The Dynamics of miR-449a/c Expression during Uterine Cycles Are Associated with Endometrial Development
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
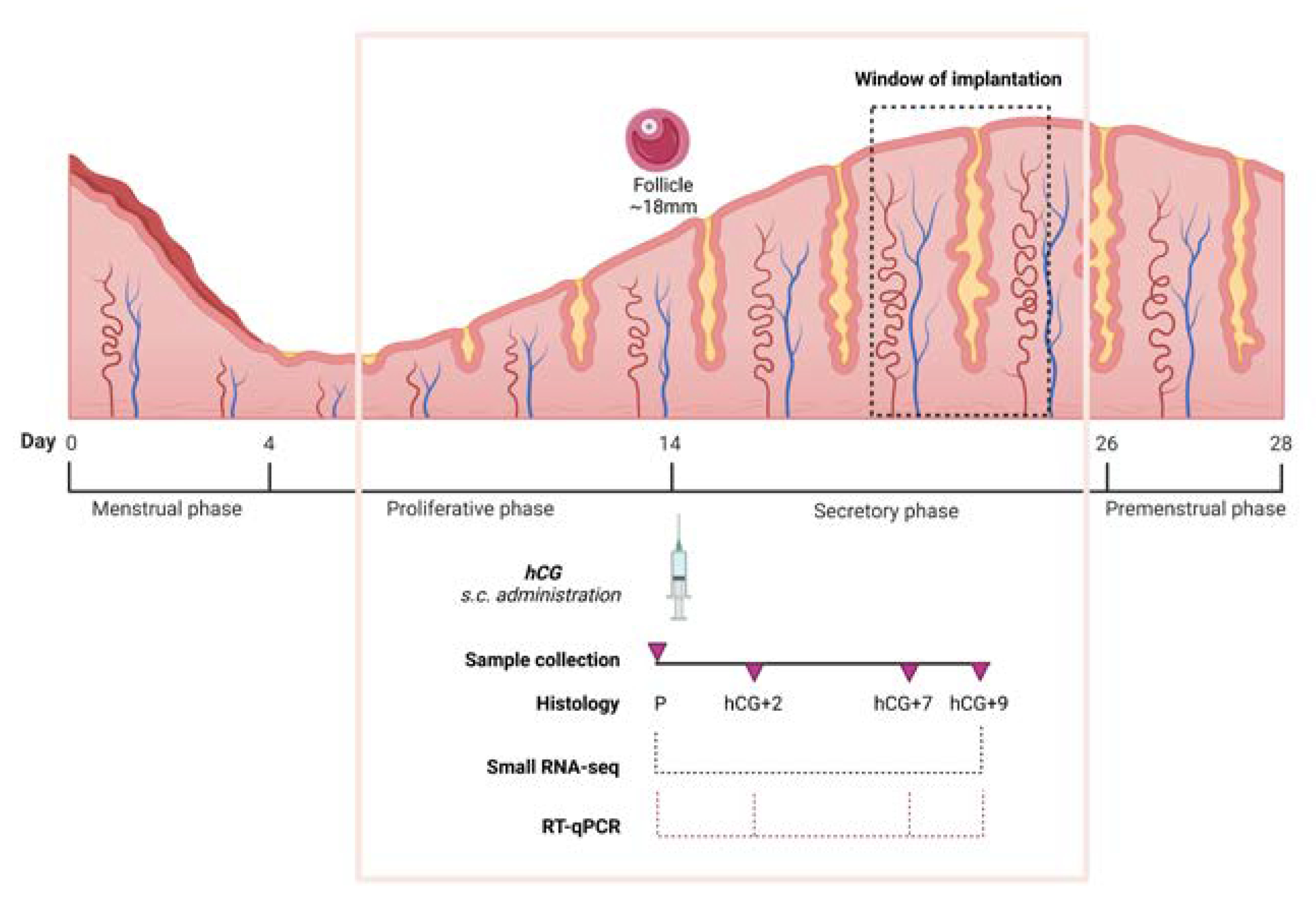
2.1. Patients, Study Design, and Samples
2.2. Histology
2.3. RNA Extraction and Quality Control
2.4. sRNA-Seq and Data Analysis
2.5. miR, isomiR and Pre-miR Expression Profiling by RT-qPCR
3. Results
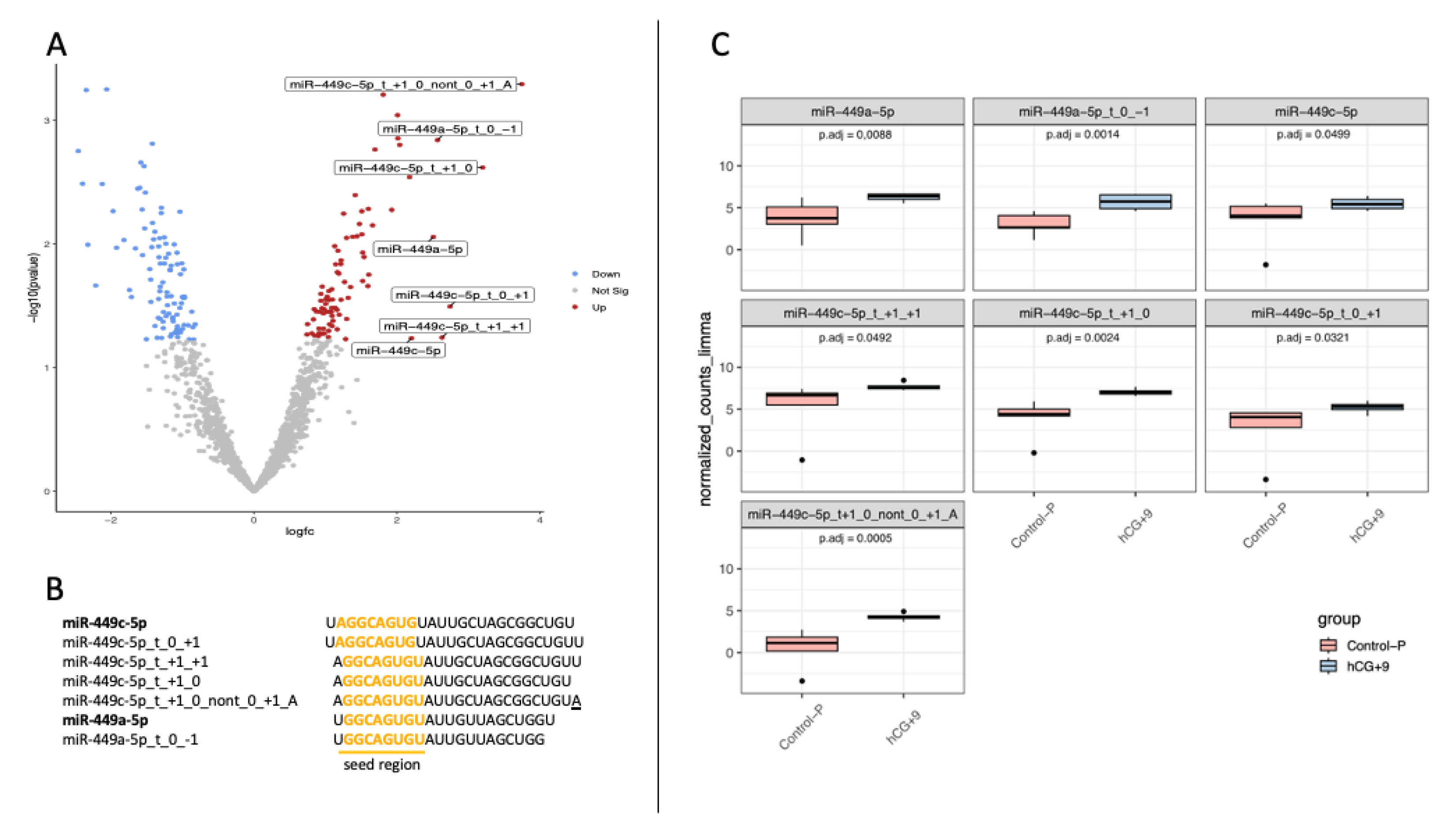
3.1. sRNA-Seq Expression Data Identified the miR-449 Family as the Most Strongly Up-Regulated in the hCG-Primed RE
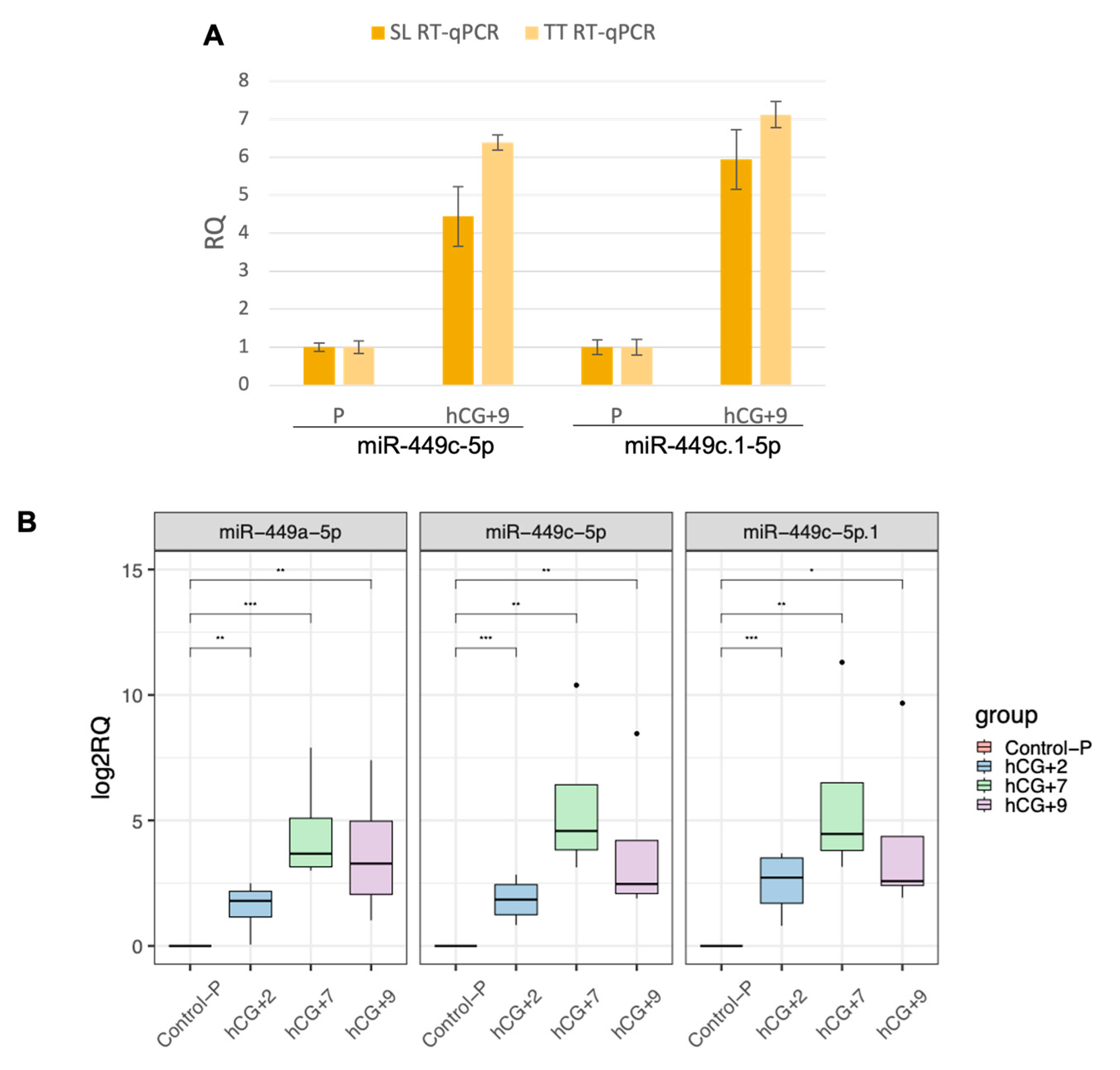
3.2. Expression of the mir-449 Family Is Dynamic throughout the Endometrial Cycle, with a Peak in the Receptive Endometrium
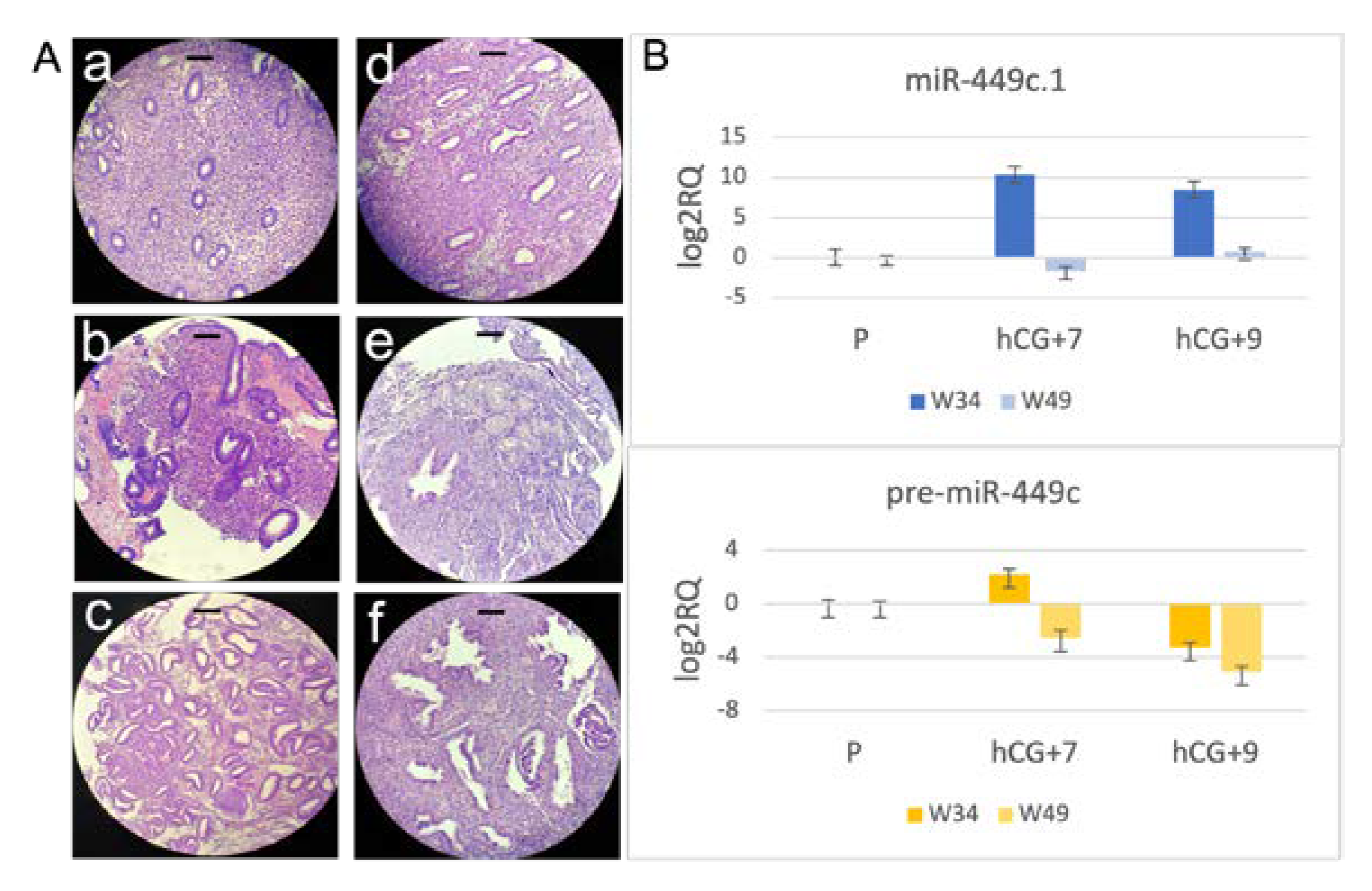
3.3. miR-449c.1 Expression Correlates with Endometrial Histological Pattern and Patient Age (Case Study)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Macklon, N.S.; Stouffer, R.L.; Giudice, L.C.; Fauser, B.C.J.M. The Science behind 25 Years of Ovarian Stimulation for in Vitro Fertilization. Endocr. Rev. 2006, 27, 170–207. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, S.; Young, S. Diagnostic and Therapeutic Options in Recurrent Implantation Failure. F1000Research 2020, 9, 208. [Google Scholar] [CrossRef]
- Koot, Y.E.M.; Teklenburg, G.; Salker, M.S.; Brosens, J.J.; Macklon, N.S. Molecular Aspects of Implantation Failure. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Horcajadas, J.A.; Pellicer, A.; Simón, C. Wide Genomic Analysis of Human Endometrial Receptivity: New Times, New Opportunities. Hum. Reprod. Update 2007, 13, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Ambros, V. An Extensive Class of Small RNAs in Caenorhabditis Elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of Mammalian MicroRNA Host Genes and Transcription Units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA Genes Are Transcribed by RNA Polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The Nuclear RNase III Drosha Initiates MicroRNA Processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor Complex Mediates the Genesis of MicroRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. Recognition and Cleavage of Primary MicroRNA Precursors by the Nuclear Processing Enzyme Drosha. EMBO J. 2005, 24, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer Recognizes the 5′ End of RNA for Efficient and Accurate Processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Jeong, K.; Kim, V.N. Genome-Wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol. Cell 2017, 66, 258–269.e5. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of MicroRNA Processing and Expression through RNA Editing by ADAR Deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. MicroRNAs as Regulators of Differentiation and Cell Fate Decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef]
- Altmäe, S.; Martinez-Conejero, J.A.; Esteban, F.J.; Ruiz-Alonso, M.; Stavreus-Evers, A.; Horcajadas, J.A.; Salumets, A. MicroRNAs MiR-30b, MiR-30d, and MiR-494 Regulate Human Endometrial Receptivity. Reprod. Sci. 2013, 20, 308–317. [Google Scholar] [CrossRef]
- Vilella, F.; Moreno-Moya, J.M.; Balaguer, N.; Grasso, A.; Herrero, M.; Martínez, S.; Marcilla, A.; Simón, C. Hsa-MiR-30d, Secreted by the Human Endometrium, Is Taken up by the Pre-Implantation Embryo and Might Modify Its Transcriptome. Development 2015, 142, 3210–3221. [Google Scholar] [CrossRef]
- Sha, A.G.; Liu, J.L.; Jiang, X.M.; Ren, J.Z.; Ma, C.H.; Lei, W.; Su, R.W.; Yang, Z.M. Genome-Wide Identification of Micro-Ribonucleic Acids Associated with Human Endometrial Receptivity in Natural and Stimulated Cycles by Deep Sequencing. Fertil. Steril. 2011, 96, 150–155.e5. [Google Scholar] [CrossRef]
- Altmäe, S.; Koel, M.; Võsa, U.; Adler, P.; Suhorutšenko, M.; Laisk-Podar, T.; Kukushkina, V.; Saare, M.; Velthut-Meikas, A.; Krjutškov, K.; et al. Meta-Signature of Human Endometrial Receptivity: A Meta-Analysis and Validation Study of Transcriptomic Biomarkers. Sci. Rep. 2017, 7, 10077. [Google Scholar] [CrossRef]
- Sigurgeirsson, B.; Åmark, H.; Jemt, A.; Ujvari, D.; Westgren, M.; Lundeberg, J.; Gidlöf, S. Comprehensive RNA Sequencing of Healthy Human Endometrium at Two Time Points of the Menstrual Cycle. Biol. Reprod. 2017, 96, 24–33. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, H.R.; Lim, E.J.; Park, M.; Yoon, J.A.; Kim, Y.S.; Kim, E.K.; Shin, J.E.; Kim, J.H.; Kwon, H.; et al. Integrative Analyses of Uterine Transcriptome and MicroRNAome Reveal Compromised LIF-STAT3 Signaling and Progesterone Response in the Endometrium of Patients with Recurrent/Repeated Implantation Failure (RIF). PLoS ONE 2016, 11, e0157696. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Shen, H.; Fan, L.J.; Guan, J.; Zheng, X.B.; Chen, X.; Liang, R.; Zhang, X.W.; Cui, Q.H.; Sun, K.K.; et al. Endometrial MicroRNA Signature during the Window of Implantation Changed in Patients with Repeated Implantation Failure. Chin. Med. J. 2017, 130, 566–573. [Google Scholar] [CrossRef]
- Simon, A.; Laufer, N. Repeated Implantation Failure: Clinical Approach. Fertil. Steril. 2012, 97, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Suhorutshenko, M.; Kukushkina, V.; Velthut-Meikas, A.; Altmäe, S.; Peters, M.; Mägi, R.; Krjutškov, K.; Koel, M.; Codoñer, F.M.; Martinez-Blanch, J.F.; et al. Endometrial Receptivity Revisited: Endometrial Transcriptome Adjusted for Tissue Cellular Heterogeneity. Hum. Reprod. 2018, 33, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Godakumara, K.; Ord, J.; Lättekivi, F.; Dissanayake, K.; Viil, J.; Boggavarapu, N.R.; Faridani, O.R.; Jääger, K.; Velthut-Meikas, A.; Jaakma, Ü.; et al. Trophoblast Derived Extracellular Vesicles Specifically Alter the Transcriptome of Endometrial Cells and May Constitute a Critical Component of Embryo-Maternal Communication. Reprod. Biol. Endocrinol. 2021, 19, 1–14. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of Massively Parallel Sequencing to MicroRNA Profiling and Discovery in Human Embryonic Stem Cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-Sequencing of Human Argonaute-Associated Small RNAs Provides Insight into MiRNA Sorting and Reveals Argonaute Association with RNA Fragments of Diverse Origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs—The Overlooked Repertoire in the Dynamic MicroRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef]
- Engkvist, M.E.; Stratford, E.W.; Lorenz, S.; Meza-Zepeda, L.A.; Myklebost, O.; Munthe, E. Analysis of the MiR-34 Family Functions in Breast Cancer Reveals Annotation Error of MiR-34b. Sci. Rep. 2017, 7, 9655. [Google Scholar] [CrossRef]
- Mercey, O.; Popa, A.; Cavard, A.; Paquet, A.; Chevalier, B.; Pons, N.; Magnone, V.; Zangari, J.; Brest, P.; Zaragosi, L.E.; et al. Characterizing IsomiR Variants within the MicroRNA-34/449 Family. FEBS Lett. 2017, 591, 693–705. [Google Scholar] [CrossRef]
- Nikolova, M.; Naydenov, M.; Glogovitis, I.; Apostolov, A.; Saare, M.; Boggavarapu, N.; Salumets, A.; Baev, V.; Yahubyan, G. Coupling MiR/IsomiR and MRNA Expression Signatures Unveils New Molecular Layers of Endometrial Receptivity. Life 2021, 11, 1391. [Google Scholar] [CrossRef] [PubMed]
- Sandbothe, M.; Buurman, R.; Reich, N.; Greiwe, L.; Vajen, B.; Gürlevik, E.; Schäffer, V.; Eilers, M.; Kühnel, F.; Vaquero, A.; et al. The MicroRNA-449 Family Inhibits TGF-β-Mediated Liver Cancer Cell Migration by Targeting SOX4. J. Hepatol. 2017, 66, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fu, H.; Liu, Q.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Downregulation of CCND1 and CDK6 by MiR-34a Induces Cell Cycle Arrest. FEBS Lett. 2008, 582, 1564–1568. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Walentek, P.; Sponer, N.; Klimke, A.; Lee, J.S.; Dixon, G.; Harland, R.; Wan, Y.; Lishko, P.; Lize, M.; et al. MiR-34/449 MiRNAs Are Required for Motile Ciliogenesis by Repressing Cp110. Nature 2014, 510, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bao, J.; Kim, M.; Yuan, S.; Tang, C.; Zheng, H.; Mastick, G.S.; Xu, C.; Yan, W. Two MiRNA Clusters, MiR-34b/c and MiR-449, Are Essential for Normal Brain Development, Motile Ciliogenesis, and Spermatogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, E2851–E2857. [Google Scholar] [CrossRef]
- Otto, T.; Candido, S.V.; Pilarz, M.S.; Sicinska, E.; Bronson, R.T.; Bowden, M.; Lachowicz, I.A.; Mulry, K.; Fassl, A.; Han, R.C.; et al. Cell Cycle-Targeting MicroRNAs Promote Differentiation by Enforcing Cell-Cycle Exit. Proc. Natl. Acad. Sci. USA 2017, 114, 10660–10665. [Google Scholar] [CrossRef]
- Lizé, M.; Pilarski, S.; Dobbelstein, M. E2F1-Inducible MicroRNA 449a/b Suppresses Cell Proliferation and Promotes Apoptosis. Cell Death Differ. 2010, 17, 452–458. [Google Scholar] [CrossRef]
- Marcet, B.; Chevalier, B.; Luxardi, G.; Coraux, C.; Zaragosi, L.E.; Cibois, M.; Robbe-Sermesant, K.; Jolly, T.; Cardinaud, B.; Moreilhon, C.; et al. Control of Vertebrate Multiciliogenesis by MiR-449 through Direct Repression of the Delta/Notch Pathway. Nat. Cell Biol. 2011, 13, 693–699. [Google Scholar] [CrossRef]
- Bao, J.; Li, D.; Wang, L.; Wu, J.; Hu, Y.; Wang, Z.; Chen, Y.; Cao, X.; Jiang, C.; Yan, W.; et al. MicroRNA-449 and MicroRNA-34b/c Function Redundantly in Murine Testes by Targeting E2F Transcription Factor-Retinoblastoma Protein (E2F-PRb) Pathway. J. Biol. Chem. 2012, 287, 21686–21698. [Google Scholar] [CrossRef]
- Bouhallier, F.; Allioli, N.; Lavial, F.; Chalmel, F.; Perrard, M.H.; Durand, P.; Samarut, J.; Pain, B.; Rouault, J.P. Role of MiR-34c MicroRNA in the Late Steps of Spermatogenesis. RNA 2010, 16, 720–731. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, D.; Wei, C.; Luo, H.; Liu, J.; Fu, R.; Cui, S. MicroRNA-34c Enhances Murine Male Germ Cell Apoptosis through Targeting ATF1. PLoS ONE 2012, 7, e33861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, M.; Liu, C.; Wang, L.; Hu, Y.; Bai, Y.; Hua, J. MIR-34c Regulates Mouse Embryonic Stem Cells Differentiation into Male Germ-like Cells through RARg. Cell Biochem. Funct. 2012, 30, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Lizé, M.; Klimke, A.; Dobbelstein, M. MicroRNA-449 in Cell Fate Determination. Cell Cycle 2011, 10, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The P53/MiR-34 Axis in Development and Disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Noyes, R.W.; Hertig, A.T.; Rock, J. Dating the Endometrial Biopsy. Am. J. Obstet. Gynecol. 1975, 122, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-Tailed RT-QPCR: A Novel Method for Highly Accurate MiRNA Quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef]
- Glogovitis, I.; Yahubyan, G.; Würdinger, T.; Koppers-Lalic, D.; Baev, V. MiRGalaxy: Galaxy-Based Framework for Interactive Analysis of MicroRNA and IsomiR Sequencing Data. Cancers 2021, 13, 5663. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem-Loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Cuman, C.; Menkhorst, E.; Winship, A.; van Sinderen, M.; Osianlis, T.; Rombauts, L.J.; Dimitriadis, E. Fetal-Maternal Communication: The Role of Notch Signalling in Embryo Implantation. Reproduction 2014, 147, R75–R86. [Google Scholar] [CrossRef]
- Loukas, I.; Skamnelou, M.; Tsaridou, S.; Bournaka, S.; Grigoriadis, S.; Taraviras, S.; Lygerou, Z.; Arbi, M. Fine-Tuning Multiciliated Cell Differentiation at the Post-Transcriptional Level: Contribution of MiR-34/449 Family Members. Biol. Rev. 2021, 96, 2321–2332. [Google Scholar] [CrossRef]
- Wang, L.; Fu, C.; Fan, H.; Du, T.; Dong, M.; Chen, Y.; Jin, Y.; Zhou, Y.; Deng, M.; Gu, A.; et al. MiR-34b Regulates Multiciliogenesis during Organ Formation in Zebrafish. Development 2013, 140, 2755–2764. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, B.; Adamiok, A.; Mercey, O.; Revinski, D.R.; Zaragosi, L.E.; Pasini, A.; Kodjabachian, L.; Barbry, P.; Marcet, B. MiR-34/449 Control Apical Actin Network Formation during Multiciliogenesis through Small GTPase Pathways. Nat. Commun. 2015, 6, 8386. [Google Scholar] [CrossRef] [PubMed]
- Gellersen, B.; Brosens, J.J. Cyclic Decidualization of the Human Endometrium in Reproductive Health and Failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef]
- Afshar, Y.; Miele, L.; Fazleabas, A.T. Notch1 Is Regulated by Chorionic Gonadotropin and Progesterone in Endometrial Stromal Cells and Modulates Decidualization in Primates. Endocrinology 2012, 153, 2884–2896. [Google Scholar] [CrossRef]
- Hassan, E.; Kojima, R.; Ozawa, F.; Yoshihara, H.; Goto, S.; Kitaori, T.; Inagaki, H.; Kato, Y.; Sugiura-Ogasawara, M. Abnormal Ciliogenesis in Decidual Stromal Cells in Recurrent Miscarriage. J. Reprod. Immunol. 2022, 150, 103486. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alonso, L.; Handfield, L.F.; Roberts, K.; Nikolakopoulou, K.; Fernando, R.C.; Gardner, L.; Woodhams, B.; Arutyunyan, A.; Polanski, K.; Hoo, R.; et al. Mapping the Temporal and Spatial Dynamics of the Human Endometrium in Vivo and in Vitro. Nat. Genet. 2021, 53, 1698–1711. [Google Scholar] [CrossRef] [PubMed]
- Damle, R.P.; Dravid, N.V.; Suryawanshi, K.H.; Gadre, A.S.; Bagale, P.S.; Ahire, N. Clinicopathological Spectrum of Endometrial Changes in Peri-Menopausal and Post-Menopausal Abnormal Uterine Bleeding: A 2 Years Study. J. Clin. Diagn. Res. 2013, 7, 2774. [Google Scholar] [CrossRef] [PubMed]
- Burger, H.G.; Hale, G.E.; Dennerstein, L.; Robertson, D.M. Cycle and Hormone Changes during Perimenopause: The Key Role of Ovarian Function. Menopause 2008, 15, 603–612. [Google Scholar] [CrossRef]
- Van den Brink, H.; Chizen, D.; Hale, G.; Baerwald, A. Age-Related Changes in Major Ovarian Follicular Wave Dynamics during the Human Menstrual Cycle. Menopause 2013, 20, 1243–1254. [Google Scholar] [CrossRef]
- Devesa-Peiro, A.; Sebastian-Leon, P.; Parraga-Leo, A.; Pellicer, A.; Diaz-Gimeno, P. Breaking the Ageing Paradigm in Endometrium: Endometrial Gene Expression Related to Cilia and Ageing Hallmarks in Women over 35 Years. Hum. Reprod. 2022, 37, 762–776. [Google Scholar] [CrossRef]
- An, X.; Liu, X.; Zhang, L.; Liu, J.; Zhao, X.; Chen, K.; Ma, H.; Li, G.; Cao, B.; Song, Y. MIR-449a Regulates Caprine Endometrial Stromal Cell Apoptosis and Endometrial Receptivity. Sci. Rep. 2017, 7, 12248. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naydenov, M.; Nikolova, M.; Apostolov, A.; Glogovitis, I.; Salumets, A.; Baev, V.; Yahubyan, G. The Dynamics of miR-449a/c Expression during Uterine Cycles Are Associated with Endometrial Development. Biology 2023, 12, 55. https://doi.org/10.3390/biology12010055
Naydenov M, Nikolova M, Apostolov A, Glogovitis I, Salumets A, Baev V, Yahubyan G. The Dynamics of miR-449a/c Expression during Uterine Cycles Are Associated with Endometrial Development. Biology. 2023; 12(1):55. https://doi.org/10.3390/biology12010055
Chicago/Turabian StyleNaydenov, Mladen, Maria Nikolova, Apostol Apostolov, Ilias Glogovitis, Andres Salumets, Vesselin Baev, and Galina Yahubyan. 2023. "The Dynamics of miR-449a/c Expression during Uterine Cycles Are Associated with Endometrial Development" Biology 12, no. 1: 55. https://doi.org/10.3390/biology12010055
APA StyleNaydenov, M., Nikolova, M., Apostolov, A., Glogovitis, I., Salumets, A., Baev, V., & Yahubyan, G. (2023). The Dynamics of miR-449a/c Expression during Uterine Cycles Are Associated with Endometrial Development. Biology, 12(1), 55. https://doi.org/10.3390/biology12010055








