Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products
Abstract
Simple Summary
Abstract
1. Introduction
2. Sources of Chitinase and COS Production Efficiency
2.1. Bacteria
2.2. Fungi
2.3. Plants
2.4. Animals
3. Chemo-Enzymic Production of COSs and Its Derivatives
3.1. Chemo-Enzymic Production of COSs
3.2. Production of COS Derivatives
4. Applications of COS
4.1. Food Additives and Functional Food
4.2. Biomaterials and Biomedicines
4.3. Plant Elicitors
5. Route to Industrialization of COSs by Bioconversion from Chitinous Waste
6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, X.; Chen, X.; Zhang, J.; Guo, W.; Jin, F.; Yan, N. Transformation of Chitin and Waste Shrimp Shells into Acetic Acid and Pyrrole. ACS Sustain. Chem. Eng. 2016, 4, 3912–3920. [Google Scholar] [CrossRef]
- Yan, N.; Chen, X. Sustainability: Don’t waste seafood waste. Nature 2015, 524, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, V. Valorization of Seafood Processing Discards: Bioconversion and Bio-Refinery Approaches. Front. Sustain. Food Syst. 2021, 5, 611835. [Google Scholar] [CrossRef]
- Mohan, K.; Muralisankar, T.; Jayakumar, R.; Rajeevgandhi, C. A study on structural comparisons of α-chitin extracted from marine crustacean shell waste. Carbohydr. Polym. Technol. Appl. 2021, 2, 100037. [Google Scholar] [CrossRef]
- Taokaew, S.; Zhang, X.; Chuenkaek, T.; Kobayashi, T. Chitin from fermentative extraction of crab shells using okara as a nutrient source and comparative analysis of structural differences from chemically extracted chitin. Biochem. Eng. J. 2020, 159, 107588. [Google Scholar] [CrossRef]
- Mahata, M.; Shinya, S.; Masaki, E.; Yamamoto, T.; Ohnuma, T.; Brzezinski, R.; Mazumder, T.K.; Yamashita, K.; Narihiro, K.; Fukamizo, T. Production of chitooligosaccharides from Rhizopus oligosporus NRRL2710 cells by chitosanase digestion. Carbohydr. Res. 2014, 383, 27–33. [Google Scholar] [CrossRef]
- Fernando, L.D.; Dickwella Widanage, M.C.; Penfield, J.; Lipton, A.S.; Washton, N.; Latgé, J.P.; Wang, P.; Zhang, L.; Wang, T. Structural Polymorphism of Chitin and Chitosan in Fungal Cell Walls From Solid-State NMR and Principal Component Analysis. Front Mol Biosci 2021, 8, 727053. [Google Scholar] [CrossRef]
- Kang, X.; Kirui, A.; Muszyński, A.; Widanage, M.C.D.; Chen, A.; Azadi, P.; Wang, P.; Mentink-Vigier, F.; Wang, T. Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 2018, 9, 2747. [Google Scholar] [CrossRef]
- Abo Elsoud, M.M.; El Kady, E.M. Current trends in fungal biosynthesis of chitin and chitosan. Bull. Natl. Res. Cent. 2019, 43, 59. [Google Scholar] [CrossRef]
- Brown, H.E.; Esher, S.K.; Alspaugh, J.A. Chitin: A “hidden figure” in the fungal cell wall. Curr. Top. Microbiol. Immunol. 2020, 425, 83–111. [Google Scholar]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, N.; Poulhazan, A.; Deligey, F.; Mentink-Vigier, F.; Marcotte, I.; Wang, T. Solid-State NMR Investigations of Extracellular Matrixes and Cell Walls of Algae, Bacteria, Fungi, and Plants. Chem. Rev. 2022, 122, 10036–10086. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Halfar, J. First evidence of chitin in calcified coralline algae: New insights into the calcification process of Clathromorphum compactum. Sci. Rep. 2014, 4, 6162. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-J.; Pan, S.-K.; Wang, H.-B.; Wu, J.-H. Preparation of chitooligosaccharides from cicada slough and their antibacterial activity. Int. J. Biol. Macromol. 2013, 62, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.-L.; Xu, Z.-Y.; Li, M.-J.; Mbuji, A.L.; Gu, M.; Zhang, L.; Gao, X.-W. Detection of Chitin Synthase Mutations in Lufenuron-Resistant Spodoptera frugiperda in China. Insects 2022, 13, 963. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.-Q.; Li, G.-Y.; Song, Q.-S.; Stanley, D.; Wei, S.-J.; Zhu, J.-Y. Genomic and transcriptomic analyses of chitin metabolism enzymes in Tenebrio molitor. Arch. Insect Biochem. Physiol. 2022, 111, e21950. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, P.; Lv, X.; Liang, Y. Insight into the structure-function relationships of the solubility of chitin/chitosan in natural deep eutectic solvents. Mater. Today Commun. 2021, 27, 102374. [Google Scholar] [CrossRef]
- Li, F.; You, X.; Li, Q.; Qin, D.; Wang, M.; Yuan, S.; Chen, X.; Bi, S. Homogeneous deacetylation and degradation of chitin in NaOH/urea dissolution system. Int. J. Biol. Macromol. 2021, 189, 391–397. [Google Scholar] [CrossRef]
- Ma, Q.; Gao, X.; Bi, X.; Han, Q.; Tu, L.; Yang, Y.; Shen, Y.; Wang, M. Dissolution and deacetylation of chitin in ionic liquid tetrabutylammonium hydroxide and its cascade reaction in enzyme treatment for chitin recycling. Carbohydr. Polym. 2020, 230, 115605. [Google Scholar] [CrossRef]
- Kaya, M.; Mujtaba, M.; Ehrlich, H.; Salaberria, A.M.; Baran, T.; Amemiya, C.T.; Galli, R.; Akyuz, L.; Sargin, I.; Labidi, J. On chemistry of γ-chitin. Carbohydr. Polym. 2017, 176, 177–186. [Google Scholar] [CrossRef]
- Moussian, B. Chitin: Structure, chemistry and biology. Adv. Exp. Med. Biol. 2019, 1142, 5–18. [Google Scholar] [PubMed]
- Jang, M.-K.; Kong, B.-G.; Jeong, Y.-I.; Lee, C.H.; Nah, J.-W. Physicochemical characterization of α-chitin, β-chitin, and γ-chitin separated from natural resources. J. Polym. Sci. Part A Polym. Chem. 2004, 42, 3423–3432. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Chakraborty, A.; Fernando, L.D.; Fang, W.; Dickwella Widanage, M.C.; Wei, P.; Jin, C.; Fontaine, T.; Latgé, J.-P.; Wang, T. A molecular vision of fungal cell wall organization by functional genomics and solid-state NMR. Nat. Commun. 2021, 12, 6346. [Google Scholar] [CrossRef]
- Mourya, V.K.; Inamdar, N.N.; Choudhari, Y.M. Chitooligosaccharides: Synthesis, characterization and applications. Polym. Sci. Ser. A 2011, 53, 583–612. [Google Scholar] [CrossRef]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015, 5, 8716. [Google Scholar] [CrossRef]
- Jiang, Y.; Fu, C.; Wu, S.; Liu, G.; Guo, J.; Su, Z. Determination of the Deacetylation Degree of Chitooligosaccharides. Mar. Drugs 2017, 15, 332. [Google Scholar] [CrossRef]
- Santos-Moriano, P.; Kidibule, P.; Míguez, N.; Fernández-Arrojo, L.; Ballesteros, A.O.; Fernández-Lobato, M.; Plou, F.J. Tailored Enzymatic Synthesis of Chitooligosaccharides with Different Deacetylation Degrees and Their Anti-Inflammatory Activity. Catalysts 2019, 9, 405. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Gao, Y.; Gao, W.; Hou, Z.; Zhu, Y. Facile Method for Surface-Grafted Chitooligosaccharide on Medical Segmented Poly(ester-urethane) Film to Improve Surface Biocompatibility. Membranes 2021, 11, 37. [Google Scholar] [CrossRef]
- Il’ina, A.V.; Varlamov, V.P. In vitro antitumor activity of heterochitooligosaccharides (Review). Appl. Biochem. Microbiol. 2015, 51, 1–10. [Google Scholar] [CrossRef]
- Rakkhumkaew, N.; Pengsuk, C. Chitosan and chitooligosaccharides from shrimp shell waste: Characterization, antimicrobial and shelf life extension in bread. Food Sci. Biotechnol. 2018, 27, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Xie, H.; Hu, S.; Xie, T.; Gong, J.; Jiang, C.; Ge, Q.; Wu, Y.; Liu, S.; Cui, Y.; et al. Preparation, Characterization, and Biochemical Activities of N-(2-Carboxyethyl)chitosan from Squid Pens. J. Agric. Food Chem. 2015, 63, 2464–2471. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.-X.; Dai, X.-Y.; Yang, W.; Xu, X.-W.; Liang, Y.-X. Antifungal activity of chitooligosaccharides against the dermatophyte Trichophyton rubrum. Int. J. Biol. Macromol. 2015, 77, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Kazami, N.; Sakaguchi, M.; Mizutani, D.; Masuda, T.; Wakita, S.; Oyama, F.; Kawakita, M.; Sugahara, Y. A simple procedure for preparing chitin oligomers through acetone precipitation after hydrolysis in concentrated hydrochloric acid. Carbohydr. Polym. 2015, 132, 304–310. [Google Scholar] [CrossRef]
- Laokuldilok, T.; Potivas, T.; Kanha, N.; Surawang, S.; Seesuriyachan, P.; Wangtueai, S.; Phimolsiripol, Y.; Regenstein, J.M. Physicochemical, antioxidant, and antimicrobial properties of chitooligosaccharides produced using three different enzyme treatments. Food Biosci. 2017, 18, 28–33. [Google Scholar] [CrossRef]
- Jia, X.; Rajib, M.R.; Yin, H. Recognition pattern, functional mechanism and application of chitin and chitosan oligosaccharides in sustainable agriculture. Curr. Pharm. Des. 2020, 26, 3508–3521. [Google Scholar] [CrossRef]
- He, Y.; Bose, S.K.; Wang, W.; Jia, X.; Lu, H.; Yin, H. Pre-Harvest Treatment of Chitosan Oligosaccharides Improved Strawberry Fruit Quality. Int. J. Mol. Sci. 2018, 19, 2194. [Google Scholar] [CrossRef]
- Yang, A.; Yu, L.; Chen, Z.; Zhang, S.; Shi, J.; Zhao, X.; Yang, Y.; Hu, D.; Song, B. Label-Free Quantitative Proteomic Analysis of Chitosan Oligosaccharide-Treated Rice Infected with Southern Rice Black-Streaked Dwarf Virus. Viruses 2017, 9, 115. [Google Scholar] [CrossRef]
- Mei, L.; Xu, Z.; Shi, Y.; Lin, C.; Jiao, S.; Zhang, L.; Li, P. Multivalent and synergistic chitosan oligosaccharide-Ag nanocomposites for therapy of bacterial infection. Sci. Rep. 2020, 10, 10011. [Google Scholar] [CrossRef]
- Guo, X.; Sun, T.; Zhong, R.; Ma, L.; You, C.; Tian, M.; Li, H.; Wang, C. Effects of Chitosan Oligosaccharides on Human Blood Components. Front. Pharmacol. 2018, 9, 1412. [Google Scholar] [CrossRef]
- Jafari, H.; Delporte, C.; Bernaerts, K.V.; De Leener, G.; Luhmer, M.; Nie, L.; Shavandi, A. Development of marine oligosaccharides for potential wound healing biomaterials engineering. Chem. Eng. J. Adv. 2021, 7, 100113. [Google Scholar] [CrossRef]
- Jang, D.; Lee, D.; Shin, Y.C.; Lee, J.S.; Jung, J.; Ryoo, S. Low molecular weight chitooligosaccharide inhibits infection of SARS-CoV-2 in vitro. J. Appl. Microbiol. 2022, 133, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- de Andrade, R.C.; de Araújo, N.K.; Torres-Rêgo, M.; Furtado, A.A.; Daniele-Silva, A.; de Souza Paiva, W.; de Medeiros Dantas, J.M.; da Silva, N.S.; da Silva-Júnior, A.A.; Ururahy, M.A.; et al. Production and Characterization of Chitooligosaccharides: Evaluation of Acute Toxicity, Healing, and Anti-Inflammatory Actions. Int. J. Mol. Sci. 2021, 22, 10631. [Google Scholar] [CrossRef] [PubMed]
- Jitprasertwong, P.; Khamphio, M.; Petsrichuang, P.; Eijsink, V.G.H.; Poolsri, W.; Muanprasat, C.; Rangnoi, K.; Yamabhai, M. Anti-inflammatory activity of soluble chitooligosaccharides (CHOS) on VitD3-induced human THP-1 monocytes. PLoS ONE 2021, 16, e0246381. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.I.; Ki, K.S.; Lim, D.H.; Vijayakumar, M.; Park, S.M.; Choi, S.H.; Kim, K.Y.; Im, S.K.; Park, B.Y. Novel Acinetobacter parvus HANDI 309 microbial biomass for the production of N-acetyl-β-d-glucosamine (GlcNAc) using swollen chitin substrate in submerged fermentation. Biotechnol. Biofuels 2017, 10, 59. [Google Scholar] [CrossRef][Green Version]
- Pan, M.; Li, J.; Lv, X.; Du, G.; Liu, L. Molecular engineering of chitinase from Bacillus sp. DAU101 for enzymatic production of chitooligosaccharides. Enzym. Microb. Technol. 2019, 124, 54–62. [Google Scholar] [CrossRef]
- Cui, D.; Yang, J.; Lu, B.; Shen, H. Efficient Preparation of Chitooligosaccharide with a Potential Chitosanase Csn-SH and Its Application for Fungi Disease Protection. Front. Microbiol. 2021, 12, 682829. [Google Scholar] [CrossRef]
- Wang, D.; Li, A.; Han, H.; Liu, T.; Yang, Q. A potent chitinase from Bacillus subtilis for the efficient bioconversion of chitin-containing wastes. Int. J. Biol. Macromol. 2018, 116, 863–868. [Google Scholar] [CrossRef]
- Rani, T.S.; Madhuprakash, J.; Podile, A.R. Chitinase-E from Chitiniphilus shinanonensis generates chitobiose from chitin flakes. Int. J. Biol. Macromol. 2020, 163, 1037–1043. [Google Scholar] [CrossRef]
- Zhang, A.; Gao, C.; Wang, J.; Chen, K.; Ouyang, P. An efficient enzymatic production of N-acetyl-d-glucosamine from crude chitin powders. Green Chem. 2016, 18, 2147–2154. [Google Scholar] [CrossRef]
- Mallakuntla, M.K.; Vaikuntapu, P.R.; Bhuvanachandra, B.; Das, S.N.; Podile, A.R. Transglycosylation by a chitinase from Enterobacter cloacae subsp. cloacae generates longer chitin oligosaccharides. Sci. Rep. 2017, 7, 5113. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, Y.; Ma, M.; Oh, D.-H.; Fu, X. Characterization of chitinase from Exiguobacterium antarcticum and its bioconversion of crayfish shell into chitin oligosaccharides. Food Res. Int. 2022, 158, 111517. [Google Scholar] [CrossRef] [PubMed]
- Vaikuntapu, P.R.; Mallakuntla, M.K.; Das, S.N.; Bhuvanachandra, B.; Ramakrishna, B.; Nadendla, S.R.; Podile, A.R. Applicability of endochitinase of Flavobacterium johnsoniae with transglycosylation activity in generating long-chain chitooligosaccharides. Int. J. Biol. Macromol. 2018, 117, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Behera, P.K.; Madhuprakash, J. Efficient conversion of crystalline chitin to N-acetylglucosamine and N,N’-diacetylchitobiose by the enzyme cocktail produced by Paenibacillus sp. LS1. Carbohydr. Polym. 2020, 250, 116889. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fu, X.; Yan, Q.; Guo, Y.; Liu, Z.; Jiang, Z. Cloning, expression, purification and application of a novel chitinase from a thermophilic marine bacterium Paenibacillus barengoltzii. Food Chem. 2016, 192, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Yan, Q.; Wang, J.; Yang, S.; Jiang, Z. Purification and biochemical characterization of novel acidic chitinase from Paenicibacillus barengoltzii. Int. J. Biol. Macromol. 2016, 91, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Sun, J.; Secundo, F.; Gao, X.; Xue, C.; Mao, X. Cloning, characterization and substrate degradation mode of a novel chitinase from Streptomyces albolongus ATCC 27414. Food Chem. 2018, 261, 329–336. [Google Scholar] [CrossRef]
- Tanaya Behera, H.; Mojumdar, A.; Kumari, K.; Kumar Gouda, S.; Das, S.; Ray, L. Exploration of genomic and functional features of chitinolytic bacterium Streptomyces chilikensis RC1830, isolated from Chilika Lake, India. 3 Biotech 2022, 12, 120. [Google Scholar] [CrossRef]
- Behera, H.T.; Mojumdar, A.; Das, S.R.; Jema, S.; Ray, L. Production of N-acetyl chitooligosaccharide by novel Streptomyces chilikensis strain RC1830 and its evaluation for anti-radical, anti-inflammatory, anti-proliferative and cell migration potential. Bioresour. Technol. Rep. 2020, 11, 100428. [Google Scholar] [CrossRef]
- Xu, T.; Qi, M.; Liu, H.; Cao, D.; Xu, C.; Wang, L.; Qi, B. Chitin degradation potential and whole-genome sequence of Streptomyces diastaticus strain CS1801. AMB Express 2020, 10, 29. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Ma, J.; Yan, Q.; Jiang, Z.; Yang, S. Biochemical characterization of a bifunctional chitinase/lysozyme from Streptomyces sampsonii suitable for N-acetyl chitobiose production. Biotechnol. Lett. 2020, 42, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Le, B.; Yang, S.H. Characterization of a chitinase from Salinivibrio sp. BAO-1801 as an antifungal activity and a biocatalyst for producing chitobiose. J. Basic Microbiol. 2018, 58, 848–856. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, R.; Tewari, R. Production of N-Acetylglucosamine Using Recombinant Chitinolytic Enzymes. Indian J. Microbiol. 2011, 51, 319–325. [Google Scholar] [CrossRef][Green Version]
- Thomas, R.; Fukamizo, T.; Suginta, W. Bioeconomic production of high-quality chitobiose from chitin food wastes using an in-house chitinase from Vibrio campbellii. Bioresour. Bioprocess. 2022, 9, 86. [Google Scholar] [CrossRef]
- Bai, L.; Kim, J.; Son, K.-H.; Chung, C.-W.; Shin, D.-H.; Ku, B.-H.; Kim, D.Y.; Park, H.-Y. Novel Bi-Modular GH19 Chitinase with Broad pH Stability from a Fibrolytic Intestinal Symbiont of Eisenia fetida, Cellulosimicrobium funkei HY-13. Biomolecules 2021, 11, 1735. [Google Scholar] [CrossRef] [PubMed]
- Vaikuntapu, P.R.; Rambabu, S.; Madhuprakash, J.; Podile, A.R. A new chitinase-D from a plant growth promoting Serratia marcescens GPS5 for enzymatic conversion of chitin. Bioresour. Technol. 2016, 220, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Swiontek Brzezinska, M.; Jankiewicz, U.; Kalwasińska, A.; Świątczak, J.; Żero, K. Characterization of chitinase from Streptomyces luridiscabiei U05 and its antagonist potential against fungal plant pathogens. J. Phytopathol. 2019, 167, 404–412. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Jansson, H.B.; Salinas, J.; Lopez-Llorca, L.V. Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J. Appl. Microbiol. 2008, 104, 541–553. [Google Scholar] [CrossRef]
- Rush, T.A.; Puech-Pagès, V.; Bascaules, A.; Jargeat, P.; Maillet, F.; Haouy, A.; Maës, A.Q.; Carriel, C.C.; Khokhani, D.; Keller-Pearson, M.; et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 2020, 11, 3897. [Google Scholar] [CrossRef]
- Alves, T.B.; de Oliveira Ornela, P.H.; de Oliveira, A.H.C.; Jorge, J.A.; Guimarães, L.H.S. Production and characterization of a thermostable antifungal chitinase secreted by the filamentous fungus Aspergillus niveus under submerged fermentation. 3 Biotech 2018, 8, 369. [Google Scholar] [CrossRef]
- Suresh, P.V.; Anil Kumar, P.K. Enhanced degradation of α-chitin materials prepared from shrimp processing byproduct and production of N-acetyl-d-glucosamine by thermoactive chitinases from soil mesophilic fungi. Biodegradation 2012, 23, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Production of chitinase from thermophilic Humicola grisea and its application in production of bioactive chitooligosaccharides. Int. J. Biol. Macromol. 2017, 104, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Homthong, M.; Kubera, A.; Srihuttagum, M.; Hongtrakul, V. Isolation and characterization of chitinase from soil fungi, Paecilomyces sp. Agric. Nat. Resour. 2016, 50, 232–242. [Google Scholar] [CrossRef][Green Version]
- Krolicka, M.; Hinz, S.W.A.; Koetsier, M.J.; Joosten, R.; Eggink, G.; van den Broek, L.A.M.; Boeriu, C.G. Chitinase Chi1 from Myceliophthora thermophila C1, a Thermostable Enzyme for Chitin and Chitosan Depolymerization. J. Agric. Food Chem. 2018, 66, 1658–1669. [Google Scholar] [CrossRef]
- Kidibule, P.E.; Santos-Moriano, P.; Jiménez-Ortega, E.; Ramírez-Escudero, M.; Limón, M.C.; Remacha, M.; Plou, F.J.; Sanz-Aparicio, J.; Fernández-Lobato, M. Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: Enzymatic activity and structural basis of protein specificity. Microb. Cell Factories 2018, 17, 47. [Google Scholar] [CrossRef]
- Yang, S.; Fu, X.; Yan, Q.; Jiang, Z.; Wang, J. Biochemical Characterization of a Novel Acidic Exochitinase from Rhizomucor miehei with Antifungal Activity. J. Agric. Food Chem. 2016, 64, 461–469. [Google Scholar] [CrossRef]
- Akamatsu, A.; Wong, H.L.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y.; et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 Module Is an Essential Early Component of Chitin-Induced Rice Immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef]
- Takashima, T.; Sunagawa, R.; Uechi, K.; Taira, T. Antifungal activities of LysM-domain multimers and their fusion chitinases. Int. J. Biol. Macromol. 2020, 154, 1295–1302. [Google Scholar] [CrossRef]
- Kitaoku, Y.; Umemoto, N.; Ohnuma, T.; Numata, T.; Taira, T.; Sakuda, S.; Fukamizo, T. A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: Crystal structure and ligand-binding studies. Planta 2015, 242, 895–907. [Google Scholar] [CrossRef]
- Inamine, S.; Onaga, S.; Ohnuma, T.; Fukamizo, T.; Taira, T. Purification, cDNA cloning, and characterization of LysM-containing plant chitinase from horsetail (Equisetum arvense). Biosci. Biotechnol. Biochem. 2015, 79, 1296–1304. [Google Scholar] [CrossRef]
- Kitaoku, Y.; Taira, T.; Numata, T.; Ohnuma, T.; Fukamizo, T. Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants. Plant Sci. 2022, 321, 111310. [Google Scholar] [CrossRef]
- Kuba, Y.; Takashima, T.; Uechi, K.; Taira, T. Purification, cDNA cloning, and characterization of plant chitinase with a novel domain combination from lycophyte Selaginella doederleinii. Biosci. Biotechnol. Biochem. 2018, 82, 1742–1752. [Google Scholar] [CrossRef]
- Kawamoto, D.; Takashima, T.; Fukamizo, T.; Numata, T.; Ohnuma, T. A conserved loop structure of GH19 chitinases assists the enzyme function from behind the core-functional region. Glycobiology 2022, 32, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Tabata, E.; Kashimura, A.; Wakita, S.; Ohno, M.; Sakaguchi, M.; Sugahara, Y.; Imamura, Y.; Seki, S.; Ueda, H.; Matoska, V.; et al. Protease resistance of porcine acidic mammalian chitinase under gastrointestinal conditions implies that chitin-containing organisms can be sustainable dietary resources. Sci. Rep. 2017, 7, 12963. [Google Scholar] [CrossRef] [PubMed]
- Tabata, E.; Wakita, S.; Kashimura, A.; Sugahara, Y.; Matoska, V.; Bauer, P.O.; Oyama, F. Residues of acidic chitinase cause chitinolytic activity degrading chitosan in porcine pepsin preparations. Sci. Rep. 2019, 9, 15609. [Google Scholar] [CrossRef] [PubMed]
- Wakita, S.; Sugahara, Y.; Nakamura, M.; Kobayashi, S.; Matsuda, K.; Takasaki, C.; Kimura, M.; Kida, Y.; Uehara, M.; Tabata, E.; et al. Mouse Acidic Chitinase Effectively Degrades Random-Type Chitosan to Chitooligosaccharides of Variable Lengths under Stomach and Lung Tissue pH Conditions. Molecules 2021, 26, 6706. [Google Scholar] [CrossRef]
- Ohno, M.; Kimura, M.; Miyazaki, H.; Okawa, K.; Onuki, R.; Nemoto, C.; Tabata, E.; Wakita, S.; Kashimura, A.; Sakaguchi, M.; et al. Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system. Sci. Rep. 2016, 6, 37756. [Google Scholar] [CrossRef]
- Tabata, E.; Kashimura, A.; Wakita, S.; Ohno, M.; Sakaguchi, M.; Sugahara, Y.; Kino, Y.; Matoska, V.; Bauer, P.O.; Oyama, F. Gastric and intestinal proteases resistance of chicken acidic chitinase nominates chitin-containing organisms for alternative whole edible diets for poultry. Sci. Rep. 2017, 7, 6662. [Google Scholar] [CrossRef]
- Tabata, E.; Kashimura, A.; Uehara, M.; Wakita, S.; Sakaguchi, M.; Sugahara, Y.; Yurimoto, T.; Sasaki, E.; Matoska, V.; Bauer, P.O.; et al. High expression of acidic chitinase and chitin digestibility in the stomach of common marmoset (Callithrix jacchus), an insectivorous nonhuman primate. Sci. Rep. 2019, 9, 159. [Google Scholar] [CrossRef]
- Uehara, M.; Takasaki, C.; Wakita, S.; Sugahara, Y.; Tabata, E.; Matoska, V.; Bauer, P.O.; Oyama, F. Crab-Eating Monkey Acidic Chitinase (CHIA) Efficiently Degrades Chitin and Chitosan under Acidic and High-Temperature Conditions. Molecules 2022, 27, 409. [Google Scholar] [CrossRef]
- Uehara, M.; Tabata, E.; Okuda, M.; Maruyama, Y.; Matoska, V.; Bauer, P.O.; Oyama, F. Robust chitinolytic activity of crab-eating monkey (Macaca fascicularis) acidic chitinase under a broad pH and temperature range. Sci. Rep. 2021, 11, 15470. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, W.-C.; Gao, L.; Sun, J.; Liu, Z.; Mao, X. Efficient enzymatic hydrolysis of ionic liquid pretreated chitin and its dissolution mechanism. Carbohydr. Polym. 2019, 211, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Ungkulpasvich, U.; Baramee, S.; Uke, A.; Kosugi, A. Symbiotic chitin degradation by a novel anaerobic thermophilic bacterium Hydrogenispora sp. UUS1-1 and the bacterium Tepidanaerobacter sp. GT38. Enzym. Microb. Technol. 2021, 144, 109740. [Google Scholar] [CrossRef] [PubMed]
- Taokaew, S.; Kriangkrai, W. Recent Progress in Processing Cellulose Using Ionic Liquids as Solvents. Polysaccharides 2022, 3, 671–691. [Google Scholar] [CrossRef]
- Kumar, M.; Madhuprakash, J.; Balan, V.; Kumar Singh, A.; Vivekanand, V.; Pareek, N. Chemoenzymatic production of chitooligosaccharides employing ionic liquids and Thermomyces lanuginosus chitinase. Bioresour. Technol. 2021, 337, 125399. [Google Scholar] [CrossRef]
- Ngo, D.-N.; Kim, M.-M.; Kim, S.-K. Protective effects of aminoethyl-chitooligosaccharides against oxidative stress in mouse macrophage RAW 264.7 cells. Int. J. Biol. Macromol. 2012, 50, 624–631. [Google Scholar] [CrossRef]
- Hong, S.; Ngo, D.-N.; Kim, M.-M. Inhibitory effect of aminoethyl-chitooligosaccharides on invasion of human fibrosarcoma cells. Environ. Toxicol. Pharmacol. 2016, 45, 309–314. [Google Scholar] [CrossRef]
- Karagozlu, M.Z.; Karadeniz, F.; Kong, C.-S.; Kim, S.-K. Aminoethylated chitooligomers and their apoptotic activity on AGS human cancer cells. Carbohydr. Polym. 2012, 87, 1383–1389. [Google Scholar] [CrossRef]
- Ngo, D.-H.; Ngo, D.-N.; Vo, T.-S.; Ryu, B.; Van Ta, Q.; Kim, S.-K. Protective effects of aminoethyl-chitooligosaccharides against oxidative stress and inflammation in murine microglial BV-2 cells. Carbohydr. Polym. 2012, 88, 743–747. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Xing, L.; Sun, F.; Yang, Z.; Wang, F.; Tan, H. Effects of the anti-angiogenic carbohydrate-peptide conjugate, chitooligosaccharide-ES2 on endothelial cells and tumor-bearing mice. Carbohydr. Polym. 2019, 208, 302–313. [Google Scholar] [CrossRef]
- Yu, Y.; Sun, F.; Zhang, C.; Wang, Z.; Liu, J.; Tan, H. Study on glyco-modification of endostatin-derived synthetic peptide endostatin2 (ES2) by soluble chitooligosaccharide. Carbohydr. Polym. 2016, 154, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Benjakul, S.; Huda, N.; Xu, C.; Wu, P. Preparation and characterization of squid pen chitooligosaccharide–epigallocatechin gallate conjugates and their antioxidant and antimicrobial activities. RSC Adv. 2020, 10, 33196–33204. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.-S.; Ngo, D.-H.; Bach, L.G.; Ngo, D.-N.; Kim, S.-K. The free radical scavenging and anti-inflammatory activities of gallate-chitooligosaccharides in human lung epithelial A549 cells. Process Biochem. 2017, 54, 188–194. [Google Scholar] [CrossRef]
- Vo, T.-S.; Ngo, D.-H.; Kim, S.-K. Gallic acid-grafted chitooligosaccharides suppress antigen-induced allergic reactions in RBL-2H3 mast cells. Eur. J. Pharm. Sci. 2012, 47, 527–533. [Google Scholar] [CrossRef]
- Ryu, B.; Kim, S.-Y.; Vo, T.-S.; Kim, W.-S.; Kim, D.G.; Kim, S.-K. Characterization of the in vitro effects of gallic acid-grafted-chitooligosaccharides in the suppression of AGS human gastric cancer cell proliferation. RSC Adv. 2017, 7, 24561–24568. [Google Scholar] [CrossRef]
- Yu, J.; Wang, Q.; Zhang, H.; Qin, X.; Chen, H.; Corke, H.; Hu, Z.; Liu, G. Increased stability of curcumin-loaded pickering emulsions based on glycated proteins and chitooligosaccharides for functional food application. LWT 2021, 148, 111742. [Google Scholar] [CrossRef]
- Jiang, Y.; Fu, C.; Liu, G.; Guo, J.; Su, Z. Cholesterol-lowering effects and potential mechanisms of chitooligosaccharide capsules in hyperlipidemic rats. Food Nutr. Res. 2018, 62, 1446. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Han, J.; Yu, Y.; Li, X.; Wang, Y.; Tian, H.; Guo, S.; Jin, S.; Luo, T.; Qin, S. Chitosan oligosaccharide decreases very-low-density lipoprotein triglyceride and increases high-density lipoprotein cholesterol in high-fat-diet-fed rats. Exp. Biol. Med. 2011, 236, 1064–1069. [Google Scholar] [CrossRef]
- Huang, L.; Chen, J.; Cao, P.; Pan, H.; Ding, C.; Xiao, T.; Zhang, P.; Guo, J.; Su, Z. Anti-Obese Effect of Glucosamine and Chitosan Oligosaccharide in High-Fat Diet-Induced Obese Rats. Mar. Drugs 2015, 13, 2732–2756. [Google Scholar] [CrossRef]
- Zong, C.; Yu, Y.; Song, G.; Luo, T.; Li, L.; Wang, X.; Qin, S. Chitosan oligosaccharides promote reverse cholesterol transport and expression of scavenger receptor BI and CYP7A1 in mice. Exp. Biol. Med. 2012, 237, 194–200. [Google Scholar] [CrossRef]
- Choi, C.-R.; Kim, E.-K.; Kim, Y.-S.; Je, J.-Y.; An, S.-H.; Lee, J.D.; Wang, J.H.; Ki, S.S.; Jeon, B.-T.; Moon, S.-H.; et al. Chitooligosaccharides decreases plasma lipid levels in healthy men. Int. J. Food Sci. Nutr. 2012, 63, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Huang, G.; Yang, Q.; Guo, J.; Su, Z. The effect of chitooligosaccharides on oleic acid-induced lipid accumulation in HepG2 cells. Saudi Pharm. J. 2016, 24, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Rahman, M.A.; Kim, S.W.; Baek, Y.M.; Hwang, H.J.; Oh, J.Y.; Hwang, H.S.; Lee, S.H.; Yun, J.W. Chitosan oligosaccharides inhibit adipogenesis in 3T3-L1 adipocytes. J. Microbiol. Biotechnol. 2008, 18, 80–87. [Google Scholar] [PubMed]
- Šimůnek, J.; Brandysová, V.; Koppová, I. The antimicrobial action of chitosan, low molar mass chitosan, and chitooligosaccharides on human colonic bacteria. Folia Microbiol. 2012, 57, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Šimůnek, J.; Koppová, I.; Filip, L.; Tishchenko, G.; BeŁżecki, G. The antimicrobial action of low-molar-mass chitosan, chitosan derivatives and chitooligosaccharides on bifidobacteria. Folia Microbiol. 2010, 55, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.-W.; Liu, C.-P.; Wu, C.; Wang, S.-L. Applied development of crude enzyme from Bacillus cereus in prebiotics and microbial community changes in soil. Carbohydr. Polym. 2013, 92, 2141–2148. [Google Scholar] [CrossRef]
- Vela Gurovic, M.S.; Dello Staffolo, M.; Montero, M.; Debbaudt, A.; Albertengo, L.; Rodríguez, M.S. Chitooligosaccharides as novel ingredients of fermented foods. Food Funct. 2015, 6, 3437–3443. [Google Scholar] [CrossRef]
- Yang, F.; Luan, B.; Sun, Z.; Yang, C.; Yu, Z.; Li, X. Application of chitooligosaccharides as antioxidants in beer to improve the flavour stability by protecting against beer staling during storage. Biotechnol. Lett. 2017, 39, 305–310. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Wang, T.; Guo, X.; Luan, J.; Sun, Y.; Li, X. The use of chitooligosaccharide in beer brewing for protection against beer-spoilage bacteria and its influence on beer performance. Biotechnol. Lett. 2016, 38, 629–635. [Google Scholar] [CrossRef]
- Jing, B.; Cheng, G.; Li, J.; Wang, Z.A.; Du, Y. Inhibition of Liver Tumor Cell Metastasis by Partially Acetylated Chitosan Oligosaccharide on A Tumor-Vessel Microsystem. Mar. Drugs 2019, 17, 415. [Google Scholar] [CrossRef]
- Dou, J.; Ma, P.; Xiong, C.; Tan, C.; Du, Y. Induction of apoptosis in human acute leukemia HL-60 cells by oligochitosan through extrinsic and intrinsic pathway. Carbohydr. Polym. 2011, 86, 19–24. [Google Scholar] [CrossRef]
- Zhai, X.; Yuan, S.; Yang, X.; Zou, P.; Li, L.; Li, G.; Shao, Y.; Abd El-Aty, A.M.; Hacımüftüoğlu, A.; Wang, J. Chitosan Oligosaccharides Induce Apoptosis in Human Renal Carcinoma via Reactive-Oxygen-Species-Dependent Endoplasmic Reticulum Stress. J. Agric. Food Chem. 2019, 67, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Mattaveewong, T.; Wongkrasant, P.; Chanchai, S.; Pichyangkura, R.; Chatsudthipong, V.; Muanprasat, C. Chitosan oligosaccharide suppresses tumor progression in a mouse model of colitis-associated colorectal cancer through AMPK activation and suppression of NF-κB and mTOR signaling. Carbohydr. Polym. 2016, 145, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Li, H.; Qiao, J.; Yang, Y.; Wang, Y.; Liu, W.; Han, B. Potential Analysis and Preparation of Chitosan Oligosaccharides as Oral Nutritional Supplements of Cancer Adjuvant Therapy. Int. J. Mol. Sci. 2019, 20, 920. [Google Scholar] [CrossRef] [PubMed]
- Johansen, A.Z.; Carretta, M.; Thorseth, M.-L.; Khan, S.; Fjæstad, K.Y.; Brøchner, C.B.; Linder, H.; Ankjærgaard, C.; Donia, M.; Chen, I.; et al. Chitooligosaccharides Improve the Efficacy of Checkpoint Inhibitors in a Mouse Model of Lung Cancer. Pharmaceutics 2022, 14, 1406. [Google Scholar] [CrossRef]
- Yeo, I.J.; Lee, C.-K.; Han, S.-B.; Yun, J.; Hong, J.T. Roles of chitinase 3-like 1 in the development of cancer, neurodegenerative diseases, and inflammatory diseases. Pharmacol. Ther. 2019, 203, 107394. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Su, Z.; Li, Y.; Zhang, X.; You, Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal Transduct. Target. Ther. 2020, 5, 201. [Google Scholar] [CrossRef]
- Theile, S.; Johansen, J.S.; Nielsen, D.L.; Jensen, B.V.; Hansen, C.P.; Hasselby, J.P.; Eiríksson, S.V.; Chen, I.M. A Randomized Placebo-Controlled Phase 2 Study of Gemcitabine and Capecitabine with or without T-ChOS as Adjuvant Therapy in Patients with Resected Pancreatic Cancer (CHIPAC). Pharmaceutics 2022, 14, 509. [Google Scholar] [CrossRef]
- Zhou, Q.; Cui, L.; Ren, L.; Wang, P.; Deng, C.; Wang, Q.; Fan, X. Preparation of a multifunctional fibroin-based biomaterial via laccase-assisted grafting of chitooligosaccharide. Int. J. Biol. Macromol. 2018, 113, 1062–1072. [Google Scholar] [CrossRef]
- Ailincai, D.; Rosca, I.; Morariu, S.; Mititelu-Tartau, L.; Marin, L. Iminoboronate-chitooligosaccharides hydrogels with strong antimicrobial activity for biomedical applications. Carbohydr. Polym. 2022, 276, 118727. [Google Scholar] [CrossRef]
- Yusof, N.; Ali Abdul, M.; Hassan, O.; Abu Bakar Che, A.; John Yew Huat, T. Effect of Chitosan Oligosaccharides on the Growth of Bifidobacterium Species. Malays. J. Appl. Sci. 2016, 1, 13–23. [Google Scholar]
- Wu, M.; Li, J.; An, Y.; Li, P.; Xiong, W.; Li, J.; Yan, D.; Wang, M.; Zhong, G. Chitooligosaccharides Prevents the Development of Colitis-Associated Colorectal Cancer by Modulating the Intestinal Microbiota and Mycobiota. Front. Microbiol. 2019, 10, 2101. [Google Scholar] [CrossRef] [PubMed]
- Han, F.S.; Yang, S.J.; Lin, M.B.; Chen, Y.Q.; Yang, P.; Xu, J.M. Chitooligosaccharides promote radiosensitivity in colon cancer line SW480. World J. Gastroenterol. 2016, 22, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Han, F.-S.; Cui, B.-H.; You, X.-F.; Xing, Y.-F.; Sun, X.-W. Anti-proliferation and radiosensitization effects of chitooligosaccharides on human lung cancer line HepG2. Asian Pac. J. Trop. Med. 2015, 8, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Fang, I.M.; Yang, C.-M.; Yang, C.-H. Chitosan oligosaccharides prevented retinal ischemia and reperfusion injury via reduced oxidative stress and inflammation in rats. Exp. Eye Res. 2015, 130, 38–50. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Yang, J.; Yin, H.; Wang, W.; Lu, H.; Du, Y. Nitric oxide production and its functional link with OIPK in tobacco defense response elicited by chitooligosaccharide. Plant Cell Rep. 2011, 30, 1153–1162. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-Induced Dimerization Activates a Plant Immune Receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Han, Z.; Gong, X.; Zhang, H.; Chai, J. Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP. Structure 2016, 24, 1192–1200. [Google Scholar] [CrossRef]
- Ramakrishna, B.; Sarma, P.V.S.R.N.; Ankati, S.; Bhuvanachandra, B.; Podile, A.R. Elicitation of defense response by transglycosylated chitooligosaccharides in rice seedlings. Carbohydr. Res. 2021, 510, 108459. [Google Scholar] [CrossRef]
- Sun, G.; Yang, Q.; Zhang, A.; Guo, J.; Liu, X.; Wang, Y.; Ma, Q. Synergistic effect of the combined bio-fungicides ε-poly-l-lysine and chitooligosaccharide in controlling grey mould (Botrytis cinerea) in tomatoes. Int. J. Food Microbiol. 2018, 276, 46–53. [Google Scholar] [CrossRef]
- Lan, W.; Wang, W.; Yu, Z.; Qin, Y.; Luan, J.; Li, X. Enhanced germination of barley (Hordeum vulgare L.) using chitooligosaccharide as an elicitor in seed priming to improve malt quality. Biotechnol. Lett. 2016, 38, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, K.; Liu, S.; Xing, R.; Yu, H.; Chen, X.; Li, P. Size effects of chitooligomers on the growth and photosynthetic characteristics of wheat seedlings. Carbohydr. Polym. 2016, 138, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Li, K.; Liu, S.; He, X.; Zhang, X.; Xing, R.; Li, P. Effect of Sulfated Chitooligosaccharides on Wheat Seedlings (Triticum aestivum L.) under Salt Stress. J. Agric. Food Chem. 2016, 64, 2815–2821. [Google Scholar] [CrossRef] [PubMed]
- Dzung, N.A.; Khanh, V.T.P.; Dzung, T.T. Research on impact of chitosan oligomers on biophysical characteristics, growth, development and drought resistance of coffee. Carbohydr. Polym. 2011, 84, 751–755. [Google Scholar] [CrossRef]
- Zong, H.; Li, K.; Liu, S.; Song, L.; Xing, R.; Chen, X.; Li, P. Improvement in cadmium tolerance of edible rape (Brassica rapa L.) with exogenous application of chitooligosaccharide. Chemosphere 2017, 181, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Kidibule, P.E.; Costa, J.; Atrei, A.; Plou, F.J.; Fernandez-Lobato, M.; Pogni, R. Production and characterization of chitooligosaccharides by the fungal chitinase Chit42 immobilized on magnetic nanoparticles and chitosan beads: Selectivity, specificity and improved operational utility. RSC Adv. 2021, 11, 5529–5536. [Google Scholar] [CrossRef]
- Li, J.; Mao, X. Construction of an Immobilized Enzyme Membrane Reactor for Efficient and Sustainable Conversion of Ionic Liquid/Ultrasound-Pretreated Chitin. ACS Sustain. Chem. Eng. 2022, 10, 7536–7544. [Google Scholar] [CrossRef]
- Busscher Guuske, F.; ZajÍCovÁ, V.; HnÁTkovÁ, T.; Schmeets Alex Alois, J.; BeneŠ, H. Enzymatic Crop Protection and Process for Preparing Biological Crop Protection Composition. WO 2021/148575 A1, 21 January 2021. [Google Scholar]
- Wong John, M. Methods of Pest Control. US 2018/0317499 A1, 12 January 2018. [Google Scholar]
- Reuter Christopher, J.; Mackenzie, S. Composition and Method for Enhancing Chitin-Containing Fertilizers. WO 2019/079031 A2, 3 October 2018. [Google Scholar]
- Fu, X.; Liang, C.; Lyu, J.; Lyu, A.; Yao, Y.A.N.; Guo, S. Exiguobacterium Antarcticum DW2 and Method for Preparing Chitosan Oligosaccharide by Using Same. CN 113862192 A, 27 October 2021. [Google Scholar]
- Luo, X.; Deng, J.; Lu, D.; Li, Z.; Shi, D.A.N.; Mao, H. Enzymatic Green process of Producing Chitin Oligosaccharide, Astaxanthin, Proteins and Calcium Powder by Using Shrimp Shells. CN 110628854 A, 20 September 2019. [Google Scholar]
- Yang, L.; Liu, C.; Jiang, M.; Wu, J.; Shen, N.; Huang, W.; Zhu, Y.; Xu, J.; Wang, J. Strain of Paenibacillus Chitinolyticus and Application of Paenibacillus Chitinolyticus. CN 110699276 A, 30 September 2019. [Google Scholar]
- Hu, Y.; Lyu, M.; Cai, J.U.N. Method for Preparing Chitooligosaccharide by Utilizing Waste Mycelia from Citric Acid Fermentation. CN 104975057 A, 15 July 2015. [Google Scholar]
- Cogollo-Herrera, K.; Bonfante-Álvarez, H.; De Ávila-Montiel, G.; Barros, A.H.; González-Delgado, Á.D. Techno-economic sensitivity analysis of large scale chitosan production process from shrimp shell wastes. Chem. Eng. Trans. 2018, 70, 2179–2184. [Google Scholar] [CrossRef]
- Gómez-Ríos, D.; Barrera-Zapata, R.; Ríos-Estepa, R. Comparison of process technologies for chitosan production from shrimp shell waste: A techno-economic approach using Aspen Plus®. Food Bioprod. Process. 2017, 103, 49–57. [Google Scholar] [CrossRef]
- Moreno-Sader, K.A.; Martinez-Consuegra, J.D.; González-Delgado, Á.D. Development of a biorefinery approach for shrimp processing in North-Colombia: Process simulation and sustainability assessment. Environ. Technol. Innov. 2021, 22, 101461. [Google Scholar] [CrossRef]
- Zuorro, A.; Moreno-Sader, K.A.; González-Delgado, Á.D. Evaluating the feasibility of a pilot-scale shrimp biorefinery via techno-economic analysis. J. Clean. Prod. 2021, 320, 128740. [Google Scholar] [CrossRef]
- Yang, H.; Gözaydın, G.; Nasaruddin, R.R.; Har, J.R.G.; Chen, X.; Wang, X.; Yan, N. Toward the Shell Biorefinery: Processing Crustacean Shell Waste Using Hot Water and Carbonic Acid. ACS Sustain. Chem. Eng. 2019, 7, 5532–5542. [Google Scholar] [CrossRef]
- Moreno-Sader, K.A.; Martínez-Consuegra, J.; González-Delgado, Á.D. An integrated biorefinery approach via material recycle/reuse networks for the extraction of value-added components from shrimp: Computer-aided simulation and environmental assessment. Food Bioprod. Process. 2021, 127, 443–453. [Google Scholar] [CrossRef]

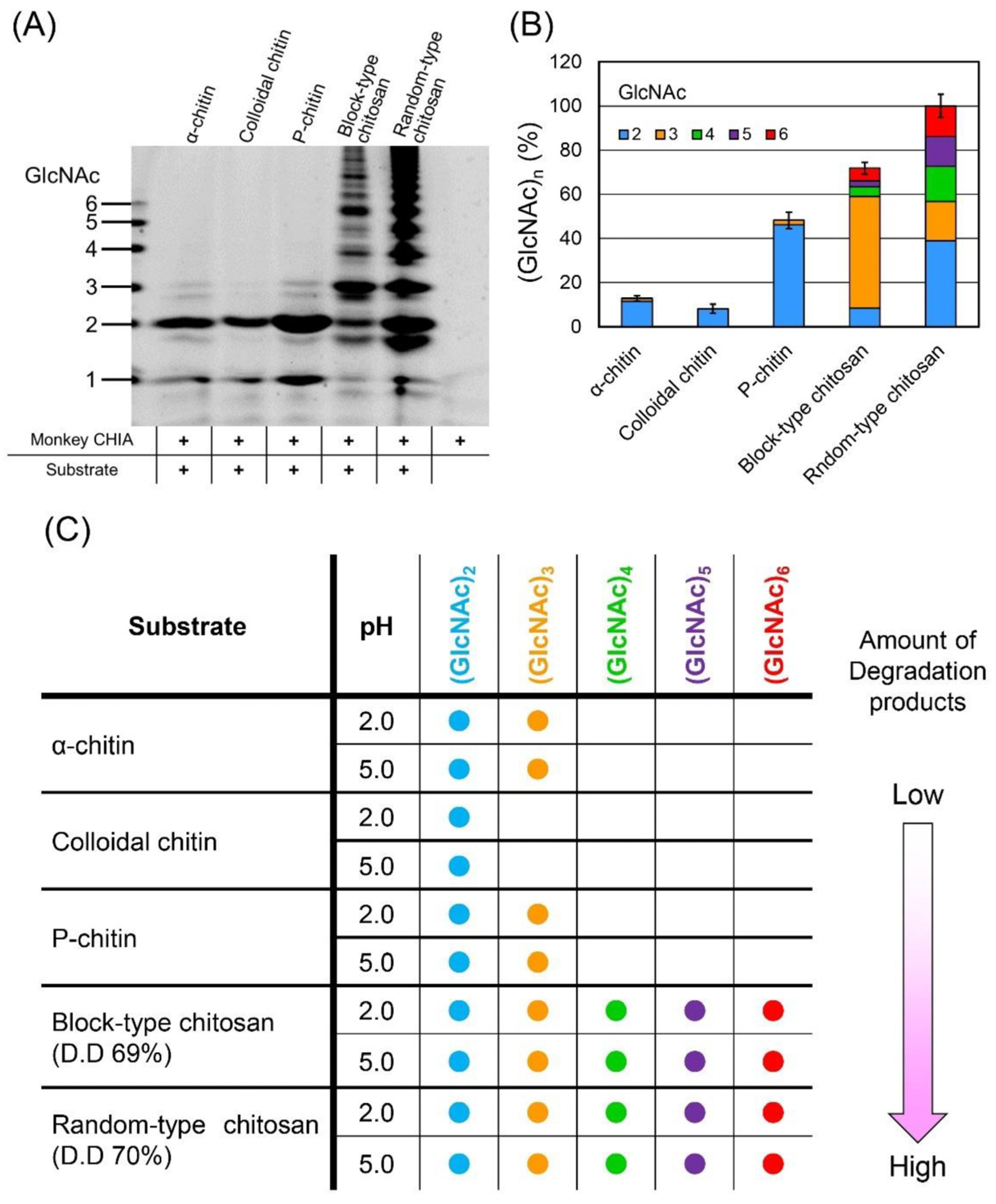

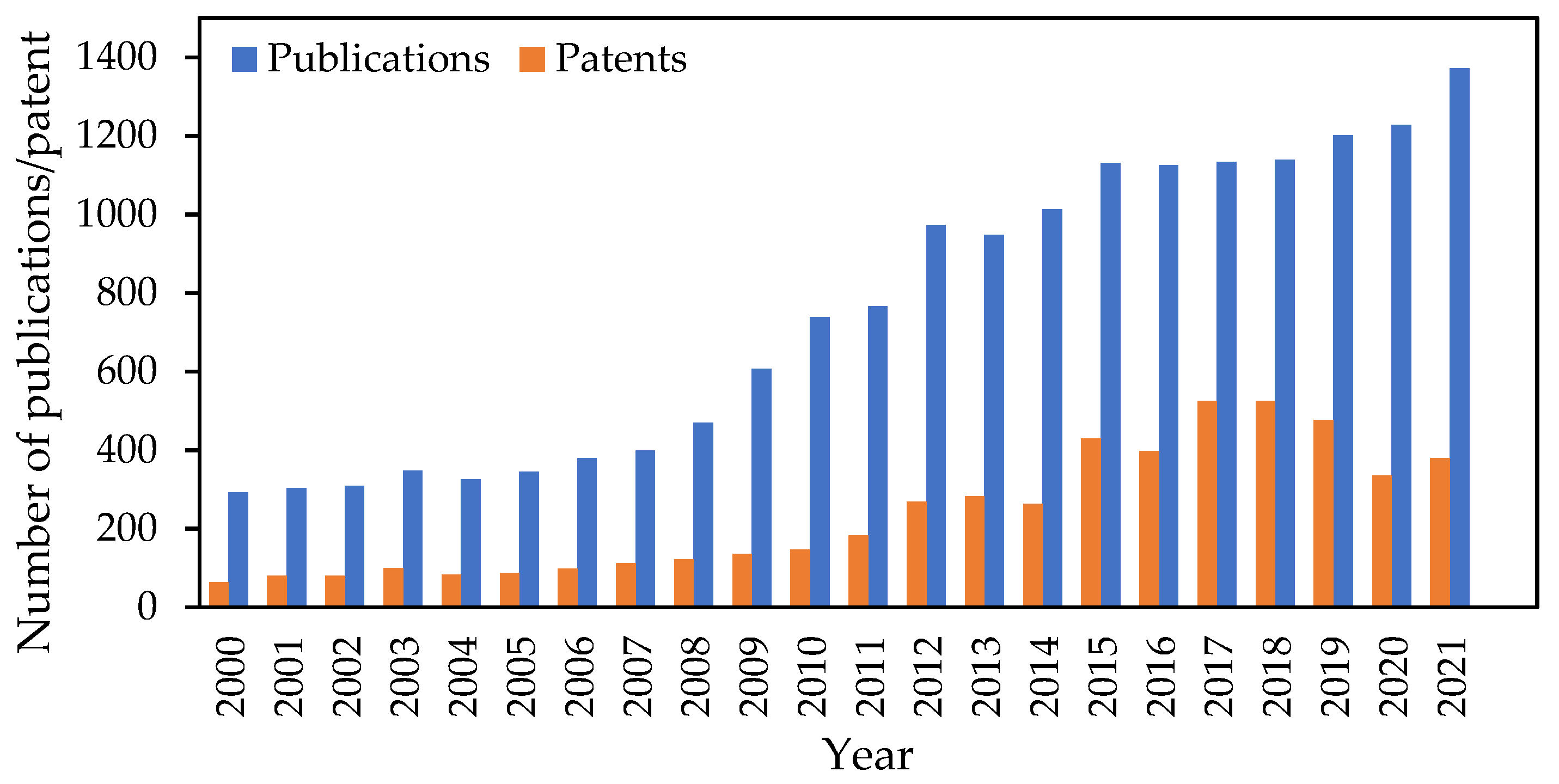
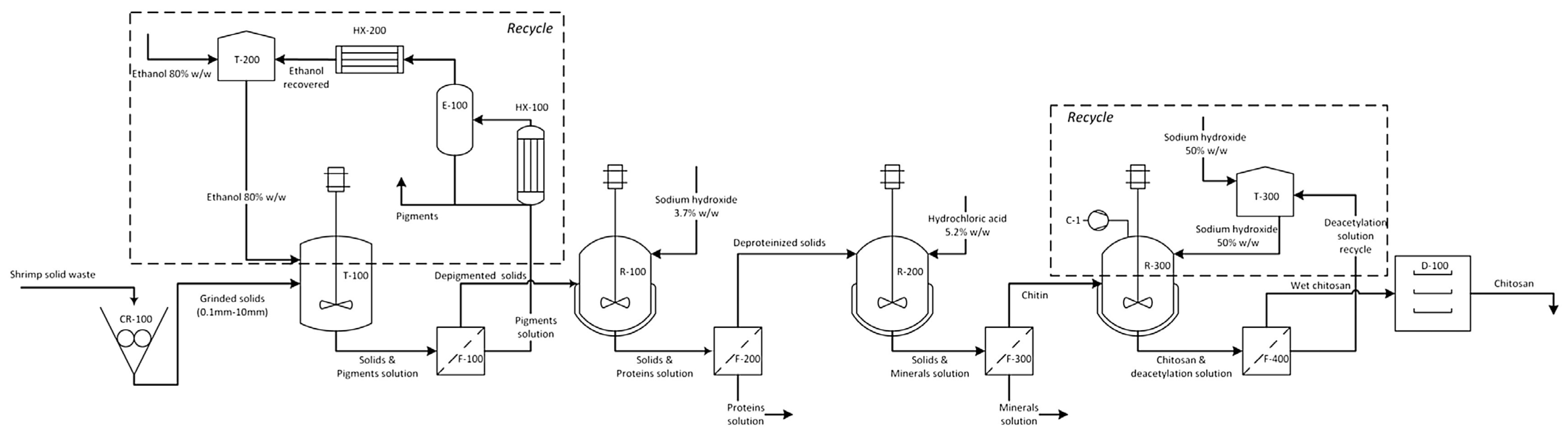
| Chitinase Source | Substrate | Yield of (GlcNAc)n (mg/g) 1 | References |
|---|---|---|---|
| Bacillus subtilis | α-chitin, β-chitin, crude crab shell powder, chitosan | 163 mg GlcNAc/g | [46,48] |
| Bacillus atrophaeus BSS | Colloidal chitosan | 806 mg (GlcNAc)2–6/g | [47] |
| Chitiniphilus shinanonensis | Shrimp/ squid pen flakes | 10.6 mg (GlcNAc)/g 62 mg (GlcNAc)/g | [49] |
| Chitinolyticbacter meiyuanensis SYBC-H1 | Shrimp chitin powder | 982 mg (GlcNAc)/g | [50] |
| Enterobacter cloacae subsp. cloacae | Colloidal chitin | 0.405 mg (GlcNAc)/g 1.06 mg (GlcNAc)2/g | [51] |
| Exiguobacterium antarcticum | Crayfish shell chitin | 761 mg (GlcNAc)1–2/g | [52] |
| Flavobacterium johnsoniae UW101 | Colloidal chitin | 59 mg (GlcNAc)2/g 47 mg (GlcNAc)3/g | [53] |
| Paenibacillus sp LS 1 | Colloidal chitin (α, β) | 53 mg (GlcNAc)1–2/g 721 mg (GlcNAc)1–2/g | [54] |
| Paenicibacillus barengoltzii | Crab shell, colloidal | 720 mg (GlcNAc)2/g | [55,56] |
| Streptomyces albolongus | Colloidal chitin | 2.8 mg (GlcNAc)/g | [57] |
| Streptomyces chilikensis RC1830 | Colloidal chitin | 761 mg (GlcNAc)1–2/g | [58,59] |
| Streptomyces diastaticus CS1801 | Colloidal chitin | 18.5 mg (GlcNAc)1–5/g | [60] |
| Streptomyces sampsonii XY 2–7 | Shrimp powder | 720 mg (GlcNAc)1–2/g | [61] |
| Salinivibrio BAO-1801 | Shrimp shell | 105 mg (GlcNAc)/g 715 mg (GlcNAc)2/g | [62] |
| Thermomyces lanuginosus | Shrimp shell | 80 mg (GlcNAc)/g 720 mg (GlcNAc)2/g | [63] |
| Vibrio campbellii (formerly V. harveyi) | Shrimp flakes | 200 mg (GlcNAc)2/g | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taokaew, S.; Kriangkrai, W. Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products. Biology 2023, 12, 87. https://doi.org/10.3390/biology12010087
Taokaew S, Kriangkrai W. Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products. Biology. 2023; 12(1):87. https://doi.org/10.3390/biology12010087
Chicago/Turabian StyleTaokaew, Siriporn, and Worawut Kriangkrai. 2023. "Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products" Biology 12, no. 1: 87. https://doi.org/10.3390/biology12010087
APA StyleTaokaew, S., & Kriangkrai, W. (2023). Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products. Biology, 12(1), 87. https://doi.org/10.3390/biology12010087







