; Yang, X.; Gao, N. Ribosome Profiling Reveals Genome-Wide Cellular Translational Regulation upon Heat Stress in Escherichia Coli. Genom. Proteom. Bioinformatics. 2017, 15, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, T. Codon Usage and TRNA Content in Unicellular and Multicellular Organisms. Mol. Biol. Evol. 1985, 2, 13–34. [Google Scholar] [CrossRef]
- Wright, F. The “effective Number of Codons” Used in a Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Sørensen, M.A.; Kurland, C.G.; Pedersen, S. Codon Usage Determines Translation Rate in Escherichia Coli. J. Mol. Biol. 1989, 207, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Komar, A.A. The Yin and Yang of Codon Usage. Hum. Mol. Genet. 2016, 25, R77–R85. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Silke, J.R.; Xia, X. An Improved Estimation of TRNA Expression to Better Elucidate the Coevolution between TRNA Abundance and Codon Usage in Bacteria. Sci. Rep. 2019, 9, 3184. [Google Scholar] [CrossRef]
- Rodnina, M.V. Translation in Prokaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032664. [Google Scholar] [CrossRef]
- Higgs, P.G.; Ran, W. Coevolution of Codon Usage and TRNA Genes Leads to Alternative Stable States of Biased Codon Usage. Mol. Biol. Evol. 2008, 25, 2279–2291. [Google Scholar] [CrossRef]
- Wang, S.E.; Brooks, A.E.S.; Poole, A.M.; Simoes-Barbosa, A. Determinants of Translation Efficiency in the Evolutionarily-Divergent Protist Trichomonas Vaginalis. BMC Mol. Cell Biol. 2020, 21, 54. [Google Scholar] [CrossRef]
- Cambray, G.; Guimaraes, J.C.; Arkin, A.P. Evaluation of 244,000 Synthetic Sequences Reveals Design Principles to Optimize Translation in Escherichia Coli. Nat. Biotechnol. 2018, 36, 1005. [Google Scholar] [CrossRef]
- Hia, F.; Takeuchi, O. The Effects of Codon Bias and Optimality on MRNA and Protein Regulation. Cell. Mol. Life Sci. 2021, 78, 1909–1928. [Google Scholar] [CrossRef] [PubMed]
- Frumkin, I.; Lajoie, M.J.; Gregg, C.J.; Hornung, G.; Church, G.M.; Pilpel, Y. Codon Usage of Highly Expressed Genes Affects Proteome-Wide Translation Efficiency. Proc. Natl. Acad. Sci. USA 2018, 115, E4940–E4949. [Google Scholar] [CrossRef] [PubMed]
- Jeacock, L.; Faria, J.; Horn, D. Codon Usage Bias Controls MRNA and Protein Abundance in Trypanosomatids. Elife 2018, 7, e32496. [Google Scholar] [CrossRef] [PubMed]
- Thanaraj, T.A.; Argos, P. Ribosome-Mediated Translational Pause and Protein Domain Organization. Protein Sci. 1996, 5, 1594–1612. [Google Scholar] [CrossRef]
- Wen, J.-D.; Lancaster, L.; Hodges, C.; Zeri, A.-C.; Yoshimura, S.H.; Noller, H.F.; Bustamante, C.; Tinoco, I. Following Translation by Single Ribosomes One Codon at a Time. Nature 2008, 452, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Pop, C.; Rouskin, S.; Ingolia, N.T.; Han, L.; Phizicky, E.M.; Weissman, J.S.; Koller, D. Causal Signals between Codon Bias, MRNA Structure, and the Efficiency of Translation and Elongation. Mol. Syst. Biol. 2014, 10, 770. [Google Scholar] [CrossRef]
- Shah, P.; Ding, Y.; Niemczyk, M.; Kudla, G.; Plotkin, J.B. XRate-Limiting Steps in Yeast Protein Translation. Cell 2013, 153, 1589. [Google Scholar] [CrossRef]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, Y.H.; Segal, E. Genome-Wide Measurement of RNA Secondary Structure in Yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef]
- Chiaruttini, C.; Guillier, M. On the Role of MRNA Secondary Structure in Bacterial Translation. Wiley Interdiscip. Rev. RNA 2020, 11, e1579. [Google Scholar] [CrossRef]
- Mauger, D.M.; Joseph Cabral, B.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. MRNA Structure Regulates Protein Expression through Changes in Functional Half-Life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, D.; Wu, F.; Standage-Beier, K.; Chen, X.; Wu, K.; Green, A.A.; Wang, X. Predictable Control of RNA Lifetime Using Engineered Degradation-Tuning RNAs. Nat. Chem. Biol. 2021, 17, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.K.; Kushner, S.R. Regulation of MRNA Decay in Bacteria. Annu. Rev. Microbiol. 2016, 70, 25–44. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Translation and Protein Quality Control: Codon Optimality, Bias and Usage in Translation and MRNA Decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Victor, M.P.; Acharya, D.; Begum, T.; Ghosh, T.C. The Optimization of MRNA Expression Level by Its Intrinsic Properties—Insights from Codon Usage Pattern and Structural Stability of MRNA. Genomics 2019, 111, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Gorochowski, T.E.; Ignatova, Z.; Bovenberg, R.A.L.; Roubos, J.A. Trade-Offs between TRNA Abundance and MRNA Secondary Structure Support Smoothing of Translation Elongation Rate. Nucleic Acids Res. 2015, 43, 3022–3032. [Google Scholar] [CrossRef] [PubMed]
- Likhoshvai, V.A.; Matushkin, Y.G. Differentiation of Single-Cell Organisms According to Elongation Stages Crucial for Gene Expression Efficacy. FEBS Lett. 2002, 516, 87–92. [Google Scholar] [CrossRef]
- Vladimirov, N.V.; Likhoshvai, V.A.; Matushkin, Y.G. Correlation of Codon Biases and Potential Secondary Structures with MRNA Translation Efficiency in Unicellular Organisms. Mol. Biol. 2007, 41, 843–850. [Google Scholar] [CrossRef]
- Korenskaia, A.E.; Matushkin, Y.G.; Lashin, S.A.; Klimenko, A.I. Bioinformatic Assessment of Factors Affecting the Correlation between Protein Abundance and Elongation Efficiency in Prokaryotes. Int. J. Mol. Sci. 2022, 23, 11996. [Google Scholar] [CrossRef]
- Сoкoлoв, B.C.; Лихoшвай, B.A.; Матушкин, Ю.Г. Экспрессия Генoв и Втoричные Структуры в Мрнк в Разных Видах Mycoplasma. Вавилoвский Журнал Генетики Селекции 2013, 17, 639–650. [Google Scholar]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Genotypic and Phenotypic Diversity of Ralstonia Pickettii and Ralstonia Insidiosa Isolates from Clinical and Environmental Sources Including High-Purity Water. Diversity in Ralstonia Pickettii. BMC Microbiol. 2011, 11, 194. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia Pickettii in Environmental Biotechnology: Potential and Applications. J. Appl. Microbiol. 2007, 103, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.L.; Holland, B.W.; Carr, J.K.; Bond, W.W.; Favero, M.S. Effect of Disinfectants on Pseudomonads Colonized on the Interior Surface of PVC Pipes. Am. J. Public Health 1990, 80, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.T.S.; Lee, S.E. A Rare Case of Ralstonia Mannitolilytica Infection in an End Stage Renal Patient on Maintenance Dialysis during Municipal Water Contamination. Pak. J. Med. Sci. 2017, 33, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, E. Hospital Infections Caused by Contaminated Fluids. Lancet 1972, 1, 1338. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Ralstonia Pickettii: A Persistent Gram-Negative Nosocomial Infectious Organism. J. Hosp. Infect. 2006, 62, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Asaad, A.M.; Al-Ayed, M.S.Z.; Qureshi, M.A. Emergence of Unusual Nonfermenting Gram-Negative Nosocomial Pathogens in a Saudi Hospital. Jpn. J. Infect. Dis. 2013, 66, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.L.; Bland, L.A.; Favero, M.S.; McNeil, M.M.; Davis, B.J.; Mackel, D.C.; Gravelle, C.R. Factors Associated with Pseudomonas Pickettii Intrinsic Contamination of Commercial Respiratory Therapy Solutions Marketed as Sterile. Appl. Environ. Microbiol. 1985, 50, 1343–1348. [Google Scholar] [CrossRef]
- De Baere, T.; Steyaert, S.; Wauters, G.; Des Vos, P.; Goris, J.; Coenye, T.; Suyama, T.; Verschraegen, G.; Vaneechoutte, M. Classification of Ralstonia Pickettii Biovar 3/’thomasii’ Strains (Pickett 1994) and of New Isolates Related to Nosocomial Recurrent Meningitis as Ralstonia Mannitolytica Sp. Nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 547–558. [Google Scholar] [CrossRef]
- Orme, J.; Rivera-Bonilla, T.; Loli, A.; Blattman, N.N. Native Valve Endocarditis Due to Ralstonia Pickettii: A Case Report and Literature Review. Case Rep. Infect. Dis. 2015, 2015, 324675. [Google Scholar] [CrossRef]
- Shankar, M.; Rampure, S.; Siddini, V.; Ballal, H.S. Outbreak of Ralstonia Mannitolilytica in Hemodialysis Unit: A Case Series. Indian J. Nephrol. 2018, 28, 323–326. [Google Scholar] [CrossRef]
- O’Connor, T.J.; Adepoju, Y.; Boyd, D.; Isberg, R.R. Minimization of the Legionella Pneumophila Genome Reveals Chromosomal Regions Involved in Host Range Expansion. Proc. Natl. Acad. Sci. USA 2011, 108, 17856, Erratum in Proc. Natl. Acad. Sci. USA 2011, 108, 14733–14740. [Google Scholar] [CrossRef] [PubMed]
- Said, M.; van Hougenhouck-Tulleken, W.; Naidoo, R.; Mbelle, N.; Ismail, F. Outbreak of Ralstonia Mannitolilytica Bacteraemia in Patients Undergoing Haemodialysis at a Tertiary Hospital in Pretoria, South Africa. Antimicrob. Resist. Infect. Control. 2020, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.N.; Sahu, C.; Aggarwal, A. Ralstonia Mannitolilytica Bacteraemia and Gastroenteritis in a Patient with Rheumatoid Arthritis: An Emerging Nosocomial Infection. Rheumatology 2020, 60, e195–e196. [Google Scholar] [CrossRef]
- Suzuki, M.; Nishio, H.; Asagoe, K.; Kida, K.; Suzuki, S.; Matsui, M.; Shibayama, K. Genome Sequence of a Carbapenem-Resistant Strain of Ralstonia Mannitolilytica. Genome Announc. 2015, 3, 00405-15. [Google Scholar] [CrossRef]
- Safni, I.; Cleenwerck, I.; De Vos, P.; Fegan, M.; Sly, L.; Kappler, U. Polyphasic Taxonomic Revision of the Ralstonia Solanacearum Species Complex: Proposal to Emend the Descriptions of Ralstonia Solanacearum and Ralstonia Syzygii and Reclassify Current R. Syzygii Strains as Ralstonia Syzygii Subsp. Syzygii Subsp. Nov., R.S. Int. J. Syst. Evol. Microbiol. 2014, 64, 3087–3103. [Google Scholar] [CrossRef]
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia Solanacearum Species Complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef]
- Remenant, B.; de Cambiaire, J.C.; Cellier, G.; Jacobs, J.M.; Mangenot, S.; Barbe, V.; Lajus, A.; Vallenet, D.; Medigue, C.; Fegan, M.; et al. Ralstonia Syzygii, the Blood Disease Bacterium and Some Asian R. Solanacearum Strains Form a Single Genomic Species despite Divergent Lifestyles. PLoS ONE 2011, 6, e24356. [Google Scholar] [CrossRef]
- Poehlein, A.; Kusian, B.; Friedrich, B.; Daniel, R.; Bowien, B. Complete Genome Sequence of the Type Strain Cupriavidus Necator N-1. J. Bacteriol. 2011, 193, 5017. [Google Scholar] [CrossRef]
- Pohlmann, A.; Fricke, W.F.; Reinecke, F.; Kusian, B.; Liesegang, H.; Cramm, R.; Eitinger, T.; Ewering, C.; Pötter, M.; Schwartz, E.; et al. Genome Sequence of the Bioplastic-Producing “Knallgas” Bacterium Ralstonia Eutropha H16. Nat. Biotechnol. 2006, 24, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Ralstonia Solanacearum GMI1000 Genome. BMC Genom. 2003, 9, 10. [Google Scholar]
- Likhoshvai, A.; Matushkin, Y.G. Nucleotide Composition-Based Prediction of Gene Expression Efficacy. Mol. Biol. 2000, 34, 397–405. [Google Scholar] [CrossRef]
- Sokolov, V.; Zuraev, B.; Lashin, S.; Matushkin, Y. Web Application for Automatic Prediction of Gene Translation Elongation Efficiency. J. Integr. Bioinform. 2015, 12, 256. [Google Scholar] [CrossRef][Green Version]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial Genome Analysis: The COG Approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.R. Statistical Methods for Detecting Molecular Adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Mustafin, Z.; Mukhin, A.; Afonnikov, D.; Matushkin, Y.; Lashin, S. OrthoWeb-Web Application for Macroand Microevolutionary Analysis of Genes. In Bioinformatics of Genome Regulation and Structure/Systems Biology (BGRS/SB-2020); Institute of Cytology and Genetics: Novosibirsk, Russia, 2020; pp. 228–229. [Google Scholar]
- Kryazhimskiy, S.; Plotkin, J.B. The Population Genetics of DN/DS. PLOS Genet. 2008, 4, e1000304. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Mustafin, Z.S.; Lashin, S.A.; Matushkin, Y.G.; Gunbin, K.V.; Afonnikov, D.A. Orthoscape: A Cytoscape Application for Grouping and Visualization KEGG Based Gene Networks by Taxonomy and Homology Principles. BMC Bioinform. 2017, 18, 1–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Man, O.; Pilpel, Y. Differential Translation Efficiency of Orthologous Genes Is Involved in Phenotypic Divergence of Yeast Species. Nat. Genet. 2007, 39, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Zeiss, D.R.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic Evaluation of Ralstonia Solanacearum Cold Shock Protein Peptide (Csp22)-Induced Responses in Solanum Lycopersicum. Front. Plant Sci. 2022, 12, 803104. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, M.C.; Theodorou, E.C.; Kyriakidis, D.A. Involvement of AtoSC Two-Component System in Escherichia Coli Flagellar Regulon. Amino Acids 2012, 43, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, T.A.; Wu, H.; Chen, Z.; Li, S.; Li, H.; Zhang, C.; Zhou, Y.; Lu, C.; Lu, C. Proposal to Classify Ralstonia Solanacearum Phylotype I Strains as Ralstonia Nicotianae Sp. Nov., and a Genomic Comparison between Members of the Genus Ralstonia. Front. Microbiol. 2023, 13, 1135872. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Rocha, E.P.C. The Systemic Imprint of Growth and Its Uses in Ecological (Meta)Genomics. PLoS Genet. 2010, 6, e1000808. [Google Scholar] [CrossRef]
- Weissman, J.L.; Hou, S.; Fuhrman, J.A. Estimating Maximal Microbial Growth Rates from Cultures, Metagenomes, and Single Cells via Codon Usage Patterns. Proc. Natl. Acad. Sci. USA 2021, 118, e2016810118. [Google Scholar] [CrossRef]
- Baroukh, C.; Zemouri, M.; Genin, S. Trophic Preferences of the Pathogen Ralstonia Solanacearum and Consequences on Its Growth in Xylem Sap. Microbiologyopen 2022, 11, e1240. [Google Scholar] [CrossRef]
- Boy, C.; Lesage, J.; Alfenore, S.; Guillouet, S.E.; Gorret, N. Investigation of the Robustness of Cupriavidus Necator Engineered Strains during Fed-Batch Cultures. AMB Express 2021, 11, 151. [Google Scholar] [CrossRef]
- Guo, A.; Xu, Y.; Mowery, J.; Nagy, A.; Bauchan, G.; Nou, X. Ralstonia Insidiosa Induces Cell Aggregation of Listeria Monocytogenes. Food Control 2016, 67, 303–309. [Google Scholar] [CrossRef]
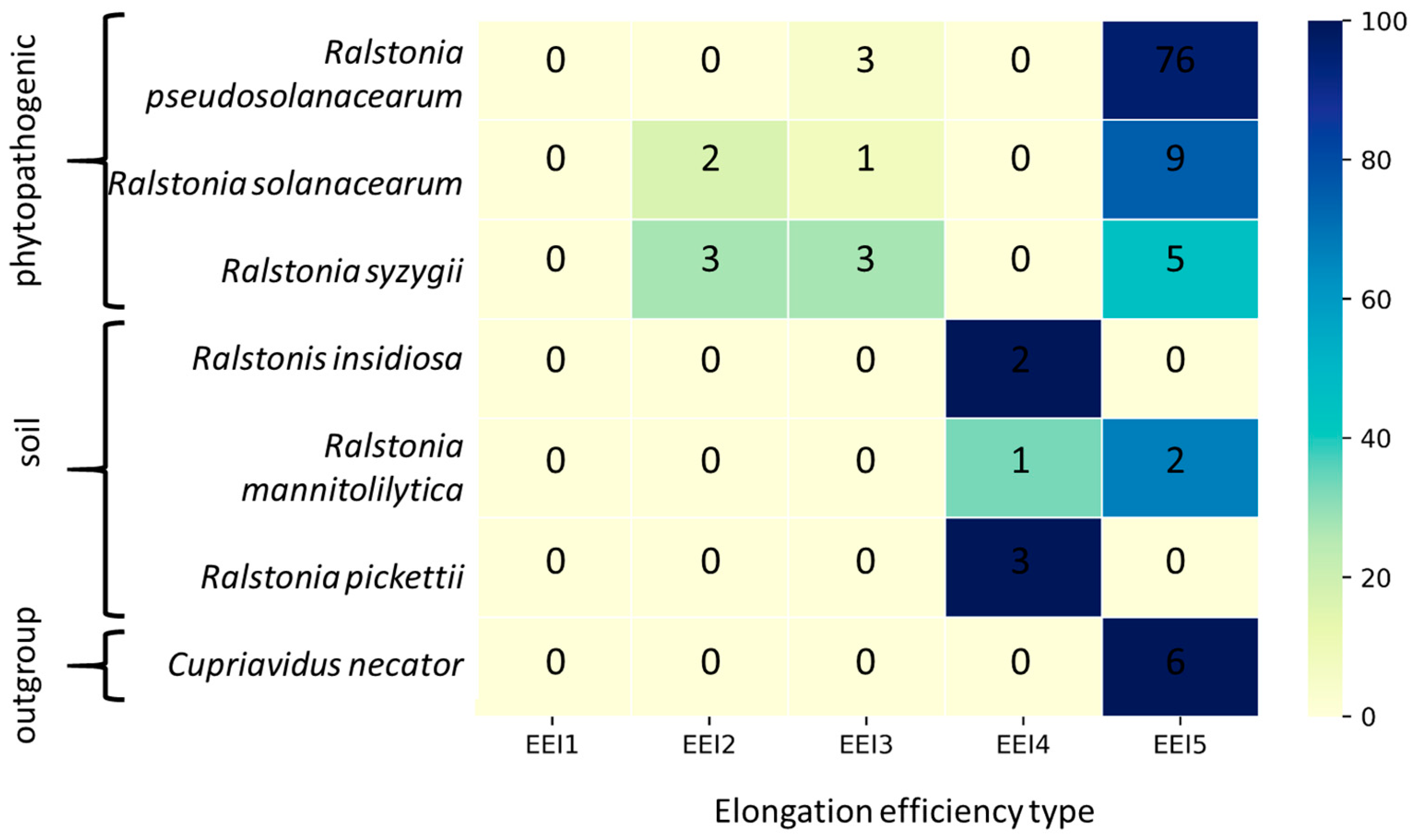
| EEI Type | Codon Composition | The Number of mRNA Hairpins | Energy of Potential Secondary Structures in mRNA |
|---|---|---|---|
| EEI1 | + | ||
| EEI2 | + | ||
| EEI3 | + | ||
| EEI4 | + | + | |
| EEI5 | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korenskaia, A.Y.; Matushkin, Y.G.; Mustafin, Z.S.; Lashin, S.A.; Klimenko, A.I. Bioinformatic Analysis Reveals the Role of Translation Elongation Efficiency Optimisation in the Evolution of Ralstonia Genus. Biology 2023, 12, 1338. https://doi.org/10.3390/biology12101338
Korenskaia AY, Matushkin YG, Mustafin ZS, Lashin SA, Klimenko AI. Bioinformatic Analysis Reveals the Role of Translation Elongation Efficiency Optimisation in the Evolution of Ralstonia Genus. Biology. 2023; 12(10):1338. https://doi.org/10.3390/biology12101338
Chicago/Turabian StyleKorenskaia, Aleksandra Y., Yury G. Matushkin, Zakhar S. Mustafin, Sergey A. Lashin, and Alexandra I. Klimenko. 2023. "Bioinformatic Analysis Reveals the Role of Translation Elongation Efficiency Optimisation in the Evolution of Ralstonia Genus" Biology 12, no. 10: 1338. https://doi.org/10.3390/biology12101338
APA StyleKorenskaia, A. Y., Matushkin, Y. G., Mustafin, Z. S., Lashin, S. A., & Klimenko, A. I. (2023). Bioinformatic Analysis Reveals the Role of Translation Elongation Efficiency Optimisation in the Evolution of Ralstonia Genus. Biology, 12(10), 1338. https://doi.org/10.3390/biology12101338








