Potential Utilization of Bacterial Consortium of Symbionts Marine Sponges in Removing Polyaromatic Hydrocarbons and Heavy Metals, Review
Simple Summary
Abstract
1. Introduction
2. Polycyclic of Aromatic and Heavy Metals Bioremediation Analysis Instrument
3. Bacterial Performance in Pollutant Bioremediation
4. Process and Mechanism of Pollutant Bioremediation by Marine Sponge Symbiont Bacteria
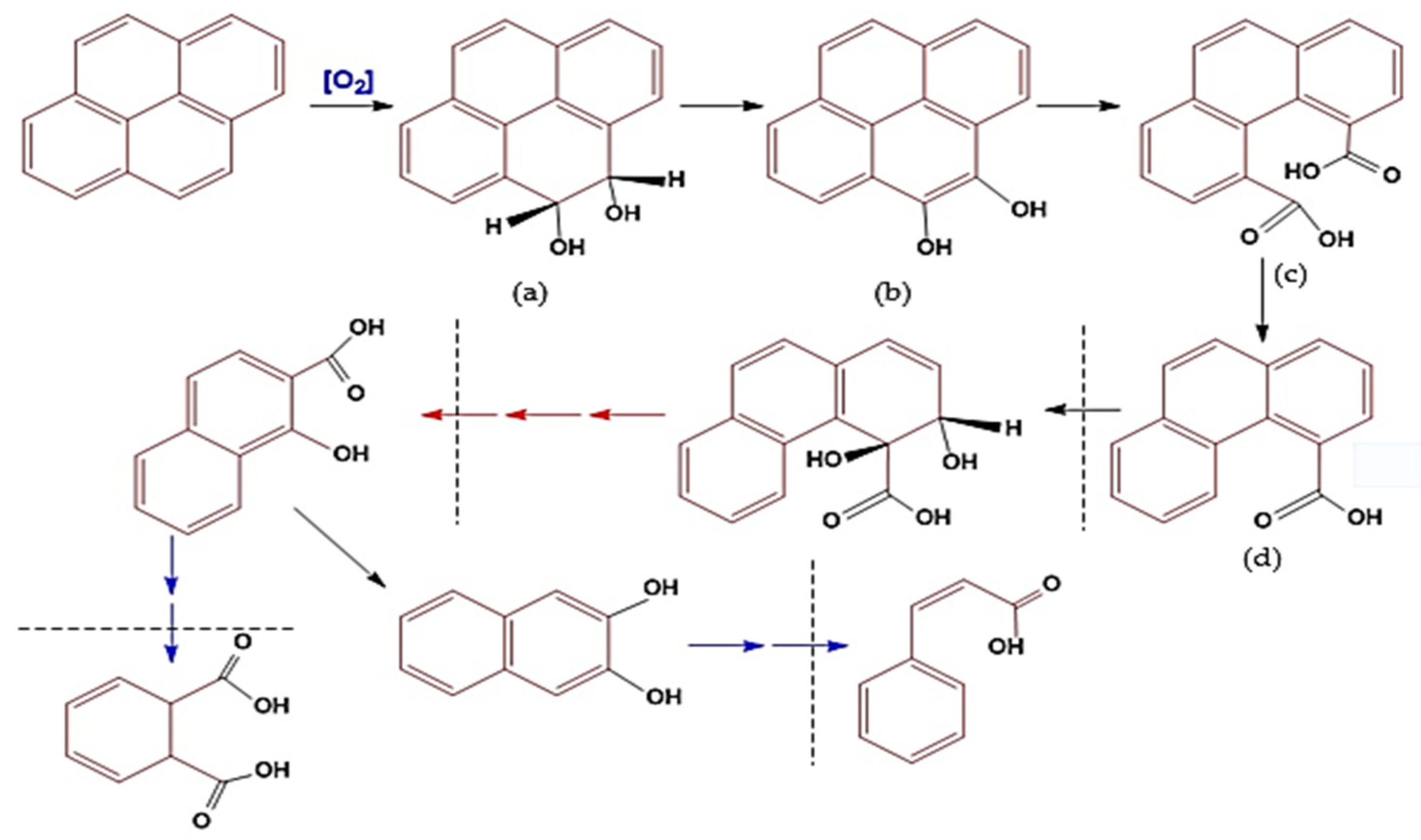
4.1. Processes and Mechanisms of Biodegradation of PAHs
4.2. Process and Mechanism of Heavy Metal Bioabsorption
5. Parameters of Pollutant Bioremediation
5.1. Biodegradation of PAHs
5.2. Heavy Metal Bioadsorption
6. Development and Formulation of Remediator Bacteria Consortium
6.1. Hydrocarbonoclastic Bacteria
6.2. Metalloclastic Bacteria
6.3. Metallo-Hydrocarbonoclastic Bacteria
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gad, A.K.; Midway, S.R. Relationship of Microplastics to Body Size for Two Estuarine Fishes. Microplastics 2022, 1, 211–220. [Google Scholar] [CrossRef]
- Akoto, O.; Azuure, A.A.; Adotey, K.D. Pesticide residues in water, sediment and fish from Tono Reservoir and their health risk implications. SpringerPlus 2016, 5, 1849. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, N.; Kratofil Krehula, L.; Ptiček Siročić, A.; Hrnjak-Murgić, Z. Analysis of recycled PET bottles products by pyrolysis-gas chromatography. Polym. Degrad. Stab. 2013, 98, 972–979. [Google Scholar] [CrossRef]
- Ye, J.; Song, Y.; Liu, Y.; Zhong, Y. Assessment of medical waste generation, associated environmental impact, and management issues after the outbreak of COVID-19: A case study of the Hubei Province in China. PLoS ONE 2022, 17, e0259207. [Google Scholar] [CrossRef]
- Abraham, N.A.; Offiong, N.A.O.; Ukafia, O.P.; Akpan, P.E. Source Apportionment of Polycyclic Aromatic Hydrocarbons (PAHs) in a Tropical Estuarine Epipelic Sediment and Its Associated Bacterial Degrading Potentials. Curr. J. of Appl. Sci. Technol. 2018, 32, 1–11. [Google Scholar] [CrossRef]
- Gran, S.A.; Ramos, Z.J.; Fuentes, E.; Bravo, D.; Pérez, D.J.M. Effect of co-contamination by PAHs and heavy metals on bacterial communities of diesel contaminated soils of south shetland islands, antarctica. Microorganisms 2020, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Septiningsih, E.; Kaseng, E.S.; Herlinah, H.; Sahrijanna, A.; Sahabuddin, S.; Asaf, R.; Athirah, A.; Isnawan, B.H.; Samidjo, G.S.; et al. Investigation of Global Trends of Pollutants in Marine Ecosystems around Barrang Caddi Island, Spermonde Archipelago Cluster: An Ecological Approach. Toxics 2022, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Asgari, A.; Nabizadeh, R.; Mahvi, A.H.; Nasseri, S.; Dehghani, M.H.; Nazmara, S.; Yaghmaeian, K. Biodegradation of total petroleum hydrocarbons from acidic sludge produced by rerefinery industries of waste oil using invessel composting. J. Env. Health Sci. Eng. 2017, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Medic, A.; Lješević, M.; Inui, H.; Stojanovi, K.; Karadzi, I.; Beskoski, V.; Koji, I. Efficient biodegradation of petroleum: N -alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa sanai with multidegradative capacity. RSC Adv. 2020, 10, 14060–14070. [Google Scholar] [CrossRef]
- Al-Dhabaan, F.A. Morphological, biochemical and molecular identification of petroleum hydrocarbons biodegradation bacteria isolated from oil polluted soil in Dhahran, Saud Arabia. Saudi. J. Biol. Sci. 2019, 26, 1247–1252. [Google Scholar] [CrossRef]
- Akinde, S.B.; Iwuozor, C.C. Alkane Degradative Potentials of Bacteria Isolated From the Deep Atlantic Ocean of the Gulf of Guinea. J. Bioremediat. Biodegrad. 2012, 3, 135. [Google Scholar] [CrossRef]
- Khabouchi, I.; Khadhar, S.; Chaouachi, D.; Chekirbene, A.; Doumenq, P. Study of organic pollution in superficial sediments of Meliane river catchment area: Aliphatic and polycyclic aromatic hydrocarbons. Enviroment Monit. Assess 2020, 192, 283. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Noor, A.; La-Nafie, N.; Djide, M.N. Sponge Role In Alleviating Oil Pollution Through Sludge Reduction, A Preliminary Approach. Int. J. Appl. Chem. 2015, 11, 427–441. [Google Scholar]
- Marzuki, I.; Pratama, I.; Heryani, H.I.; Paserangi, I.; Kamaruddin, M.; Chaerul, M.; Ahmad, R. The Identification and Distribution Components of Polycyclic Aromatic Hydrocarbon Contaminants at the Port of Paotere, Makassar, South Sulawesi. In Proceedings of the The 1st International Conference on Biotechnology and Food Sciences, Surabaya, Indonesia, 11 September 2020; Volume 679. [Google Scholar]
- Zhang, B.; Zhang, L.; Zhang, X. Bioremediation of petroleum hydrocarbon-contaminated soil by petroleum degrading bacteria immobilized on biochar. RSC Adv. 2019, 9, 35304–35311. [Google Scholar] [CrossRef]
- Košnář, Z.; Částková, T.; Wiesnerová, L.; Praus, L.; Jablonský, I.; Koudela, M.; Tlustoš, P. Comparing the removal of polycyclic aromatic hydrocarbons in soil after different bioremediation approaches in relation to the extracellular enzyme activities. J. Env. Sci. 2019, 76, 249–258. [Google Scholar] [CrossRef]
- Sibero, M.T.; Igarashi, Y.; Radjasa, O.K.; Sabdono, A.; Trianto, A.; Zilda, D.S.; Wijaya, Y.J. Sponge-associated fungi from a mangrove habitat in Indonesia: Species composition, antimicrobial activity, enzyme screening and bioactive profiling. Int. Aquat. Res. 2019, 11, 173–186. [Google Scholar] [CrossRef]
- Bendouz, M.; Dionne, D.; Tran, L.H.; Coudert, L.; Mercier, G.; Blais, J.F. Polycyclic Aromatic Hydrocarbon Oxidation from Concentrates Issued from an Attrition Process of Polluted Soil Using the Fenton Reagent and Permanganate. Water Air Soil Pollut. 2017, 228, 114–127. [Google Scholar] [CrossRef]
- Bojes, H.K.; Pope, P.G. Characterization of EPA’s 16 priority pollutant polycyclic aromatic hydrocarbons (PAHs) in tank bottom solids and associated contaminated soils at oil exploration and production sites in Texas. Regul. Toxicol. Pharmacol. 2007, 47, 288–295. [Google Scholar] [CrossRef]
- Abdel-monem, N.M.; Abdel-azeem, A.M.; Ghareeb, D.A.; Nabil-adam, A. Pretreatment Hepatoprotective Effect of the Marine Fungus Derived from Sponge on Hepatic Toxicity Induced by Heavy Metals in Rats. Biomed. Res. Int. 2013, 2013, 510879. [Google Scholar] [CrossRef]
- Baquiran, J.I.P.; Nada, M.A.L.; Posadas, N.; Manogan, D.P.; Cabaitan, P.C.; Conaco, C. Population structure and microbial community diversity of two common tetillid sponges in a tropical reef lagoon. Peer J. 2020, 4, e9017. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Dragh, M.A.; Li, S.; Alhujaily, A.; Abbood, H.A.; Zhang, X.; Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018, 44, 71–76. [Google Scholar] [CrossRef]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and Bioremediation of Heavy Metals Contaminated Ecosystem from Tannery Wastewater: A Review. J. Toxicol. 2018, 2018, 2568038. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lyu, C.; Li, Y. Analysis of Factors Influencing Plant–Microbe Combined Remediation of Soil Contaminated by Polycyclic Aromatic Hydrocarbons. Sustainability 2021, 13, 10695. [Google Scholar] [CrossRef]
- Zaimee, M.Z.A.; Sarjadi, M.S.; Rahman, M.L. Heavy metals removal from water by efficient adsorbents. Water 2021, 13, 2659. [Google Scholar] [CrossRef]
- Bisht, S.; Pandey, P.; Bhargava, B.; Sharma, S.; Kumar, V.; Krishan, D. Bioremediation of polyaromatic hydrocarbons (PAHs) using rhizosphere technology. Braz. J. Microbiol. 2015, 46, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Shityakov, S. New Trends in Bioremediation Technologies Toward Environment-Friendly Society: A Mini-Review. Front. Bioeng. Biotechnol. 2021, 9, 666858. [Google Scholar] [CrossRef]
- Khan, M.N.; Ullah, H.; Naeem, S.; Uddin, J.; Hamid, Y.; Ahmad, W.; Ding, J. Remediation of Emerging Heavy Metals from Water Using Natural Adsorbent: Adsorption Performance and Mechanistic Insights. Sustainability 2021, 13, 8817. [Google Scholar] [CrossRef]
- Duran, R.; Cravo, L.C. Role of environmental factors and microorganisms in determining the fate of polycyclic aromatic hydrocarbons in the marine environment. FEMS Microbiol. Rev. 2016, 40, 814–830. [Google Scholar] [CrossRef]
- Hou, L.; Majumder, E.L.W. Potential for and distribution of enzymatic biodegradation of polystyrene by environmental microorganisms. Materials 2021, 14, 503. [Google Scholar] [CrossRef]
- Marzuki, I.; Chaerul, M.; Erniati, E.; Asmeati, A.; Paserangi, I. Biodegradation of aliphatic waste components of oil sludge used micro symbiont of Sponge Niphates sp. In Proceedings of the 3rd International Conference on Marine Science, Towards Sustainable Marine Resources and Environment, Bogor City, Indonesia, 4 September 2019. [Google Scholar]
- Marzuki, I.; Daris, L.; Nisaa, K.; Emelda, A. The power of biodegradation and bio-adsorption of bacteria symbiont sponges sea on waste contaminated of polycyclic aromatic hydrocarbons and heavy metals. In Proceedings of the International Conference on Fisheries and Marine Research, North Maluku, Indonesia, 13–14 July 2020. [Google Scholar]
- Nikel, P.I.; Silva-Rocha, R.; Benedetti, I.; De-Lorenzo, V. The private life of environmental bacteria: Pollutant biodegradation at the single cell level. Environ. Microbiol. 2014, 16, 628–642. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. Biosurfactant-enhanced bioremediation of aged polycyclic aromatic hydrocarbons (PAHs) in creosote contaminated soil Fisseha. Chemosphere 2016, 144, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Kui-Ma, X.; Ling-Wu, L.; Fam, H. Heavy metal ions affecting the removal of polycyclic aromatic hydrocarbons by fungi with heavy-metal resistance. Appl. Microbiol. Biotechnol. 2014, 98, 9817–9827. [Google Scholar]
- Yu, Y.; Zhang, Y.; Zhao, N.; Guo, J.; Xu, W.; Ma, M.; Li, X. Remediation of crude oil-polluted soil by the bacterial rhizosphere community of suaeda salsa revealed by 16S rRNA genes. Int. J. Environ. Res. Public Health 2020, 17, 1471. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Zeng, G.-M.; Niu, Q.-Y.; Liu, Y.; Zhou, L.; Jiang, L.-H.; Tang, Z.-F.; Xu, P.; Zhang, C.; Cheng, M. Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: A mini review. Bioresour. Technol. 2017, 224, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Elenga-Wilson, P.S.; Kayath, C.A.; Mokemiabeka, N.S.; Nzaou, S.A.E.; Nguimbi, E.; Ahombo, G. Profiling of Indigenous Biosurfactant-Producing Bacillus Isolates in the Bioremediation of Soil Contaminated by Petroleum Products and Olive Oil. Int. J. Microbiol. Hindawi. 2021, 221, 9565930. [Google Scholar] [CrossRef] [PubMed]
- Essumang, D.K.; Togoh, G.K.; Chokky, L. Pesticide residues in the water and Fish (lagoon tilapia) samples from lagoons in Ghana. Bull. Chem. Soc. Ethiop. 2009, 23, 19–27. [Google Scholar] [CrossRef]
- Li, Q.; Liu, J.; Gadd, G.M. Fungal bioremediation of soil co-contaminated with petroleum hydrocarbons and toxic metals. Appl. Microbiol. Biotechnol. 2020, 104, 8999–9008. [Google Scholar] [CrossRef]
- Medaura, M.C.; Guivernau, M.; Moreno-Ventas, X.; Prenafeta-Boldú, F.X.; Viñas, M. Bioaugmentation of Native Fungi, an Efficient Strategy for the Bioremediation of an Aged Industrially Polluted Soil With Heavy Hydrocarbons. Front. Microbiol. 2021, 12, 626436. [Google Scholar] [CrossRef]
- Yetti, E.; Thontowi, A.; Yopi, Y.; Lisdiyanti, P. Screening of Marine bacteria capable of degrading various polyaromatic hydrocarbons. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2015, 10, 121–127. [Google Scholar] [CrossRef][Green Version]
- Kamaruddin, M.; Marzuki, I.; Burhan, A.; Ahmad, R. Screening acetylcholinesterase inhibitors from marine-derived actinomycetes by simple chromatography. In Proceedings of the 1st International Conference on Biotechnology and Food Sciences, Surabaya, Indonesia, 11 September 2020. [Google Scholar]
- Agrawal, N.; Verma, P.; Shahi, S.K. Degradation of polycyclic aromatic hydrocarbons (phenanthrene and pyrene) by the ligninolytic fungi Ganoderma lucidum isolated from the hardwood stump. Bioresour. Bioprocess. 2018, 5, 11. [Google Scholar] [CrossRef]
- Atagana, H.I. Biodegradation of PAHs by fungi in contaminated-soil containing cadmium and nickel ions. Afr. J. Biotechnol. 2009, 8, 5780–5789. [Google Scholar]
- Cao, H.; Wang, C.; Liu, H.; Jia, W.; Sun, H. Enzyme activities during Benzo[a]pyrene degradation by the fungus Lasiodiplodia theobromae isolated from a polluted soil. Sci. Rep. 2020, 10, 865. [Google Scholar] [CrossRef] [PubMed]
- Armus, R.; Selry, C.; Marzuki, I.; Hasan, H.; Syamsia; Sapar, A. Investigation of Potential Marine Bacterial Isolates in Biodegradation Methods on Hydrocarbon Contamination. In Proceedings of the 2nd Workshop on Engineering, Education, Applied Sciences and Technology (WEAST), Makassar, Indonesia, 5 October 2020. [Google Scholar]
- Obire, O.; Aleruchi, O.; Wemedo, S. Fungi in Biodegradation of Polycyclic Aromatic Hydrocarbons in Oilfield Wastewater. Acta Sci Microbiol. 2020, 3, 220–224. [Google Scholar]
- Omoni, V.T.; Lagb, A.J.; Ibeto, C.N.; Semple, K.T. Effects of biological pre-treatment of lignocellulosic waste with white-rot fungi on the stimulation of 14C-phenanthrene catabolism in soils. Int. Biodeterior Biodegrad. 2021, 165, 105324. [Google Scholar] [CrossRef]
- Saraswath, A.; Hallberg, R. Degradation of pyrene by indigenous fungi from a former gasworks site. FEMS Microbiol. Lett. 2002, 210, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Nedoroda, V.; Trokhymenko, G.; Khrapko, T.; Koliehova, A. Analysis of Petroleum Biodegradation by a Bacterial Consortium of Bacillus amyloliquefaciens ssp. Plantarum and Bacillus subtilis. J. Ecol. Eng. 2021, 22, 36–42. [Google Scholar] [CrossRef]
- Bello-, A.M.; Adeleke, R.; Swanevelder, D.; Thantsha, M. Draft Genome Sequence of Pseudomonas sp. Strain 10-1B, a Polycyclic Aromatic Hydrocarbon Degrader in Contaminated Soil. Genome Announc. 2015, 3, e00325-15. [Google Scholar]
- Brzeszcz, J.; Kaszycki, P. Aerobic bacteria degrading both n-alkanes and aromatic hydrocarbons: An undervalued strategy for metabolic diversity and flexibility. Biodegradation 2018, 29, 359–407. [Google Scholar] [CrossRef]
- Crampon, M.; Cébron, A.; Portet-Koltalo, F.; Uroz, S.; Le Derf, F.; Bodilis, J. Low effect of phenanthrene bioaccessibility on its biodegradation in di_usely contaminated soil. Environ. Pollut. 2017, 225, 663–673. [Google Scholar] [CrossRef]
- Dadrasnia, A.; Usman, M.M.; Lim, K.T.; Farahiyah, F.H.; Rodzhan, N.S.M.; Karim, S.H.A.; Ismail, S. Bio-Enhancement of Petroleum Hydrocarbon Polluted Soil Using Newly Isolated Bacteria. Polycycl. Aromat. Compd. 2020, 40, 484–493. [Google Scholar] [CrossRef]
- Guo, J.; Wen, X. Performance and kinetics of benzo(a)pyrene biodegradation in contaminated water and soil and improvement of soil properties by biosurfactant amendment. Ecotoxicol. Environ. Saf. 2021, 207, 111292. [Google Scholar] [CrossRef] [PubMed]
- Hermabessiere, L.; Himber, C.; Boricaud, B.; Kazour, M.; Amara, R.; Cassone, A.-L.; Laurentie, M.; Paul-Pont, I.; Soudant, P.; Dehaut, A.; et al. Optimization, performance, and application of a pyrolysis-GC/MS method for the identification of microplastics. Anal. Bioanal. Chem. 2018, 410, 6663–6676. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Sinardi, S.; Pratama, I.; Chaerul, M.; Paserangi, I.; Kamaruddin, M. Performance of sea sponges micro symbionts as a biomaterial in biodegradation naphthalene waste of modified. In Proceedings of the 5th International Seminar on Sustainable Urban Development, Jakarta, Indonesia, 5 August 2020. [Google Scholar]
- Freeman, C.J.; Easson, C.G.; Fiore, C.L.; Thacker, R.W. Sponge–Microbe Interactions on Coral Reefs: Multiple Evolutionary Solutions to a Complex Environment. Front. Mar. Sci. 2021, 8, 705053. [Google Scholar] [CrossRef]
- Zahirnejad, M.; Ziarati, P.; Asgarpanah, J. The Efficiency of Bio-adsorption of Heavy Metals from Pharmaceutical Effluent by Rumex crispus L. Seed. J. Pharm. Health Sci. 2017, 5, 231–243. [Google Scholar]
- Abass, O.K.; Zhuo, M.; Zhang, K. Concomitant degradation of complex organics and metals recovery from fracking wastewater: Roles of nano zerovalent iron. Chem. Eng. J. 2017, 328, 159–171. [Google Scholar] [CrossRef]
- Lavy, A.; Keren, R.; Haber, M.; Schwartz, I.; Ilan, M. Implementing sponge physiological and genomic information to enhance the diversity of its culturable associated bacteria. FEMS Microbiol. Ecol. 2014, 87, 486–502. [Google Scholar] [CrossRef]
- Maldonado, M.; López, A.M.; Busch, K.; Slaby, B.M.; Bayer, K.; Beazley, L.; Hentschel, U.; Kenchington, E.; Rapp, H.T. A Microbial Nitrogen Engine Modulated by Bacteriosyncytia in Hexactinellid Sponges: Ecological Implications for Deep-Sea Communities. Front. Mar. Sci. 2021, 8, 638505. [Google Scholar] [CrossRef]
- Marzuki, I.; Enryani, H.I.; Nafie, N.-L.; Dali, S. Study Biodegradation of Aromatics Pyrene Using Bacterial Isolates from the Sea and micro symbionts Sponges. Int. J. Appl. Chem. 2017, 13, 707–720. [Google Scholar]
- Marzuki, I. The Bio-adsorption Pattern Bacteria Symbiont Sponge Marine Against Contaminants Chromium and Manganese In The Waste Modification of Laboratory Scale. Indo Chim. Acta. 2020, 13, 1–9. [Google Scholar] [CrossRef]
- Cajthaml, T.; Erbanová, P.; Kollmann, A.; Novotný, C.; Sasek, V.; Mougin, C. Degradation of PAHs by ligninolytic enzymes of Irpex lacteus. Folia. Microbiol. 2008, 53, 289–294. [Google Scholar] [CrossRef]
- Kadri, T.; Rouissi, T.; Brar, S.K.; Cledon, M.; Sarma, S.; Verma, M. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: A review. J. Env. Sci. 2017, 51, 52–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, X.; Huang, Y.; Zhou, F.; Zhao, Q.; Zhang, W.; Chen, G.; Gao, Y. Isolation of Petroleum Degraders and Petroleum-Degradation Characteristics of Crude Enzymes from Providencia rettgeri L1. Polish J. Envirin. Stud. 2022, 31, 3859–3866. [Google Scholar] [CrossRef] [PubMed]
- Ohowa, B.; Kiteresi, L.I.; Wanjeri, V.W.; Mwamburi, S.M.; Tunje, S.L. Sponges as simple biomonitoring tools for trace element pollution in marine environments: Insights from a Kenyan study focused on the leaf sponge Phyllospongia foliascens. Afr. J. Mar. Sci. 2021, 43, 533–538. [Google Scholar] [CrossRef]
- Marzuki, I.; Gusty, S.; Armus, R.; Sapar, A.; Asaf, R.; Athirah, A.; Jaya, J. Secondary Metabolite Analysis and Anti-Bacteria and Fungal Activities of Marine Sponge Methanol Extract Based on Coral Cover. In Proceedings of the 6th International Conference on Basic Sciences, Ambon City, Indonesia, 4–5 November 2020. [Google Scholar]
- Marzuki, I.; Nisaa, K.; Asaf, R.; Paena, M.; Athirah, A.; Susianingsih, E.; Nurhidayah, N.; Kadriah, I.A.K.; Kamaruddin, K.; Sahabuddin, S.; et al. Comparison of Pyrene Biodegradation Using Two Types of Marine Bacterial Isolates. Sustainability 2022, 14, 9890. [Google Scholar] [CrossRef]
- Rua, C.P.J.; Oliveira, L.S.; De-Froes, A.; Tschoeke, D.A.; Soares, A.C.; Leomil, L.; Gregoracci, G.B.; Coutinho, R.; Hajdu, E.; Thompson, C.C.; et al. Microbial and Functional Biodiversity Patterns in Sponges that Accumulate Bromopyrrole Alkaloids Suggest Horizontal Gene Transfer of Halogenase Genes. Microb. Ecol. J. 2018, 76, 825–838. [Google Scholar] [CrossRef]
- Marzuki, I.; Daris, L.; Yunus, S.; Riana, A.D. Selection and characterization of potential bacteria for polycyclic aromatic biodegradation of hydrocarbons in sea sponges from Spermonde Islands, Indonesia. AACL Bioflux. 2020, 13, 3493–3506. [Google Scholar]
- Kim, H.-W.; Jo, J.H.; Kim, Y.-B.; Le, T.-K.; Cho, C.-W.; Yun, C.-H.; Chi, W.S.; Yeom, S.-J. Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 2021, 416, 126239. [Google Scholar] [CrossRef]
- Fu, X.; Wang, H.; Bai, Y.; Xue, J.; Gao, Y.; Hu, S.; Wu, T.; Sum, J. Systematic degradation mechanism and pathways analysis of the immobilized bacteria: Permeability and biodegradation, kinetic and molecular simulation. Env. Sci. Ecotechnol. 2020, 2, 100028. [Google Scholar] [CrossRef]
- Marzuki, I.; Kamaruddin, M.; Ahmad, R. Identification of marine sponges-symbiotic bacteria and their application in degrading polycyclic aromatic hydrocarbons. Biodiversitas 2021, 22, 1481–1488. [Google Scholar] [CrossRef]
- Marzuki, I.; Ahmad, R.; Kamaruddin, M.; Asaf, R.; Armus, R.; Siswanty, I. Performance of cultured marine sponges-symbiotic bacteria as a heavy metal bio-adsorption. Biodiversitas 2021, 22, 5536–5543. [Google Scholar] [CrossRef]
- Marzuki, I.; Ali, M.Y.; Syarif, H.U.; Erniati, E.; Gusty, S.; Ritnawati, R.; Daris, L.; Nisaa, K. Investigation of Biodegradable Bacteria as Bio indicators of the Presence of PAHs Contaminants in Marine Waters in the Marine Tourism Area of Makassar City. In Proceedings of the 6th International Conference on Tropical Coastal Region Eco-Development, Semarang, Indonesia, 27–28 October 2020. [Google Scholar]
- Liu, Y.F.; Mbadinga, S.M.; Gu, J.D.; Mu, B.Z. Type II chaperonin gene as a complementary barcode for 16S rRNA gene in study of Archaea diversity of petroleum reservoirs. Int. Biodeterior Biodegrad. 2017, 123, 113–120. [Google Scholar] [CrossRef]
- Su, X.M.; Bamba, A.M.; Zhang, S.; Zhang, Y.G.; Hashmi, M.Z.; Lin, H.J.; Ding, L.X. Revealing potential functions of VBNC bacteria in polycyclic aromatic hydrocarbons biodegradation. Lett. Appl. Microbiol. 2018, 66, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Syakti, A.D.; Yani, M.; Hidayati, N.V.; Siregar, A.S.; Doumenq, P.; Sudiana, I.M. The bioremediation potential of hydrocarbonoclastic bacteria isolated from a mangrove contaminated by petroleum hydrocarbons on the cilacap coast, Indonesia. Bioremediat. J. 2013, 17, 11–20. [Google Scholar] [CrossRef]
- Ahmad, M.; Wang, P.; Li, J.L.; Wang, R.; Duan, L.; Luo, X.; Irfan, M.; Peng, Z.; Yin, L.; Li, W. Impacts of bio-stimulants on pyrene degradation, prokaryotic community compositions, and functions. Environ. Pollut. 2021, 289, 117863. [Google Scholar] [CrossRef]
- Fang, H.; Shi, Y.; Zhou, M.; Niu, Q. Influence of n-Hexadecane and Naphthalene on Anaerobic Digestion: Kinetic Simulation, DOM Variation and Microbial Community Assessment. In Proceedings of the 2020 International Conference on Green Energy, Environment and Sustainable Development, Wuhan, China, 24–25 April 2020. [Google Scholar]
- Galitskaya, P.; Biktasheva, L.; Blagodatsky, S.; Selivanovskaya, S. Response of bacterial and fungal communities to high petroleum pollution in different soils. Sci. Rep. 2021, 11, 164. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, J.; Peng, C. Shift of Soil Polycyclic Aromatic Hydrocarbons (PAHs) dissipation pattern and microbial community composition due to rhamnolipid supplementation. Water Air Soil Pollut. 2019, 230, 107. [Google Scholar] [CrossRef]
- Mao, J.; Guan, W. Fungal degradation of polycyclic aromatic hydrocarbons (PAHs) by Scopulariopsis brevicaulis and its application in bioremediation of PAH-contaminated soil. Acta Agric. Scand Sect. B Soil Plant Sci. 2016, 66, 399–405. [Google Scholar]
- Sandhu, M.; Paul, A.T. Metagenomic Analysis for Taxonomic and Functional Potential of Polyaromatic Hydrocarbons (PAHs) and Polychlorinated Biphenyl ( PCB ) Degrading Bacterial Communities in Steel Industrial Soil. PLoS ONE 2022, 17, e0266808. [Google Scholar] [CrossRef]
- Tziveleka, L.-A.; Ioannou, E.; Tsiourvas, D.; Berillis, P.; Foufa, E.; Roussis, V. Collagen from the marine sponges Axinella cannabina and Suberites carnosus: Isolation and morphological, biochemical, and biophysical characterization. Mar. Drugs 2017, 15, 152. [Google Scholar] [CrossRef]
- Marzuki, I.; Nisaa, K.; Asaf, R.; Armus, R.; Kamaruddin, M.; Sapar, A.; Emelda, A. Biodegradation mechanism of naphthalene using marine sponge symbiotic bacteria. In Proceedings of the 2nd International Conference on Fisheries and Marine, Ternate, Indonesia, 15 July 2021. [Google Scholar]
- Chulalaksananukul, S.; Gadd, G.M.; Sangvanich, P.; Sihanonth, P.; Piapukiew, J.; Vangnai, A.S. Biodegradation of benzo(a)pyrene by a newly isolated Fusarium sp. FEMS Microbiol. Lett. 2006, 262, 99–106. [Google Scholar] [CrossRef]
- Govarthanan, M.; Fuzisawa, S.; Hosogai, T.; Chang, Y.C. Biodegradation of aliphatic and aromatic hydrocarbons using the filamentous fungus Penicillium sp. CHY-2 and characterization of its manganese peroxidase activity. RSC Adv. 2017, 7, 20716–20723. [Google Scholar] [CrossRef]
- Laothamteep, N.; Kawano, H.; Vejarano, F.; Minakuchi, C.S.; Shintani, M.; Nojiri, H.; Pinyakong, O. Effects of environmental factors and coexisting substrates on PAH degradation and transcriptomic responses of the defined bacterial consortium OPK. Environ. Pollut. 2021, 277, 116769. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Xu, M.; Sun, K.; Cao, L.H.; Dai, C.; Jia, Y. Biodegradation of phenanthrene by endophytic fungus Phomopsis liquidambari in vitro and in vivo. Chemosphere 2018, 203, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Haleyur, N.; Shahsavari, E.; Jain, S.S.; Koshlaf, E.; Ravindran, V.B.; Morrison, P.D.; Osborn, A.M.; Ball, A.S. Influence of bioaugmentation and biostimulation on PAH degradation in aged contaminated soils: Response and dynamics of the bacterial community. J. Environ. Manag. 2019, 238, 49–58. [Google Scholar] [CrossRef]
- Guerra, A.B.; Oliveira, J.S.; Silva-Portela, R.C.B.; Araújo, W.; Carlos, A.C.; Vasconcelos, A.T.R.; Freitas, A.T.; Domingos, Y.S.; de Farias, M.F.; Fernandes, G.J.T.; et al. Metagenome enrichment approach used for selection of oil-degrading bacteria consortia for drill cutting residue bioremediation. Environ. Pollut. 2018, 235, 869–880. [Google Scholar] [CrossRef]
- Janczuk, B.; Szymczyk, K.; Zdziennicka, A. Adsorption Properties of Hydrocarbon and Fluorocarbon Surfactants Ternary Mixture at the Water-Air Interface. Molecules 2021, 26, 4313. [Google Scholar] [CrossRef]
- Lasota, J.; Łyszczarz, Y.; Kempf, P.; Kempf, M.; Błońska, E. Effect of Species Composition on Polycyclic Aromatic Hydrocarbon (PAH) Accumulation in Urban Forest Soils of Krakow. Water. Air. Soil Pollut. 2021, 232, 74. [Google Scholar] [CrossRef]
- Lu, C.; Hong, Y.; Liu, J.; Gao, Y.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. A PAH-degrading bacterial community enriched with contaminated agricultural soil and its utility for microbial bioremediation. Environ. Pollut. 2019, 251, 773–782. [Google Scholar] [CrossRef]
- Lundstedt, S. Analysis of PAHs and Their Transformation Products in Contaminated Soil and Remedial Processes; Department of Environment and Toxicological Chemistry, University of Amsterdam: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Nzila, A. Current status of the degradation of aliphatic and aromatic petroleum hydrocarbons by thermophilic microbes and future perspectives. Int. J. Environ. Res. Public Health 2018, 15, 2782. [Google Scholar] [CrossRef]
- Pandey, P.; Kapley, A.; Brar, S.K. Editorial: Biodegradation of High Molecular Weight Polyaromatic Hydrocarbons in Different Environments. Front. Microbiol. 2021, 12, 2020–2022. [Google Scholar] [CrossRef] [PubMed]
- Sawulski, P.; Boots, B.; Clipson, N.; Doyle, E. Differential degradation of polycyclic aromatic hydrocarbon mixtures by indigenous microbial assemblages in soil. Lett. Appl. Microbiol. 2015, 61, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Wang, H.; Li, Y.; Duan, X. Impact of co-contamination by PAHs and heavy metals on micro-ecosystem in bioretention systems with soil, sand, and water treatment residuals. J. Clean. Prod. 2023, 383, 135417. [Google Scholar] [CrossRef]
- Shehu, U.; Ahmad, F.A.; Yusuf, F.; Muhammad, F.; Yakasai, H.M. Isolation and Identification of Anthracene Utilizing Proteus vulgaris from Oil Spill Contaminated Soil at NNPC Depot Kano State Nigeria. J. Adv. Biol. Biotechnol. 2021, 24, 46–53. [Google Scholar] [CrossRef]
- Smułek, W.; Sydow, M.; Matejuk, Z.J.; Kaczorek, E. Bacteria involved in biodegradation of creosote PAH–A case study of longterm contaminated industrial area. Ecotoxicol. Environ. Saf. 2020, 187, 109843. [Google Scholar] [CrossRef] [PubMed]
- Souza, H.M.d.L.; Barreto, L.R.; da Mota, A.J.; de Oliveira, L.A.; Barroso, H.d.S.; Zanotto, S.P. Tolerance to polycyclic aromatic hydrocarbons (PAHs) by filamentous fungi isolated from contaminated sediment in the Amazon region. Acta Sci. Biol. Sci. 2017, 39, 481–488. [Google Scholar] [CrossRef]
- Ray, M.; Kumar, V.; Banerjee, C.; Gupta, P.; Singh, S.; Singh, A. Investigation of biosurfactants produced by three indigenous bacterial strains, their growth kinetics and their anthracene and fluorene tolerance. Ecotoxicol. Environ. Saf. 2021, 208, 111621. [Google Scholar] [CrossRef]
- Sahu, L. Presence of Hydrocarbon Degrading Bacteria in Contaminated Soil Collected From Various Fuel Station in Bhilai, Chhattisgarh. Int. J. Res. Appl. Sci. Eng. Technol. 2021, 9, 1802–1804. [Google Scholar] [CrossRef]
- Vaezzadeh, V.; Zakaria, M.P.; Bong, C.W.; Masood, N.; Magam, M.S.; Alkhadher, S. Mangrove Oyster (Crassostrea belcheri) as a Biomonitor Species for Bioavailability of Polycyclic Aromatic Hydrocarbons (PAHs) from Sediment of the West Coast of Peninsular Malaysia. Polycycl. Aromat. Compd. 2019, 39, 470–485. [Google Scholar] [CrossRef]
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. J. 2008, 10, 1948–1963. [Google Scholar] [CrossRef]
- Wolf, D.C.; Gan, J. Influence of rhamnolipid biosurfactant and Brij-35 synthetic surfactant on 14C-pyrene mineralization in soil. Environ. Pollut. 2018, 243, 1846–1853. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Shao, Z. Polycyclic Aromatic Hydrocarbon (PAH) Degradation Pathways of the Obligate Marine PAH Degrader Cycloclasticus sp. Strain P1. App. Env. Microbiol. 2018, 84, e1261-18. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Asaf, R.; Paena, M.; Athirah, A.; Nisaa, K.; Ahmad, R.; Kamaruddin, M. Anthracene and Pyrene Biodegradation Performance of Marine Sponge Symbiont Bacteria Consortium. Molecules 2021, 26, 6851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qin, F.; Qiao, J.; Li, G.; Shen, C.; Huang, T.; Hu, Z. Draft Genome Sequence of Rhodococcus sp. Strain P14, a Biodegrader of High-Molecular-Weight Polycyclic Aromatic Hydrocarbons. J. Bacteriol. 2012, 194, 3546. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Tauler, M.; Grifoll, M. Bacterial PAH degradation in marine and terrestrial habitats. Curr. Opin. Biotechnol. 2015, 33, 95–102. [Google Scholar] [CrossRef]
- Tawniczak, T.; Woźniak-Karczewska, M.; Loibner, A.P.; Heipieper, H.J.; Chrzanowski, T. Microbial degradation of hydrocarbons-basic principles for bioremediation: A review. Molecules 2020, 25, 856. [Google Scholar] [CrossRef]
- Miao, L.L.; Qu, J.; Liu, Z.P. Hydroxylation at Multiple Positions Initiated the Biodegradation of Indeno[1,2,3-cd]Pyrene in Rhodococcus aetherivorans IcdP1. Front. Microbiol. 2020, 11, 568381. [Google Scholar] [CrossRef]
- Bodor, A.; Bounedjouma, N.; Feigl, G.; Duzs, A.; Laczi, K.; Szilágyi, A.; Rákhely, G.; Perei, K. Exploitation of extracellular organic matter from Micrococcus luteus to enhance ex situ bioremediation of soils polluted with used lubricants. J. Hazard. Mater. 2021, 417, 125996. [Google Scholar] [CrossRef]
- Zakaria, H.Y.; Hassan, A.K.M.; El-Naggar, H.A.; Abo-Senna, F.M. Biomass determination based on the individual volume of the dominant copepod species in the Western Egyptian Mediterranean Coast. Egypt J. Aquat. Res. 2018, 44, 89–99. [Google Scholar] [CrossRef]
- Çoban-Yıldız, Y.; Chiavari, G.; Fabbri, D.; Gaines, A.F.; Galletti, G.; Tuğrul, S. The chemical composition of Black Sea suspended particulate organic matter: Pyrolysis-GC/MS as a complementary tool to traditional oceanographic analyses. Mar. Chem. 2000, 69, 55–67. [Google Scholar] [CrossRef]
- Gomiero, A.; Øysæd, K.B.; Palmas, L.; Skogerbø, G. Application of GC/MS-pyrolysis to estimate the levels of microplastics in a drinking water supply system. J. Hazard. Mater. 2021, 416, 125708. [Google Scholar] [CrossRef]
- Käppler, A.; Fischer, M.; Scholz-Böttcher, B.M.; Oberbeckmann, S.; Labrenz, M.; Fischer, D.; Eichhorn, K.-J.; Voit, B. Comparison of μ-ATR-FTIR spectroscopy and py-GCMS as identification tools for microplastic particles and fibers isolated from river sediments. Anal. Bioanal. Chem. 2018, 410, 5313–5327. [Google Scholar] [CrossRef] [PubMed]
- Spini, G.; Spina, F.; Poli, A.; Blieux, A.L.; Regnier, T.; Gramellini, C.; Varese, G.C.; Puglisi, E. Molecular and Microbiological Insights on the Enrichment Procedures for the Isolation of Petroleum Degrading Bacteria and Fungi. Front. Microbiol. 2018, 9, 2543. [Google Scholar] [CrossRef] [PubMed]
- Marzuki, I.; Asdar, A.M.; Hardimas, H. Performance Analysis of bio-sorption of Heavy Metal and Biodegradation PAH of Isolates Marine Sponges Symbiont Bacteria. Indo Chim. Acta. 2021, 13, 18333. [Google Scholar]
- Tsaboula, A.; Papadakis, E.; Vryzas, Z.; Kotopoulou, A.; Kintzikoglou, K.; Papadopoulou, M.E. Assessment and Management of Pesticide Pollution at a River Basin Level Part II: Optimization of Pesticide Monitoring Networks on Surface Aquatic Ecosystems by Data Analysis Methods. Sci. Total Environ. 2019, 653, 1597–1611. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, B.K.; Paudel, K.P. Assessing the efficiency of alternative best management practices to reduce nonpoint source pollution in a rural watershed located in Louisiana, USA. Water 2019, 11, 1714. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, J.; Dick, R.P.; Li, H.; Shen, D.; Gao, Y.; Waigi, M.G.; Ling, W. Rhamnolipid influences biosorption and biodegradation of phenanthrene by phenanthrene-degrading strain Pseudomonas sp. Ph6. Environ. Pollut. 2018, 240, 359–367. [Google Scholar] [CrossRef]
- Roy, A.; Dutta, A.; Pal, S.; Gupta, A.; Sarkar, J.; Chatterjee, A.; Saha, A.; Sarkar, P.; Sar, P.; Kazy, S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour. Technol. 2018, 253, 22–32. [Google Scholar] [CrossRef]
- Liu, X.; Hu, X.; Cao, Y.; Jing, W.P.; Yu, H.J.; Guo, P.; Huang, L. Biodegradation of Phenanthrene and Heavy Metal Removal by Acid-Tolerant Burkholderia fungorum FM-2. Front. Microbiol. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Selvin, J.; Shanmugha Priya, S.; Seghal-Kiran, G.; Thangavelu, T.; Sapna-Bai, N. Sponge-associated marine bacteria as indicators of heavy metal pollution. Microbiol. Res. 2009, 164, 352–363. [Google Scholar] [CrossRef]
- Anggela, A.; Marzuki, I. Bioadsorption capacity of marine sponge symbiotic bacteria against heavy metal Contaminants. Kovalen J. Ris. Kim. 2021, 7, 12–22. (In Indonesia) [Google Scholar]
- Knobloch, S.; Jóhannsson, R.; Marteinsson, V. Bacterial diversity in the marine sponge Halichondria panicea from Icelandic waters and host-specificity of its dominant symbiont candidatus Halichondribacter symbioticus. FEMS Microbiol. Ecol. 2018, 95, 1–13. [Google Scholar]
- Ziss, E.; Friesl, H.W.; Noller, C.; Watzinger, A.; Hood, N.R. Heavy Metal City-Zen. Exploring the potential risk of heavy metal contamination of food crop plants in urban gardening contexts using a citizen science approach. EGU Gen. Assem. Conf. Abstract 2020, 13, 8626. [Google Scholar]
- White, J.R.; Patel, J.; Ottesen, A.; Arce, G.; Blackwelder, P.; Lopez, J.V. Pyrosequencing of Bacterial Symbionts within Axinella corrugata Sponges: Diversity and Seasonal Variability. PLoS ONE 2012, 7, e38204. [Google Scholar] [CrossRef]
- Parhamfar, M.; Abtahia, H.; Godinib, K.; Saeedi, R.; Sartaje, M.; Villaseñorf, J.; Coulong, F.; Kumarg, V.; Soltanighiash, T.; Radi, E.G.; et al. Biodegradation of heavy oily sludge by a two-step inoculation composting process using synergistic effect of indigenous isolated bacteria. Process Biochem. 2020, 91, 223–230. [Google Scholar] [CrossRef]
- Arroyo, A.; Provoste, F.; Rodríguez, M.; Prieto, A.L. A mechanistic model to assess the fate of naphthalene and Benzo(A)etilene in a chilean wwtp. Processes 2021, 9, 1313. [Google Scholar] [CrossRef]
- Okoro, C.C. Biosurfactant-enhanced remediation of hydrocarbon contaminated mangrove swamp. Nat. Sci. 2010, 8, 152–162. [Google Scholar] [CrossRef]
- Liu, W.J.; Duan, X.D.; Wu, L.P.; Masakorala, K. Biosurfactant Production by Pseudomonas aeruginosa SNP0614 and its Effect on Biodegradation of Petroleum. Appl. Biochem. Microbiol. 2018, 54, 155–162. [Google Scholar] [CrossRef]
- Tenea, A.G.; Vasile, G.G.; Dinu, C.; Gheorgh, S.; Pascu, L.F.; Mureseanu, M.; Ene, C. Behavior of Cd accumulation in sinapis alba L. In the presence of essential elements (Ca, Mg, Fe, Zn, Mn, Cu, Ni). Rev. Chim. 2020, 71, 378–389. [Google Scholar] [CrossRef]
- Alaboudi, A.A.; Ahmed, B.; Brodie, G. Annals of Agricultural Sciences Phytoremediation of Pb and Cd contaminated soils by using sun fl ower (Helianthus annuus) plant. Ann. Agric. Sci. 2018, 63, 123–127. [Google Scholar] [CrossRef]
- Isikhuemhen, O.S.; Mikiashvili, N.A.; Senwo, Z.N.; Ohimain, E.I. Biodegradation and Sugar Release from Canola Plant Biomass by Selected White Rot Fungi. Adv. Biol. Chem. 2014, 04, 395–406. [Google Scholar] [CrossRef]
- Al-Nasrawi, H. Role of Fungi in Bioremediation. Adv. Biotechnol. Microbiol. 2019, 12, 77–81. [Google Scholar]
- Fomina, M.; Charnock, J.M.; Hillier, S.; Alvarez, R.; Livens, F.; Gadd, G.M. Role of fungi in the biogeochemical fate of depleted uranium. Curr. Biol. 2008, 18, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Morganti, T.M.; Ribes, M.; Yahel, G.; Coma, R. Size Is the Major Determinant of Pumping Rates in Marine Sponges. Front. Physiol. 2019, 10, 1474. [Google Scholar] [CrossRef] [PubMed]
- Keller-Costa, T.; Jousset, A.; Van-Overbeek, L.; Van-Elsas, J.D.; Costa, R. The freshwater sponge Ephydatia fluviatilis harbours diverse Pseudomonas species (Gammaproteobacteria, Pseudomonadales) with broad-spectrum antimicrobial activity. PLoS ONE 2014, 9, e88429. [Google Scholar] [CrossRef]
- Shimoda, T.; Suryati, E.; Ahmad, T. Evaluation in a Shrimp Aquaculture System Using Mangroves, Oysters, and Seaweed as Biofilters Based on the Concentrations of Nutrients and Chlorophyll. JARQ 2006, 40, 189–193. [Google Scholar] [CrossRef][Green Version]
- Cecotti, M.; Coppotelli, B.M.; Mora, V.C.; Viera, M.; Morelli, I.S. Efficiency of surfactant-enhanced bioremediation of aged polycyclic aromatic hydrocarbon-contaminated soil: Link with bioavailability and the dynamics of the bacterial community. Sci. Total Environ. 2018, 634, 224–234. [Google Scholar] [CrossRef]
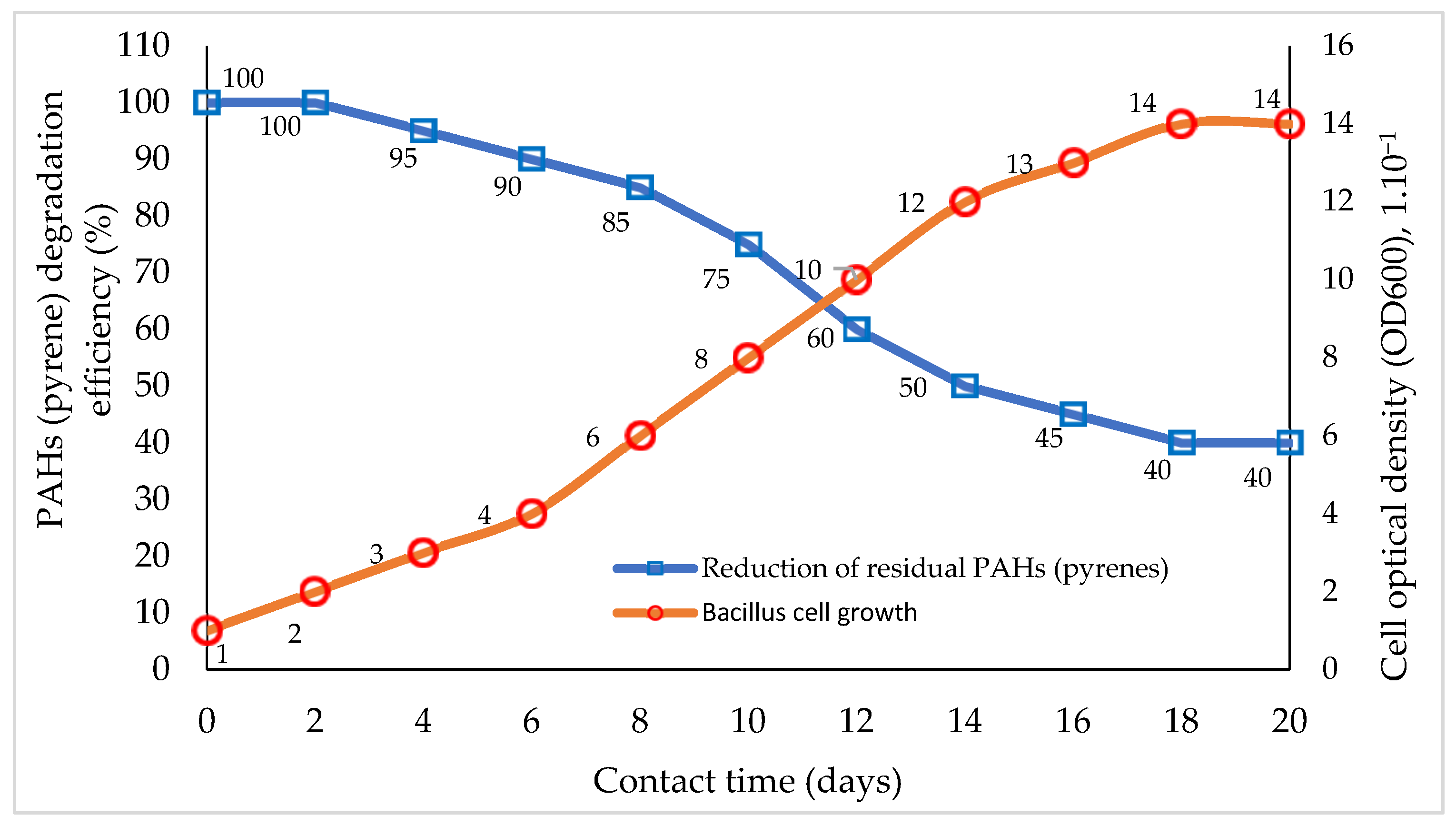
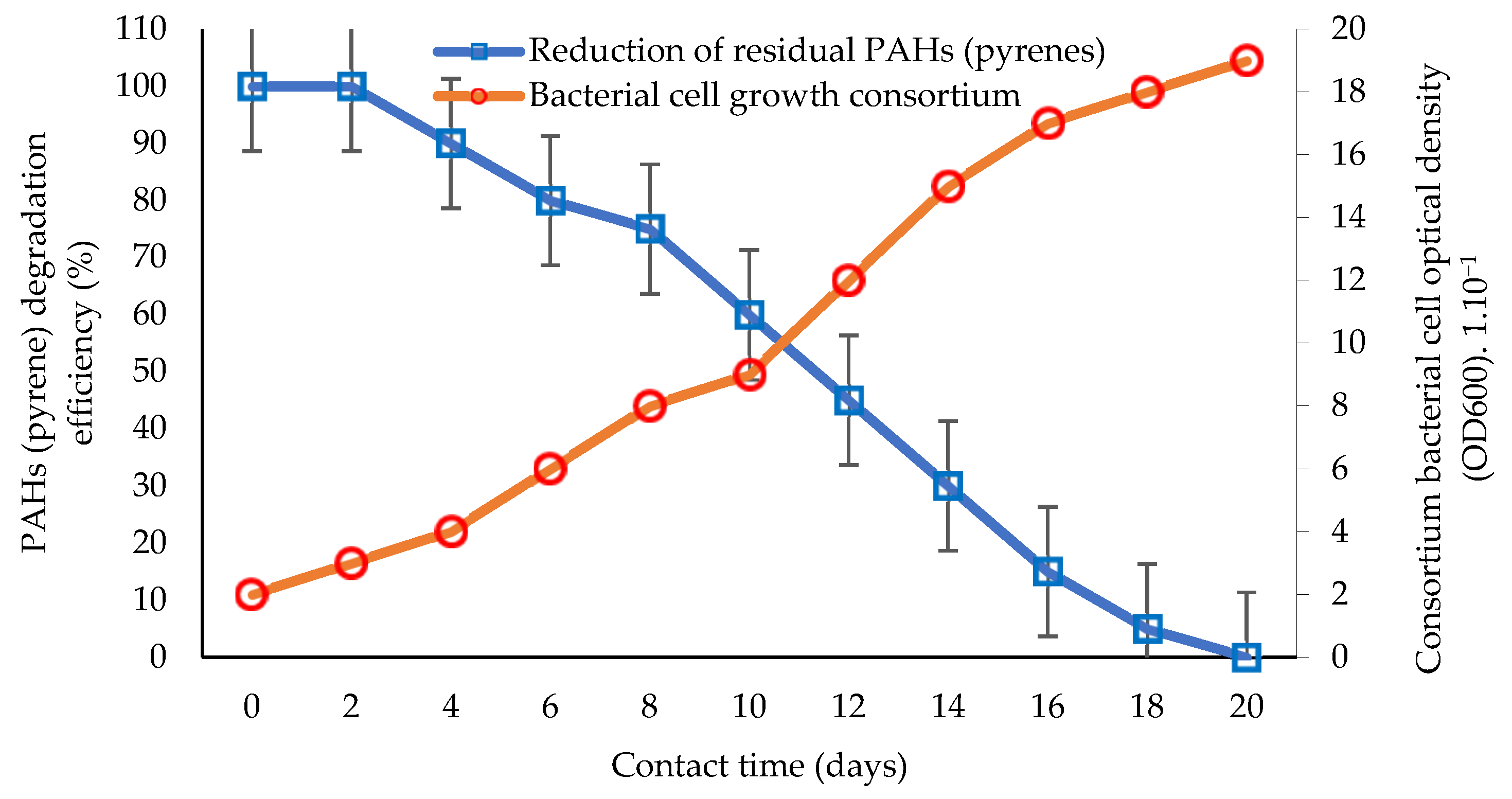
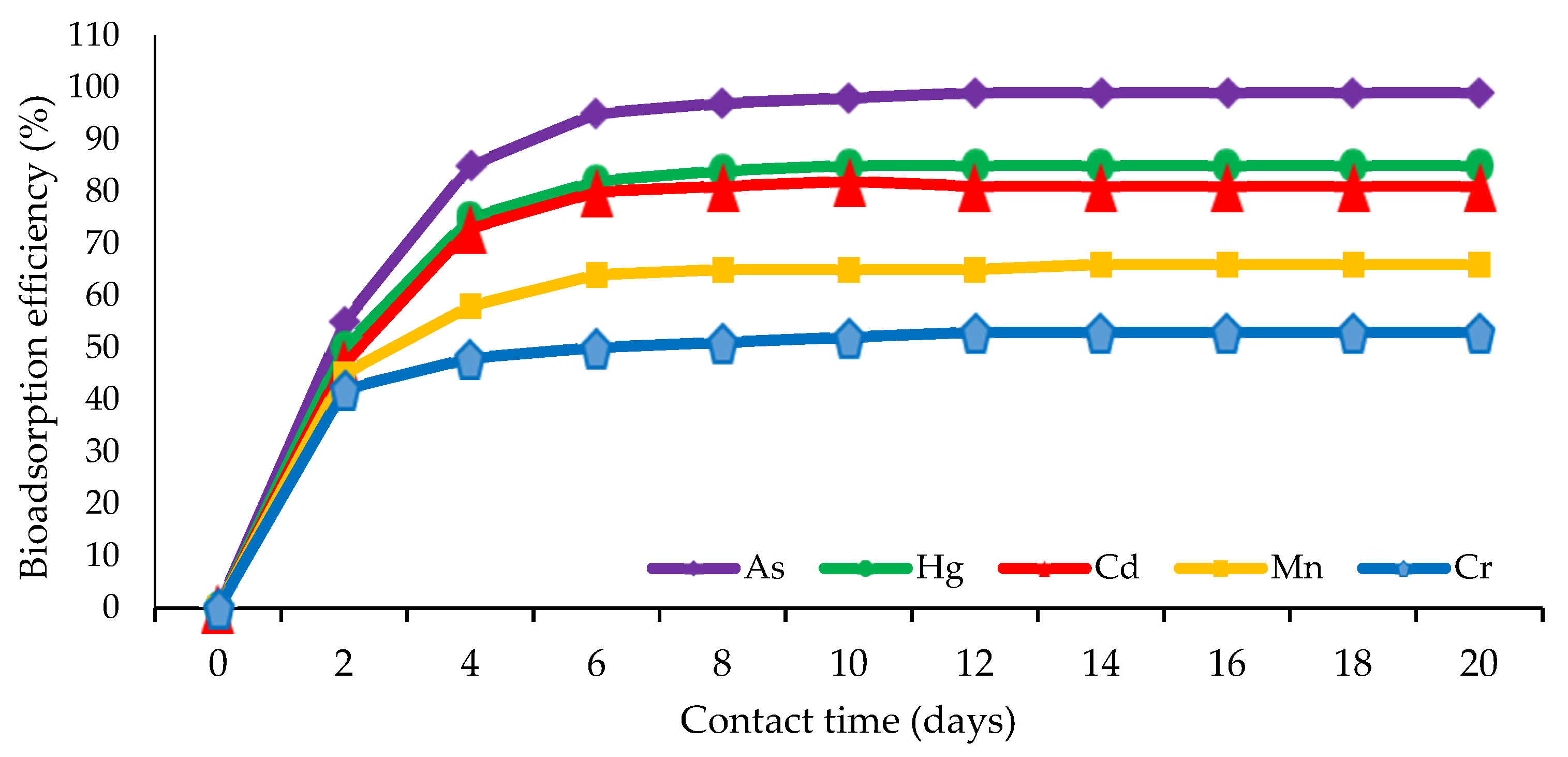
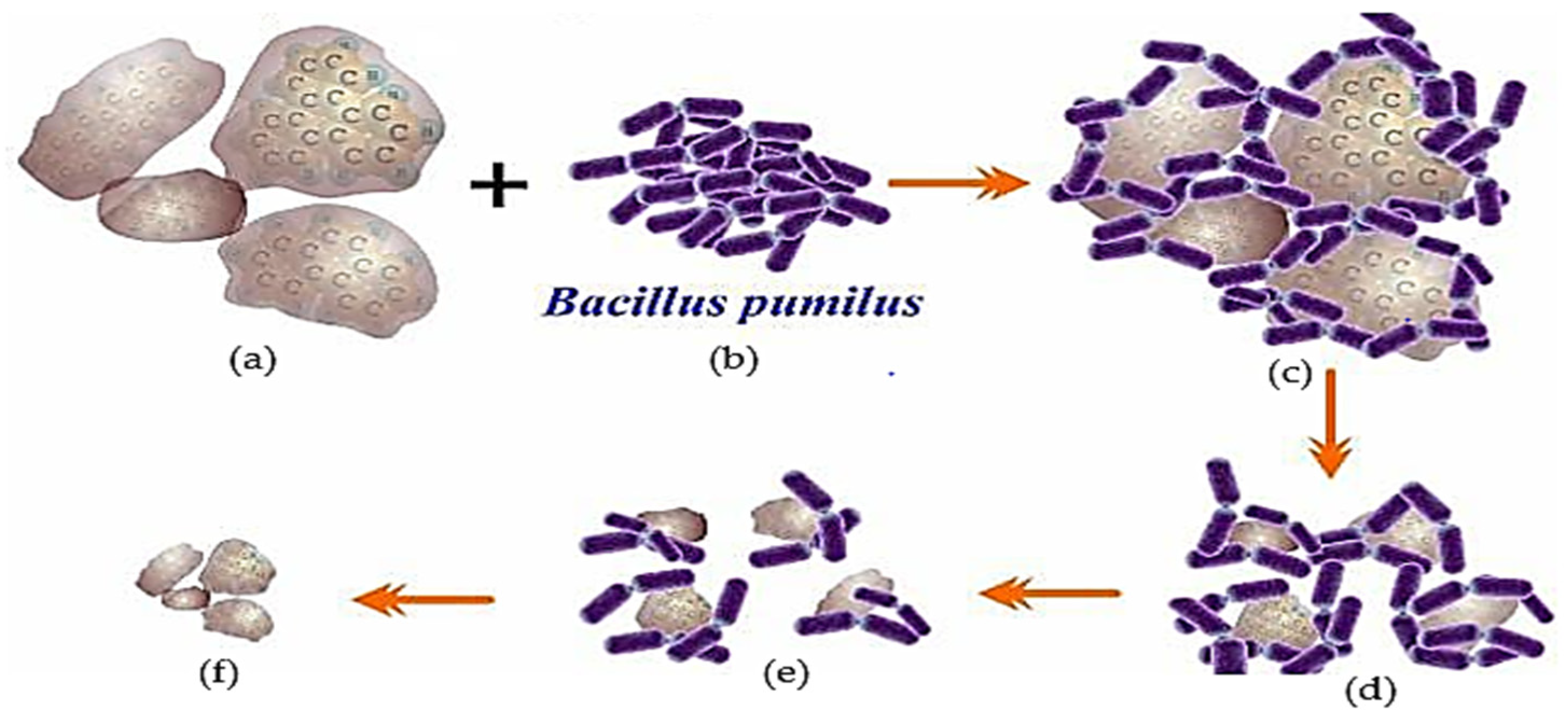

| Type of Contaminant | Bioremediation Method | Test System | Interaction Duration | Removal Efficiency | Conclusion | References |
|---|---|---|---|---|---|---|
| Pyrene (±10 mg/kg) | Biosurfactants (Biodegradation) | Soil microorganism | 10 days | ±60% | The biodegradation process can occur due to the ability of rhamnolipids to convert carbon into energy sources | [111] |
| Phenanthrene (±1.0 mg/L) | Biodegradation | Using soil adsorption reactor | more than 50 days | 90.0% | No significant effect of the observed biodegradation efficiency of surfactants | [54] |
| PAHs (±574 mg/kg) | Biodegradation | Soil microorganism | 84 days | 72.0–77.0% | The formation of surfactants marks the ongoing biodegradation process | [74] |
| Pyrene (±100 mg/L) | Biodegradation | Sphingobacterium sp. strain 21 | 30 days | ±38.3% | The biodegradation performance of pyrene increases at the contact period of 6–20 days | [71] |
| Pyrene, phenanthrene and others (±6 mg/kg) | Biosurfactants (Biodegradation) | Soil microorganism | ±35 days | 58.7% | Biosurfactants (rhamnolipids) can only carry out biodegradation until the 7th day, then the PAHs biodegradation process does not appear until the 35th day | [85] |
| Phenanthrene (±1.0 mg/L) | Biodegradation | Flake model | 14 days | 60.0% | Using Rhamnolipids as surfactants can increase the efficiency of biodegradation at a concentration of 100 mg/L | [127] |
| PAHs (±1.5 mg/g) | Biostimulation | Soil microorganism | 56 days | ±99.0% | The biostimulant effect can increase biodegradation kinetics | [94] |
| Pyrene (100 mg/L) | Biodegradation using vial reactor | Alcaligenes faecalis strain Cu4-1 | 25 days | 97.7% | The products of pyrene biodegradation via the two types of bacteria are relatively different, indicating that there are different metabolic pathways that are influenced by these types of bacteria | [64] |
| Bacillus Cereus strain MER-8 | 93.2% | |||||
| Petroleum refinery waste (±144 g/kg) | Combined biostimulation and bioaugmentation | Microorganisms in vial | 120 days | 57–75% | Modification of the method by applying a combination of biostimulation and bioaugmentation to increase remediation efficiency | [128] |
| Alkanes (initial concentration not determined) | Bioaugmentation | activated microcosm consortium | 85 days | 35–66% | The use of the adapted microcosm consortium can degrade the hydrocarbon component as a substrate to produce biosurfactants | [95] |
| Pb(II) and Cd(II) | Bioadsorption | Burkholderia fungorum FM-2 | 7 days | 50 mg/L and 400 mg/L | Bacillus fungorum strain FM-2 is tolerant to PB(II) and Cd(II) or can carry out the function of bioadsorption of heavy metals | [129] |
| Pollutant Cd and Hg | Bioadsorption | Pseudomonas sp., Salinobacter sp., Streptomyces sp., Roseobacter sp., Vibrio sp., Sac-charomonospo-ra sp. and others isolated from marine sponge Fasciospongia cavernosa | 7 days | Preliminary test | This sponge symbiotic bacteria is able to survive in habitats contaminated with heavy metals mercury and cadmium | [130] |
| Pb(II) and Cd(II) | Remediation in oxidation method | Natural adsorbent available in aquatic habitat | 2 days | 9.0 mg/g and 8.9 mg/g | Natural adsorbent found in the aquatic environment in remediating Pb(II) and Cd(II) pollutants. The isotherm data were processed using the Langmuir approach, showing that lead remediation is endothermic and cadmium is exothermic | [28] |
| Ions Co, Pb, Cu, Zn | Bioadsorption | Rumex crispus L | 7 days | 83.5–91.0% | The findings reveal that the heavy metal absorption mechanism occurs on the surface of the biosorbent to form a metal–biosorbent complex | [60] |
| Types of Hydrocarbon Contaminants | Sponge Symbiont Bacterial Species | Type of Sea Sponge | Interaction Duration | Removal Efficiency | Conclusion | References |
|---|---|---|---|---|---|---|
| Pyrene (±100 mg/L) | Bacillus licheniformis strain ATCC 9789 (Bl) | Auletta sp. | 30 days | ±39.0% | The performance and biodegradation kinetics increased during the contact period of 10–25 days, then slowed down to day 30 | [71] |
| PAHs (Anthracene and pyrene) | Bacillus pumilus strain GLB197 | Niphates sp. | 25 days | Antracene (21.9%) Pyrene (7.7%) | The consortium of three types of bacteria isolated from sea sponges can carry out the function of biodegradation of pyrene and anthracene components, but the performance is less significant, presumably due to competition for carbon as an energy source | [113] |
| Pseudomonas stutzeri strain SLG510A3-8 | Hyrtios erectus | |||||
| Acinetobacter calcoaceticus strain SLCDA 976 | Clathria (Thalysias) reinwardtii | |||||
| PAHs | Pseudomonas sp. strain Hi1 | Auleeta sp | Preliminary test on PAHs contaminated media | Observation (qualitative) | All types of sponge symbiont bacteria showed activity on media exposed to PAHs | [78] |
| Bacillus subtilis strain BAB-1684 | Clathria reinwardti | |||||
| Pseudomonas stutzeri strain RCH2 | Callyspongia sp | |||||
| Bacillus flexus strain PHCD-20 | Hyrtios erectus | |||||
| Naphthalene | Bacillus sp. | Neopetrosia sp | 25 days | ±51.4% | Both types of spongy symbiont bacteria can degrade naphthalene, characterized by several parameters, namely, the increased acidity of the interaction medium, increased optical density (OD600), smells of fermentation and gas bubbles are formed | [58] |
| Acinetobacter Calcoaceticus | Callyspongia Aerizusa | ±37.3% | ||||
| Pyrene | Sp AB1 and Sp AB2 | Hyrtios erectus(Sp A) | Preliminary test on pyrene contaminated media | The activity of the two isolates is weak | The activity of isolates against pyrene generally came from sponges whose body surface was covered with mucus. This mucus is thought to have an enzyme character | [76] |
| Sp BB1 and Sp BB2 | Clathria (Thalysias) reinwardti (Sp B) | Both isolates did not show activity | ||||
| Sp CB1 and Sp CB2 | Niphates sp. (Sp C) | Both isolates showed strong activity | ||||
| Sp DB1 and Sp DB2 | Callyspongia sp. (Sp D) | Both isolates showed moderate activity | ||||
| PAHs Naphthalene and Anthracene | Isolate Sp6. B2 | Auletta sp. | 20 days | There is biodegradation activity | The biodegradation activity of Sp6.B2 isolates against naphthalene and anthracene appeared to be more dominant than Sp8.B1 isolates. | [124] |
| Isolate Sp8. B1 | Callyspongia Aerizusa | |||||
| Aliphatic Components | Bacillus cohnii strain DSM 6307 | Niphates sp. | 25 days | Average 48.1% | GC-MS and FTIR detected new organic compounds of alcohol, aldehyde and carboxylic acid groups | [31] |
| Bacillus pumilus strain GLB197 | ||||||
| Petroleum sludge | BacillusFlexus strain PHCDB20. | Callyspongia sp. | 35 days | Identified 18 types of aliphatic comp. and 2 aromatic comps. | All hydrocarbon components in the degraded sludge are characterized by a decrease in abundance | [13] |
| Types of Hyd-rocarbon Contaminants | Sponge Symbiont Bacterial Species | Type of Sea Sponge | Interaction Duration | Removal Efficiency | Conclusion | References |
|---|---|---|---|---|---|---|
| Chromium (VI) Manganese (VII) | Acinetobacter calcoaceticus strain PHCDB14 | Callyspongia aerizusa | 15 days | ±63.2% | Both types of pollutants are absorbed to the maximum at a contact period of 3 days | [65] |
| ±66.8% | ||||||
| Cr, Zn, Cu, Fe, Co, Mn, Ag and Cd | Bacillus cohnii strains DSM 6307 | Niphates sp. | 16 days | Heavy metal pollutant removal efficiency varies | All types of heavy metals tested can be absorbed by the symbiont bacteria isolates Niphates sp. and Clathria (Thalysias) reinwardtii with varying biosorption performances. Optimum biosorption occurs at a contact period of 4 days | [77] |
| Pseudomonas stutzeri RCH2 | Clathria (Thalysias) reinwardti | |||||
| Cd2+ and As3+ | Isolate Sp6. B2 | Auletta sp. | 20 days | 83.2%, and 82.2% | Optimum biosorption occurred at a contact duration of 5 days, then weakened until the 20th day of the contact period | [124] |
| Isolate Sp8. B1 | Callyspongia Aerizusa | 99.9%, and 99.9% | ||||
| Cr(VI) and Cd(II) | Bacillus pumilus strain GLB197 | Niphates sp. | 15 days | 56.3% and 61.2% | The optimum bioadsorption of these two types of sponge symbiont bacteria against the two types of heavy metal pollutants tested occurred in the range of 3–6 days of contact | [32] |
| Pseudomonas stutzeri strain SLG510A3-8 | Hyrtios erectus | 52.7% and 57.8%. | ||||
| As3+ and Hg2+ | Bacillus licheniformis strain ATCC 9789 | Auletta sp. | 16 days | 99.9%, and 88.5%, | Biosorption takes place optimally at a contact duration of 3–6 days. Another supporting indicator is the increase in optical density (OD600) and gas bubbles detected in the interaction medium | [131] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzuki, I.; Rosmiati, R.; Mustafa, A.; Sahabuddin, S.; Tarunamulia, T.; Susianingsih, E.; Hendrajat, E.A.; Sahrijanna, A.; Muslimin, M.; Ratnawati, E.; et al. Potential Utilization of Bacterial Consortium of Symbionts Marine Sponges in Removing Polyaromatic Hydrocarbons and Heavy Metals, Review. Biology 2023, 12, 86. https://doi.org/10.3390/biology12010086
Marzuki I, Rosmiati R, Mustafa A, Sahabuddin S, Tarunamulia T, Susianingsih E, Hendrajat EA, Sahrijanna A, Muslimin M, Ratnawati E, et al. Potential Utilization of Bacterial Consortium of Symbionts Marine Sponges in Removing Polyaromatic Hydrocarbons and Heavy Metals, Review. Biology. 2023; 12(1):86. https://doi.org/10.3390/biology12010086
Chicago/Turabian StyleMarzuki, Ismail, Rosmiati Rosmiati, Akhmad Mustafa, Sahabuddin Sahabuddin, Tarunamulia Tarunamulia, Endang Susianingsih, Erfan Andi Hendrajat, Andi Sahrijanna, Muslimin Muslimin, Erna Ratnawati, and et al. 2023. "Potential Utilization of Bacterial Consortium of Symbionts Marine Sponges in Removing Polyaromatic Hydrocarbons and Heavy Metals, Review" Biology 12, no. 1: 86. https://doi.org/10.3390/biology12010086
APA StyleMarzuki, I., Rosmiati, R., Mustafa, A., Sahabuddin, S., Tarunamulia, T., Susianingsih, E., Hendrajat, E. A., Sahrijanna, A., Muslimin, M., Ratnawati, E., Kamariah, K., Nisaa, K., Herlambang, S., Gunawan, S., Santi, I. S., Isnawan, B. H., Kaseng, E. S., Septiningsih, E., Asaf, R., ... Basri, B. (2023). Potential Utilization of Bacterial Consortium of Symbionts Marine Sponges in Removing Polyaromatic Hydrocarbons and Heavy Metals, Review. Biology, 12(1), 86. https://doi.org/10.3390/biology12010086















