A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. RNA Isolation and cDNA Synthesis
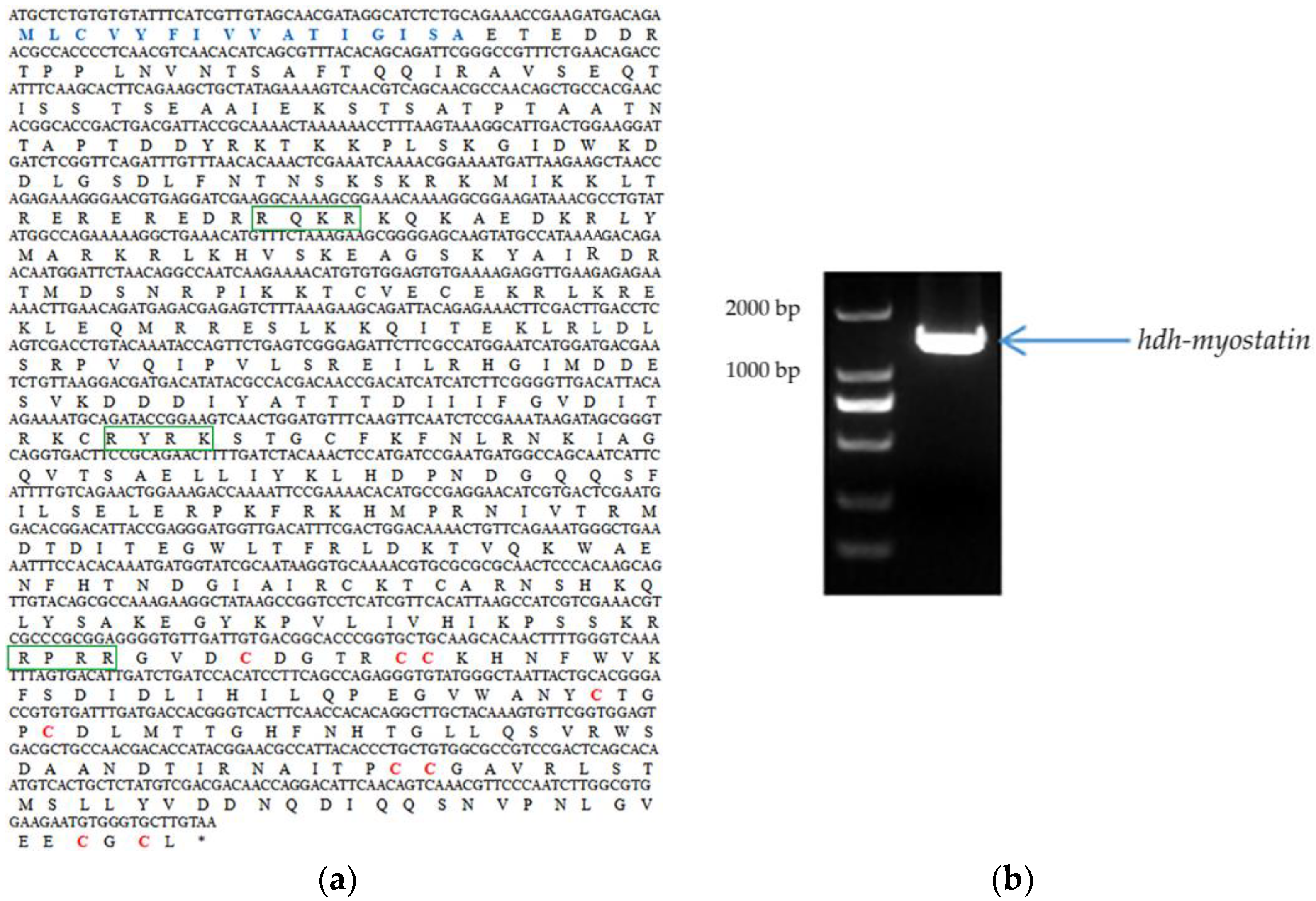
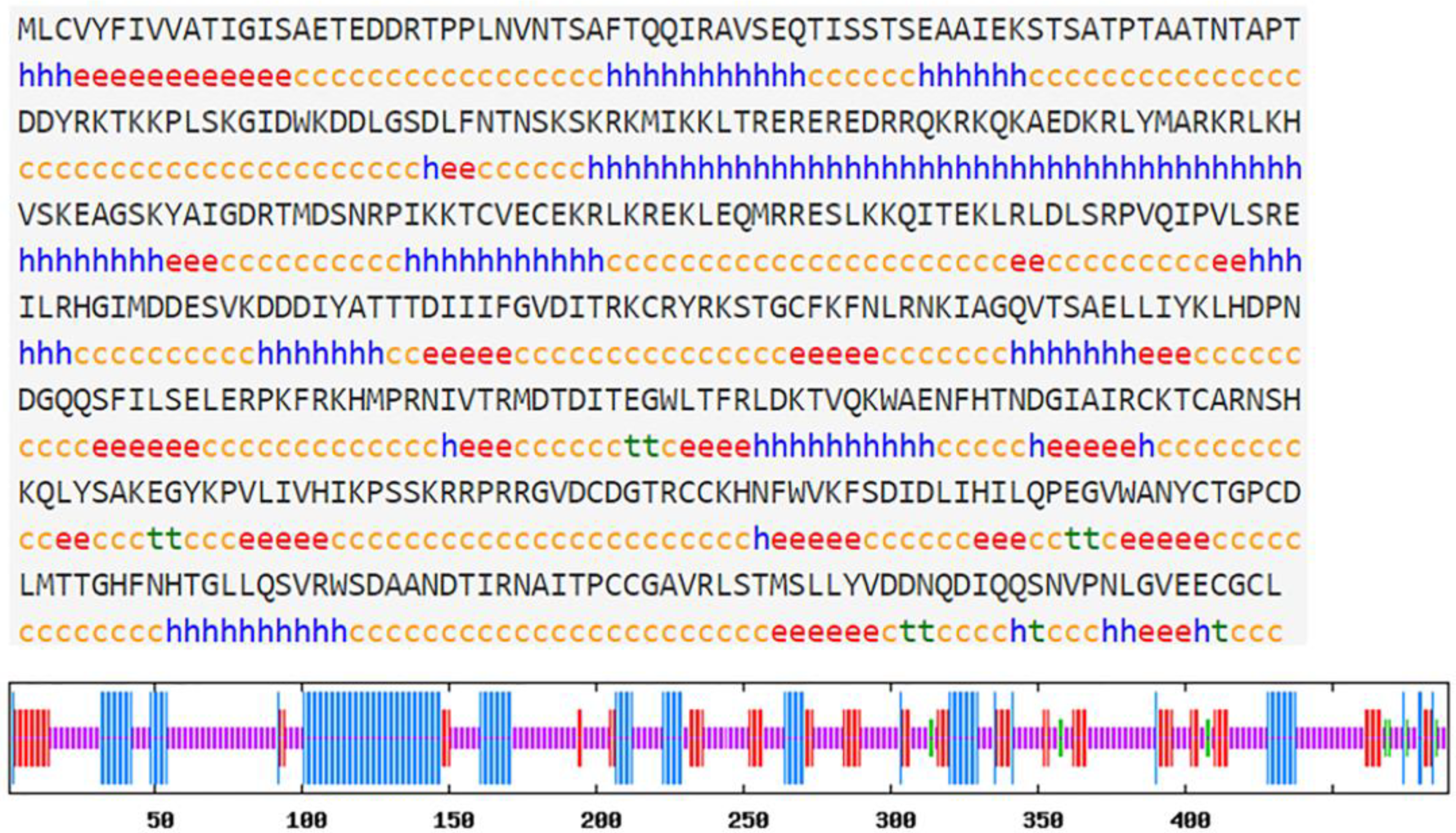
2.3. Hdh-Myostatin ORF Confirmation and Sequence Analysis
2.4. Sequence and SNP Analysis
2.5. RNA Interference of Hdh-Myostatin
2.6. Real-Time Quantitative Reverse Transcription PCR
2.7. Statistical Analysis
3. Results
3.1. Characterization of Hdh-Myostatin
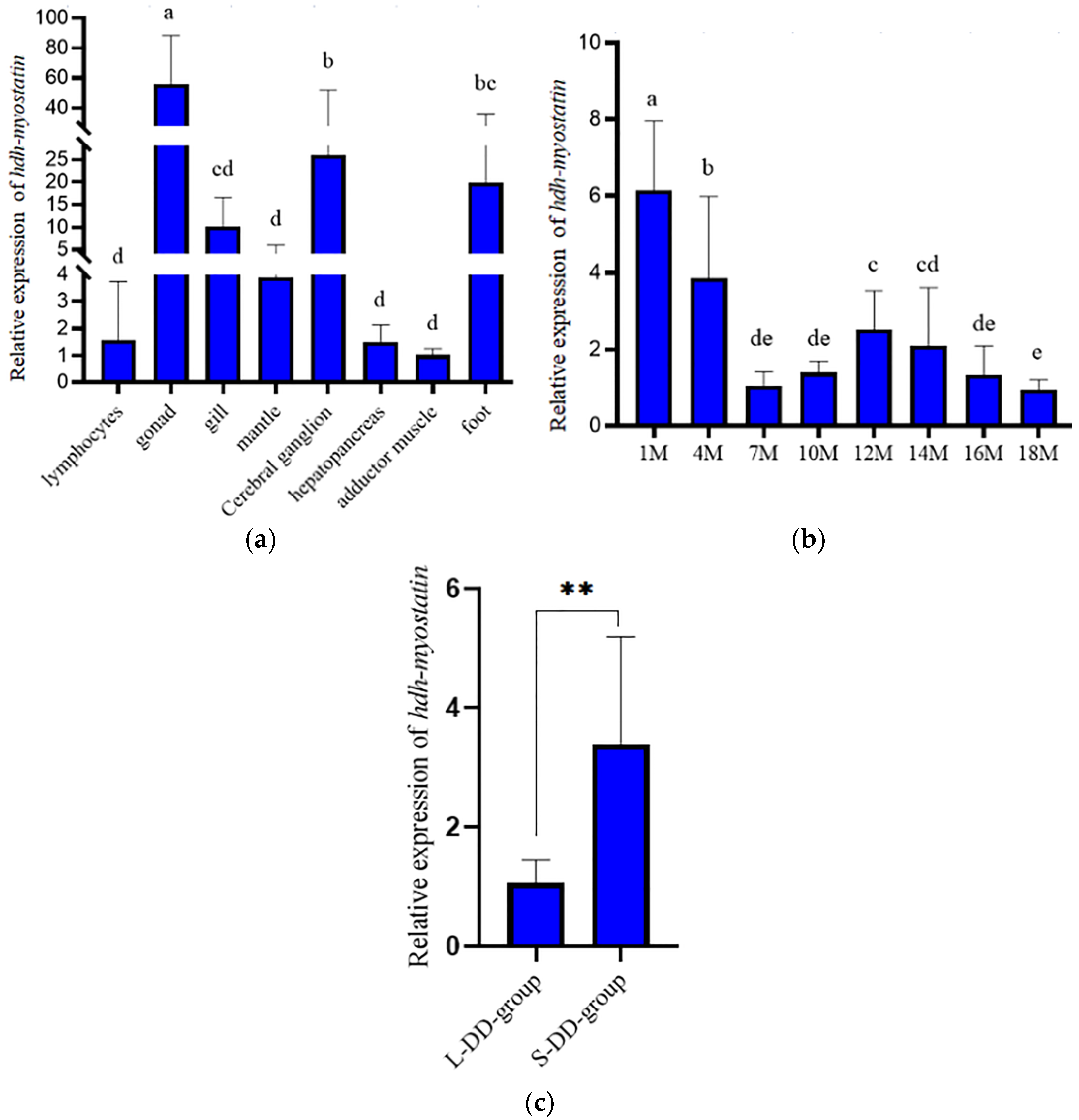
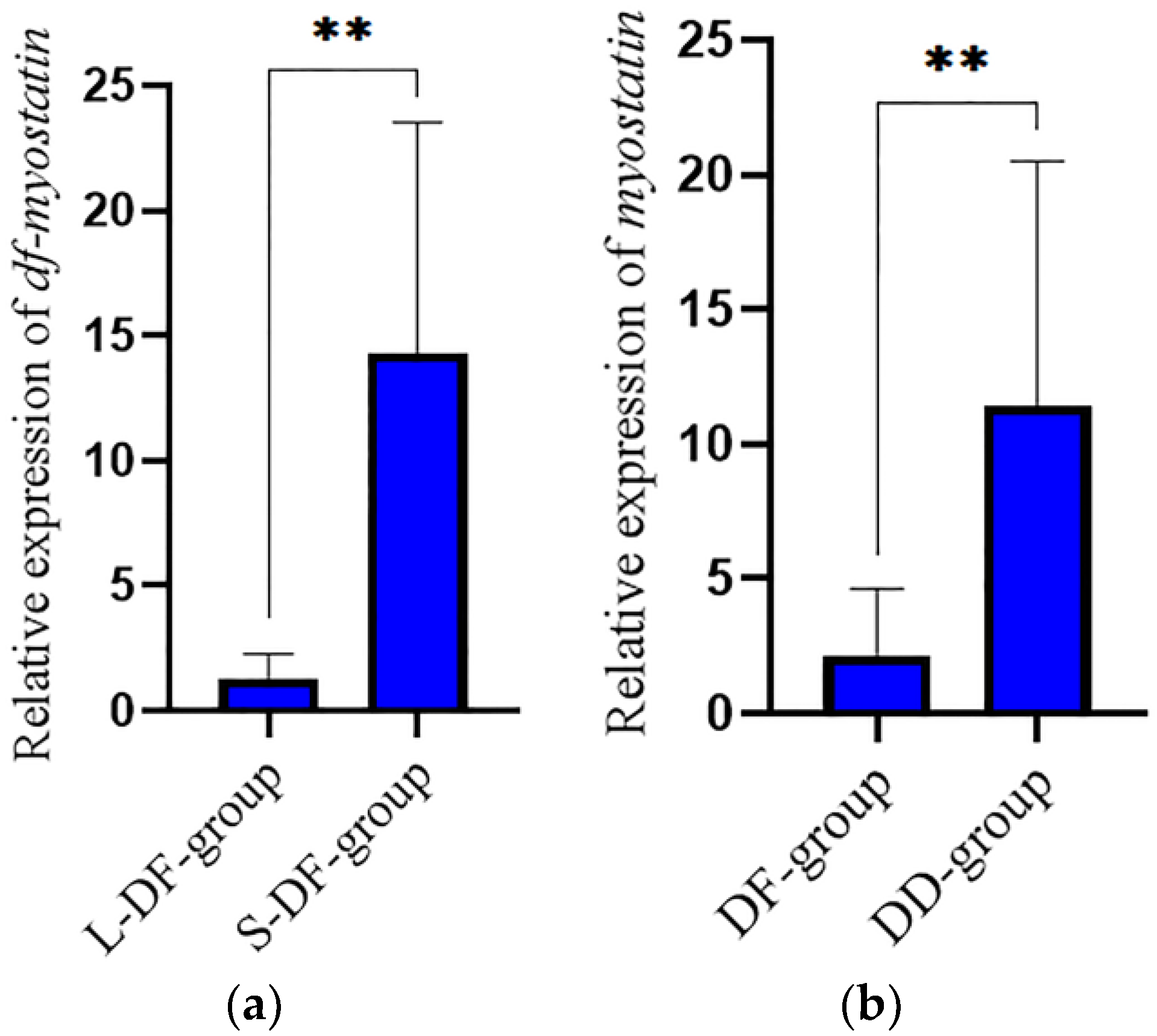
3.2. Expression Analysis of Hdh-Myostatin
3.3. Growth-Related SNP Loci in Hdh-Myostatin
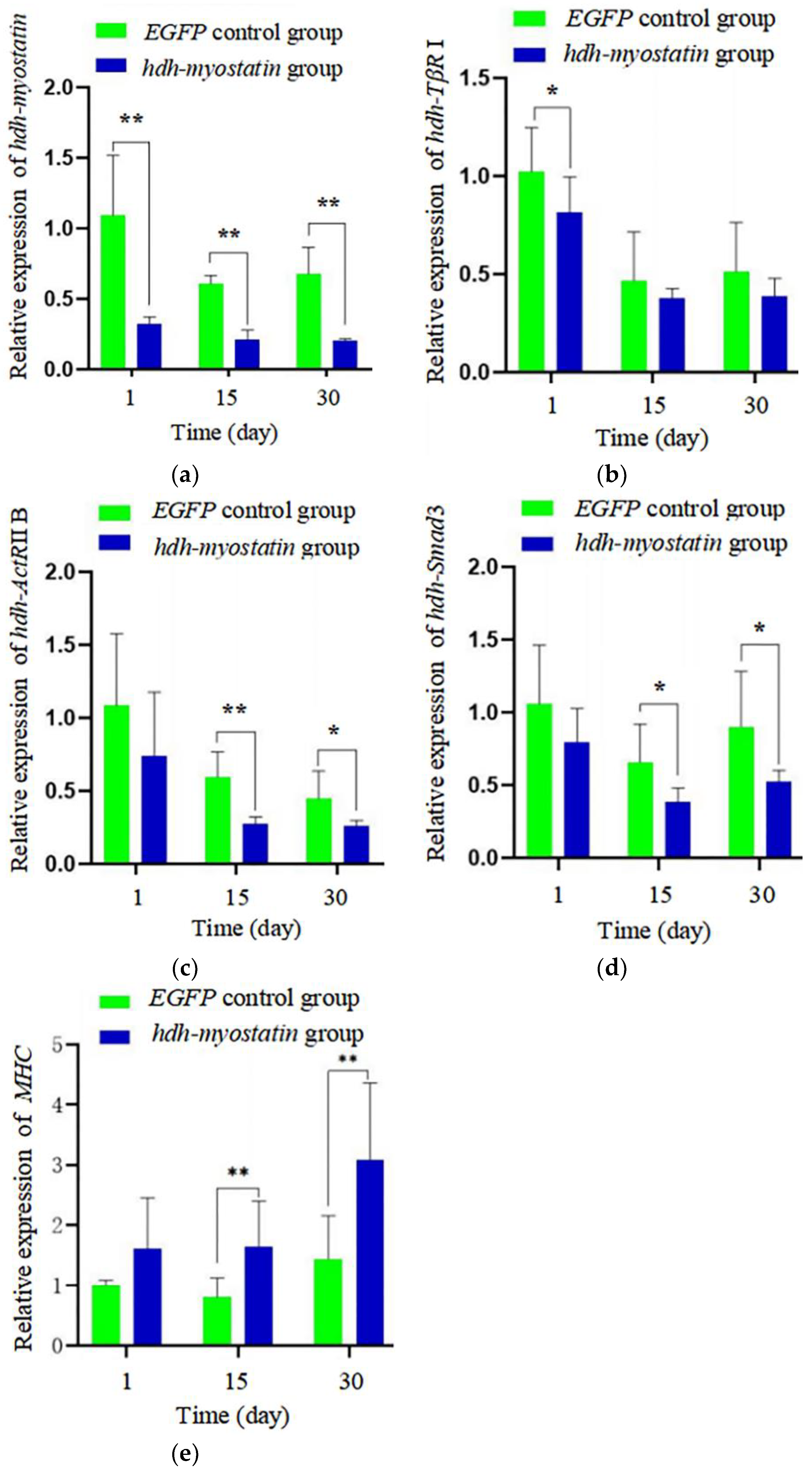
3.4. Effects of Hdh-Myostatin dsRNA Injection
3.5. Verification in the Hybrid Lvpan Abalone
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, N.; Luo, X.; Gu, Y.T.; Han, G.D.; Dong, Y.W.; You, W.W.; Ke, C.H. Assessment of the thermal tolerance of abalone based on cardiac performance in Haliotis discus hannai, H. gigantea and their interspecific hybrid. Aquaculture 2016, 465, 258–264. [Google Scholar] [CrossRef]
- Wang, X.Z.; Tang, B.; Luo, X.; Ke, C.H.; Huang, M.Q.; You, W.W.; Wang, Y. Effects of temperature, diet and genotype-induced variations on the gut microbiota of abalone. Aquaculture 2020, 524, 735269. [Google Scholar] [CrossRef]
- Guo, Y.F.; Zhao, W.W.; Gao, H.Q.; Wang, S.; Yu, P.M.; Yu, H.S.; Wang, D.; Wang, Q.; Wang, J.X.; Wang, Z.F.; et al. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2017. [Google Scholar]
- Hu, J.J.; Chen, Y.L.; Duan, X.K.; Jin, T.C.; Li, Y.; Zhang, L.J.; Liu, G.M.; Cao, M.J. Involvement of clip-domain serine protease in the anti-Vibrio immune response of abalone (Haliotis discus hannai)–Molecular cloning, characterization and functional analysis. Fish. Shellfish Immunol. 2018, 72, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Luo, X.; You, W.W.; Luo, L.; Ke, C.H. The role of hybridization in improving the immune response and thermal tolerance of abalone. Fish. Shellfish Immunol. 2014, 39, 69–77. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, J.F.; Yang, C.Y.; Chen, J.M.; Wang, W. Transcriptomic responses to thermal stress in hybrid abalone (Haliotis discus hannai ♀× H. fulgens♂). Front. Genet. 2022, 13, 1053674. [Google Scholar] [CrossRef]
- Diaz, F.; Re, A.D.; Medina, Z.; Re, G.; Valdez, G.; Valenzuela, F. Thermal preference and tolerance of green abalone Haliotis fulgens (Philippi, 1845) and pink abalone Haliotis corrugata (Gray, 1828). Aquac. Res. 2006, 37, 877–884. [Google Scholar] [CrossRef]
- You, W.W.; Guo, Q.; Fan, F.; Ren, P.; Luo, X.; Ke, C.H. Experimental hybridization and genetic identifification of Pacifific abalone Haliotis discus hannai and green abalone. Aquaculture 2015, 448, 243–249. [Google Scholar] [CrossRef]
- Lee, S.J. Targeting the myostatin signaling pathway to treat muscle loss and metabolic dysfunction. J. Clin. Investig. 2021, 131, e148372. [Google Scholar] [CrossRef]
- Ai, J.L.; Li, Z.M.; Liu, J.Y.; Shen, Y.C. Single nucleotide polymorphisms of myostatin gene and its association with growth traits in Haliotis diversicolor supertexta. J. Trop. Oceanogr. 2019, 38, 78–85. [Google Scholar]
- Esposito, P.; Picciotto, D.; Costigliolo, F.; Viazzi, F.; Verzola, D. Myostatin: Basic biology to clinical application. Adv. Clin. Chem. 2022, 106, 181–234. [Google Scholar]
- Grobet, L.; Martin, L.J.; Poncelet, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Ménissier, F.; Massabanda, J.; Fries, R.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef]
- Terova, G.; Rimoldi, S.; Bernardini, G.; Saroglia, M. Inhibition of myostatin gene expression in skeletal muscle of fish by in vivo electrically mediated dsRNA and shRNAi delivery. Mol. Biotechnol. 2013, 54, 673–684. [Google Scholar] [CrossRef]
- Sun, X.J.; Li, L.; Liu, Z.H.; Zhao, D.; Yang, A.G.; Zhou, L.Q.; Wu, B.; Tian, J.T. Molecular characterization of the myostatin gene and its regulation on muscle growth in Yesso scallop Patinopecten yessoensis. Aquaculture 2020, 520, 734982. [Google Scholar] [CrossRef]
- Kim, H.W.; Mykles, D.L.; Goetz, F.W.; Roberts, S.B. Characterization of a myostatin like gene from the bay scallop, Argopecten irradians. Biochim. Biophys. Acta 2004, 1679, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.L.; Guo, H.H.; He, Y.; Wang, H.; Zhang, L.L.; Wang, S.; Huang, X.T.; Roy, S.W.; Lu, W.; Hu, J.J.; et al. Molecular characterization of myostatin gene from Zhikong scallop Chlamys farreri (Jones et Preston 1904). Genes Genet. Syst. 2010, 85, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Acuna, G.; Gallardo-Escarate, C. The myostatin gene of Mytilus chilensis evidences a high level of polymorphism and ubiquitous transcript expression. Gene 2014, 536, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Morales-Collio, K.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C. Myostatin gene in the northern Chilean scallop Argopecten purpuratus (ApMSTN) reveals differences between wild and hatchery-bred populations. J. Mollus. Stud. 2014, 80, 169–176. [Google Scholar] [CrossRef]
- Niu, D.H.; Wang, L.; Bai, Z.Y.; Xie, S.M.; Zhao, H.G.; Li, J.L. Identification and expression characterization of the myostatin (MSTN) gene and association analysis with growth traits in the razor clam Sinonovacula constricta. Gene 2015, 555, 297–304. [Google Scholar] [CrossRef]
- Fan, S.G.; Xu, Y.H.; Liu, B.S.; He, W.Y.; Zhang, B.; Su, J.Q.; Yu, D.H. Molecular characterization and expression analysis of the myostatin gene and its association with growth traits in Noble scallop (Chlamys nobilis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 212, 24–31. [Google Scholar] [CrossRef]
- Carrera-Naipil, C.; Valenzuela-Muñoz, V.; Valdés, J.A.; Molina, A.; Gallardo-Escárate, C. RNA interference in Haliotis rufescens myostatin evidences upregulation of insulin signaling pathway. Agri Gene 2016, 1, 93–99. [Google Scholar] [CrossRef]
- De-Caestecker, M. The transforming growth factor-β superfamily of receptors. Cytokine Growth Factor Rev. 2004, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sartori, R.; Gregorevic, P.; Sandri, M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends. Endocrinol. Metab. 2014, 25, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.M.; Zhao, Y.P.; Zhao, Y.; Deng, S.L.; Yu, K. Regulation of Myostatin on the Growth and Development of Skeletal Muscle. Front. Cell. Dev. Biol. 2021, 9, 785712. [Google Scholar] [CrossRef]
- Morelos, R.M.; Ramírez, J.L.; García-Gasca, A.; Ibarra, A.M. Expression of the myostatin gene in the adductor muscle of the Pacific lion-paw scallop Nodipecten subnodosus in association with growth and environmental conditions. J. Exp. Zool. A Ecol. Genet. Physiol. 2015, 323, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Meng, X.Y.; Song, B.; Qiu, X.M.; Liu, H.Y. SNPs in the myostatin gene of the mollusk Chlamys farreri: Association with growth traits. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 155, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.Z.; Yu, F.; Wu, Y.Y.; Zhang, Y.F.; Lu, C.K.; Wang, Y.; Huang, Z.K.; Lu, Y.S.; Chen, N.; Luo, X.; et al. Identification of growth-related SNPs and genes in the genome of the Pacific abalone (Haliotis discus hannai) using GWAS. Aquaculture 2021, 541, 736820. [Google Scholar] [CrossRef]
- Huang, J.F.; Luo, X.; Huang, M.Q.; Liu, G.M.; You, W.W.; Ke, C.H. Identification and characteristics of muscle growth-related microRNA in the Pacific abalone, Haliotis discus hannai. BMC Genom. 2018, 19, 915. [Google Scholar] [CrossRef]
- Mc Pherron, A.C.; Lawler, A.M.; Lee, S. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.M.; Li, Y.J.; Xu, Y.F.; Wang, L.; Ma, X.; Dong, C.J.; Zhao, X.L.; Tian, X.; Li, X.J.; Kong, X.H. The roles of two myostatins and immune effects after inhibition in Qi river crucian carp (Carassius auratus). Fish. Shellfish Immunol. 2020, 98, 710–719. [Google Scholar] [CrossRef]
- Amali, A.A.; Lin, C.; Chen, Y.H.; Wang, W.L.; Gong, H.Y.; Yueh Lee, C.; Lin Ko, Y.; Khan Lu, J.; Her, G.M.; Chen, T.T.; et al. Up-regulation of muscle-specific transcription factors during embryonic somitogenesis of zebrafish (Danio rerio) by knock-down of myostatin-1. Dev. Dyn. 2004, 229, 847–856. [Google Scholar] [CrossRef]
- Garikipati, D.K.; Gahr, S.A.; Rodgers, B.D. Identification, characterization, and quantitative expression analysis of rainbow trout myostatin-1a and myostatin-1b genes. J. Endocrinol. 2006, 190, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.G.; Tan, X.G.; Zhang, P.J.; Ma, J.K.; Zhang, Y.Q.; Xu, P.; Xu, Y.L. Characterization of amphioxus GDF8/11 gene, an archetype of vertebrate MSTN and GDF11. Dev. Genes. Evol. 2007, 217, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Huynh, T.V.; Lee, S.J. Paracrine and endocrine modes of myostatin action. J. Appl. Physiol. 2016, 120, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Czaja, W.; Nakamura, Y.K.; Li, N.; Eldridge, J.A.; DeAvila, D.M.; Thompson, T.B.; Rodgers, B.D. Myostatin regulates pituitary development and hepatic IGF1. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E1036–E1049. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Hu, L.; Liu, M.; Shi, X.H.; Zheng, Y. Detection of the MSTN gene expression in Hailan chicken. Shanghai J. Anim. Husb. Vet. Med. 2005, 2, 25–26. [Google Scholar]
- Li, S.L.; Zhou, Z.C.; Dong, Y.; Sun, H.J.; Gao, S.; Chen, Z.; Yang, A.F.; Liu, W.D.; Wang, Q.Z. Molecular characterization, expression analysis of the myostatin gene and its association with growth traits in sea cucumber (Apostichopus japonicus). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2016, 201, 12–20. [Google Scholar] [CrossRef]
- Fu, Q.; Guo, H.H.; Feng, L.Y.; Li, X.; Zhang, L.L.; Wang, S.; Hu, X.L.; Bao, Z.M. Association of myostatin variants with growth traits of Zhikong Scallop (Chlamys farreri). J. Ocean. Univ. China 2016, 15, 145–151. [Google Scholar] [CrossRef]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef]
- Hu, S.W.; Ni, W.; Sai, W.; Qiao, J.; Wang, P.Y.; Sheng, J.L.; Chen, C.F. Knockdown of myostatin expression by RNAi enhances muscle growth in transgenic sheep. PLoS ONE 2013, 8, e58521. [Google Scholar] [CrossRef]
- Acosta, J.; Carpio, Y.; Borroto, I.; González, O.; Estrada, M.P. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J. Biotechnol. 2005, 119, 324–331. [Google Scholar] [CrossRef]
- Zhuo, R.Q.; Zhou, T.T.; Yang, S.P.; Chan, S.F. Characterization of a molt-related myostatin gene (FmMstn) from the banana shrimp Fenneropenaeus merguiensis. Gen. Comp. Endocrinol. 2017, 248, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Yan, H.S.; Huang, R.; Tsai, H.J. Transfer of a foreign gene to Japanese abalone (Haliotis diversicolor supertexta) by direct testis-injection. Aquaculture 2006, 253, 249–258. [Google Scholar] [CrossRef]
- Wrana, L.; Attisano, L.; Wieser, R.; Ventura, F.; Massagué, J. Mechanism of activation of the TGF-beta receptor. Nature 1994, 370, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Walsh, F.S.; Celeste, A.J. Myostatin: A modulator of skeletal-muscle stem cells. Biochem. Soc. Trans. 2005, 33, 1513–1517. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, X.Q.; Wei, Z.Y.; Yang, M.M.; Zhou, X.Y.; Lei, J.R.; Bai, C.L.; Su, G.H.; Liu, X.F.; Yang, L.; et al. Myostatin Deficiency Enhances Antioxidant Capacity of Bovine Muscle via the SMAD-AMPK-G6PD Pathway. Oxid. Med. Cell. Longev. 2022, 2022, 3497644. [Google Scholar] [CrossRef] [PubMed]
- Sartori, R.; Milan, G.; Patron, M.; Mammucari, C.; Blaauw, B.; Abraham, R.; Sandri, M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell. Physiol. 2009, 296, C1248–C1257. [Google Scholar] [CrossRef]
- Trendelenburg, A.U.; Meyer, A.; Rohner, D.; Boyle, J.; Hatakeyama, S.; Glass, D.J. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Cell. Physiol. 2009, 296, C1258–C1270. [Google Scholar] [CrossRef]

| Primer | Sequence (5′-3′) |
|---|---|
| hdh-myostatin-F | AGTGTATTGGCAAGTCGTGA |
| hdh-myostatin-R | CAACGGCAGTATAGTAGGTCAA |
| hdh-myostatin-dsF | TAATACGACTCACTATAGGGGCCGGTCCTCATCGTTCAC |
| hdh-myostatin-dsR | TAATACGACTCACTATAGGGTTACAAGCACCCACATTCTTCCAC |
| EGFP-dsF | TAATACGACTCACTATAGGGGTGCCCATCCTGGTCGAGCT |
| EGFP-dsR | TAATACGACTCACTATAGGGTGCACGCTGCCGTCCTCGAT |
| hdh-myostatin-qF | TGAGTCGGGAGATTCTTCGC |
| hdh-myostatin-qR | TGATGATGTCGGTTGTCGTG |
| hdh-TβR I-qF | ACCATCACACCATGACACAG |
| hdh-TβR I-qR | GCCACACCTCACCGTACCTC |
| hdh-ActR IIB-qF | GCTGGTAATGAAGGGCTG |
| hdh-ActR IIB-qR | AGTCGTGATGGGAAGTTG |
| hdh-Smad3-qF | GTTTGCCGAGTGTCTCAGTG |
| hdh-Smad3-qR | CCCTGGTGGTATCTTGCAGA |
| MHC-qF | GACCCCAACGACCCTGATAT |
| MHC-qR | TCTTCTCCCTTGGTGCTCTG |
| β-actin-qF | GGTATCCTCACCCTCAAGT |
| β-actin-qR | GGGTCATCTTTTCACGGTTG |
| 18S rRNA-qF | TTCCCAGTAAGCGTCAGTCATC |
| 18S rRNA-qR | CGAGGGTCTCACTAAACCATTC |
| Locus | Genotype | Sample Number | Shell Length (mm) | Shell Width (mm) | Total Weight (g) | Muscle Weight (g) |
|---|---|---|---|---|---|---|
| C-6G | CC | 102 | 74.37 ± 9.68 a | 49.85 ± 6.40 a | 42.54 ± 16.13 a | 17.58 ± 7.58 a |
| GC | 83 | 75.52 ± 9.53 a | 50.66 ± 5.90 a | 43.82 ± 16.07 a | 17.96 ± 7.86 a | |
| GG | 32 | 68.34 ± 7.84 b | 47.15 ± 5.31 b | 33.48 ± 10.20 b | 13.59 ± 5.05 b | |
| T-115C | TT | 102 | 74.38 ± 9.66 a | 49.93 ± 6.38 ab | 42.52 ± 16.06 a | 17.44 ± 7.60 a |
| CT | 82 | 75.16 ± 9.69 ab | 50.45 ± 6.11 a | 43.56 ± 16.42 a | 18.03 ± 7.97 a | |
| CC | 34 | 69.72 ± 8.42 c | 47.78 ± 5.39 b | 35.03 ± 11.39 b | 14.09 ± 5.39 b | |
| A-117G | AA | 102 | 74.55 ± 9.65 a | 50.02 ± 6.36 ab | 42.89 ± 16.00 a | 17.65 ± 7.60 a |
| GA | 81 | 75.07 ± 9.71 a | 50.41 ± 6.13 a | 43.36 ± 16.42 a | 17.91 ± 7.94 a | |
| GG | 34 | 69.72 ± 8.42 b | 47.78 ± 5.39 b | 35.03 ± 11.39 b | 14.09 ± 5.39 b | |
| C-282T | CC | 128 | 74.18 ± 9.85 a | 49.97 ± 6.52 a | 42.55 ± 16.47 a | 17.45 ± 7.69 a |
| TC | 71 | 74.16 ± 9.51 a | 49.88 ± 5.65 a | 41.98 ± 15.28 ab | 17.28 ± 7.62 a | |
| TT | 20 | 69.95 ± 8.00 a | 47.12 ± 5.29 a | 34.39 ± 11.65 b | 14.10 ± 5.78 a | |
| G-288A | GG | 187 | 74.56 ± 9.15 a | 49.92 ± 5.97 a | 42.40 ± 15.41 a | 17.43 ± 7.41 a |
| AG | 29 | 69.49 ± 11.35 b | 48.48 ± 7.29 a | 37.37 ± 18.03 a | 15.44 ± 8.29 a | |
| C-414A | CC | 101 | 74.95 ± 9.63 a | 50.36 ± 6.36 a | 43.53 ± 16.01 a | 17.84 ± 7.56 a |
| AC | 82 | 74.03 ± 9.56 ab | 49.59 ± 5.93 ab | 41.38 ± 15.84 ab | 17.01 ± 7.72 ab | |
| AA | 35 | 69.67 ± 8.97 b | 47.64 ± 5.79 b | 35.26 ± 13.00 b | 14.51 ± 6.32 b | |
| T-437C | TT | 118 | 74.01 ± 10.14 a | 49.83 ± 6.58 a | 42.25 ± 16.67 a | 17.37 ± 7.78 a |
| CT | 80 | 74.41 ± 9.45 a | 49.90 ± 5.82 a | 41.86 ± 15.01 a | 17.16 ± 7.44 ab | |
| CC | 19 | 69.13 ± 5.67 b | 47.10 ± 4.37 a | 33.28 ± 9.26 b | 13.47 ± 4.57 b | |
| G-897A | GG | 179 | 73.09 ± 9.68 a | 49.28 ± 6.16 a | 40.53 ± 15.75 a | 16.71 ± 7.61 a |
| AG | 37 | 76.87 ± 8.81 b | 51.41 ± 5.88 a | 46.45 ± 15.92 b | 18.79 ± 7.31 a | |
| G-1278A | GG | 143 | 74.50 ± 9.57 a | 50.09 ± 5.98 a | 42.15 ± 15.16 a | 17.20 ± 7.15 a |
| AG | 69 | 73.52 ± 9.50 ab | 49.52 ± 6.51 a | 41.70 ± 16.99 a | 17.43 ± 8.30 a | |
| AA | 8 | 65.00 ± 8.68 b | 44.86 ± 5.58 b | 31.15 ± 14.02 a | 12.28 ± 6.34 a |
| Indicator | EGFP Control Group (N = 39) | hdh-Myostatin Experimental Group (N = 38) |
|---|---|---|
| Initial shell length (mm) | 35.48 ± 1.08 a | 35.19 ± 1.10 a |
| Initial shell width (mm) | 23.45 ± 0.92 a | 23.51 ± 0.78 a |
| Initial total weight (g) | 5.76 ± 0.48 a | 5.69 ± 0.51 a |
| Final shell length (mm) | 37.32 ± 1.17 a | 37.45 ± 1.48 a |
| Final shell width (mm) | 24.83 ± 0.81 a | 25.25 ± 0.92 a |
| Final total weight (g) | 6.57 ± 0.59 a | 6.69 ± 0.52 a |
| increment of shell length (mm) | 1.87 ± 0.46 a | 2.23 ± 0.66 b |
| increment of shell width (mm) | 1.51 ± 0.39 a | 1.75 ± 0.48 a |
| increment of total weight (g) | 0.81 ± 0.31 a | 1.08 ± 0.40 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhou, M.; Chen, J.; Ke, C. A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai. Biology 2023, 12, 14. https://doi.org/10.3390/biology12010014
Huang J, Zhou M, Chen J, Ke C. A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai. Biology. 2023; 12(1):14. https://doi.org/10.3390/biology12010014
Chicago/Turabian StyleHuang, Jianfang, Mingcan Zhou, Jianming Chen, and Caihuan Ke. 2023. "A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai" Biology 12, no. 1: 14. https://doi.org/10.3390/biology12010014
APA StyleHuang, J., Zhou, M., Chen, J., & Ke, C. (2023). A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai. Biology, 12(1), 14. https://doi.org/10.3390/biology12010014





