Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. Morphologic Evaluation
2.3. Cytogenetic Analysis
2.4. Next-generation Sequencing
3. Results
3.1. Concurrent SF3B1 and PHF6 Mutations Occur at Low Frequency in Myeloid Neoplasms
3.2. Concurrent SF3B1 and PHF6 Mutations Occur in a Variety of Myeloid Neoplasms
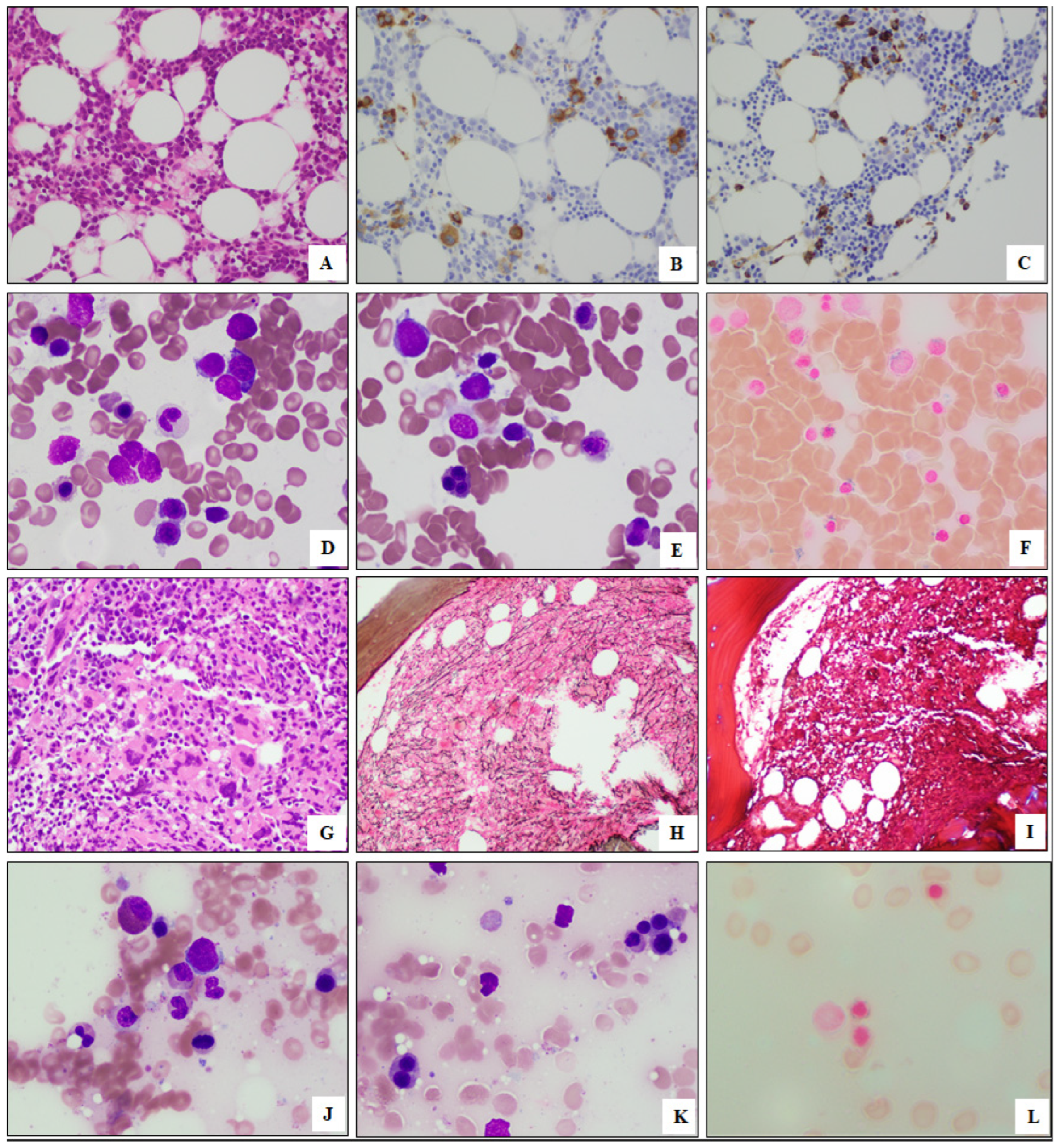
3.3. Morphologic Findings of Myeloid Neoplasms with SF3B1 and PHF6 Mutations
3.4. Cytogenetic Findings of Myeloid Neoplasms with SF3B1 and PHF6 Mutations
3.5. Molecular Findings of Myeloid Neoplasms with SF3B1 and PHF6 Mutations
3.6. Clinical Findings of Myeloidoutcomes of Myeloid Neoplasms with SF3B1 and PHF6 Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McClure, R.F.; Ewalt, M.D.; Crow, J.; Temple-Smolkin, R.L.; Pullambhatla, M.; Sargent, R.; Kim, A.S. Clinical Significance of DNA Variants in Chronic Myeloid Neoplasms: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 717–737. [Google Scholar] [CrossRef] [PubMed]
- Schwind, S.; Jentzsch, M.; Kubasch, A.S.; Metzeler, K.H.; Platzbecker, U. Myelodysplastic syndromes: Biological and therapeutic consequences of the evolving molecular aberrations landscape. Neoplasia 2021, 23, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, C.; Zhang, L.; Schaar, D.G. Molecular Mutations and Their Cooccurrences in Cytogenetically Normal Acute Myeloid Leukemia. Stem Cells Int. 2017, 2017, 6962379. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Venable, E.R.; Chen, D.; Chen, C.P.; Bessonen, K.R.; Nguyen, P.L.; Oliveira, J.L.; Reichard, K.K.; Hoyer, J.D.; Althoff, S.D.; Roh, D.J.; et al. Pathologic Spectrum and Molecular Landscape of Myeloid Disorders Harboring SF3B1 Mutations. Am. J. Clin. Pathol. 2021, 156, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Nagata, Y.; Makishima, H.; Sanada, M.; Shiozawa, Y.; Kon, A.; Yoshizato, T.; Sato-Otsubo, A.; Kataoka, K.; Shiraishi, Y.; et al. Somatic PHF6 mutations in 1760 cases with various myeloid neoplasms. Leukemia 2016, 30, 2270–2273. [Google Scholar] [CrossRef]
- Kurzer, J.H.; Weinberg, O.K. PHF6 Mutations in Hematologic Malignancies. Front. Oncol. 2021, 11, 704471. [Google Scholar] [CrossRef]
- Patel, K.P.; Ruiz-Cordero, R.; Chen, W.; Routbort, M.J.; Floyd, K.; Rodriguez, S.; Galbincea, J.; Barkoh, B.A.; Hatfield, D.; Khogeer, H.; et al. Ultra-Rapid Reporting of GENomic Targets (URGENTseq): Clinical Next-Generation Sequencing Results within 48 hours of Sample Collection. J. Mol. Diagn. 2019, 21, 89–98. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627, quiz 3699. [Google Scholar] [CrossRef] [PubMed]
- Loghavi, S.; Zuo, Z.; Ravandi, F.; Kantarjian, H.M.; Bueso-Ramos, C.; Zhang, L.; Singh, R.R.; Patel, K.P.; Medeiros, L.J.; Stingo, F.; et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J. Hematol. Oncol. 2014, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Jimma, T.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. SF3B1 mutations in primary myelofibrosis: Clinical, histopathology and genetic correlates among 155 patients. Leukemia 2012, 26, 1135–1137. [Google Scholar] [CrossRef]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1-mutant MDS as a distinct disease subtype: A proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef]
- Zhao, L.P.; Daltro de Oliveira, R.; Marcault, C.; Soret, J.; Gauthier, N.; Verger, E.; Cassinat, B.; Kiladjian, J.; Benajiba, L. SF3B1 mutations in the driver clone increase the risk of evolution to myelofibrosis in patients with myeloproliferative neopalsms (MPN). Blood 2020, 136, 1. [Google Scholar]
- Boiocchi, L.; Hasserjian, R.P.; Pozdnyakova, O.; Wong, W.J.; Lennerz, J.K.; Le, L.P.; Dias-Santagata, D.; Iafrate, A.J.; Hobbs, G.S.; Nardi, V. Clinicopathological and molecular features of SF3B1-mutated myeloproliferative neoplasms. Hum. Pathol. 2019, 86, 1–11. [Google Scholar] [CrossRef]
- Palomo, L.; Meggendorfer, M.; Hutter, S.; Twardziok, S.; Adema, V.; Fuhrmann, I.; Fuster-Tormo, F.; Xicoy, B.; Zamora, L.; Acha, P.; et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood 2020, 136, 1851–1862. [Google Scholar] [CrossRef]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood 2011, 118, 6904–6908. [Google Scholar] [CrossRef]
- Dalton, W.B.; Helmenstine, E.; Pieterse, L.; Li, B.; Gocke, C.D.; Donaldson, J.; Xiao, Z.; Gondek, L.P.; Ghiaur, G.; Gojo, I.; et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 2020, 4, 1192–1196. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.P.; Gonen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- Chien, K.S.; Kanagal-Shamanna, R.; Naqvi, K.; Sasaki, K.; Alvarado, Y.; Takahashi, K.; Borthakur, G.M.; Jabbour, E.; Routbor, M.; Pierce, S.A.; et al. The impact of PHF6 mutations in myelodysplastic syndromes, chronic myelomonocytic leukemia, and acute myeloid leukemia. Blood 2019, 134, 1436–1439. [Google Scholar] [CrossRef]


| Case | Age | Sex | WBC | Hb | Platelet | LDH | Karyotype | SF3B1 | PHF6 | SCT | FU (m) | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDS | ||||||||||||
| 1 | 69 | M | 8.2 | 10.2 | 234 | NA | inv(17)(p13q23) [20/20] | K666M | K21fs, R116*, F198fs, C242Y, G291E, C297R, C297Y | No | 39 | Died |
| 2 | 51 | M | 8.9 | 12.1 | 116 | 204 | del(11q)[9/20] | R625C, D781G | C305S, L324fs | Yes | 14 | CR |
| 3 | 56 | M | 7.0 | 9.8 | 510 | 207 | 46,XY[20] | E783K | C212G | Yes | 155 | Died |
| 4 | 77 | M | 2.0 | 8.6 | 244 | 130 | 46,XY[20] | S779P | C280fs | No | 38 | MDS |
| 5 | 77 | M | 6.3 | 10.2 | 153 | 124 | del(13)(q12q14) [3/19] | K666T | Q7* | No | 65 | Died |
| 6 | 84 | M | 11.1 | 7.4 | 88 | 568 | 46,XY[20] | K666N | R319* | No | 3 | Died |
| 7 | 74 | M | 1.7 | 9.8 | 41 | 159 | add(1)(p36.1) [2/20] | R625C | H229fs, C283* | Yes | 39 | CR |
| 8 | 62 | M | 7.2 | 9.9 | 336 | 441 | 46,XY[20] | K700E | R225* | Yes | 71 | Died |
| 9 | 68 | M | 3.1 | 7.6 | 63 | 177 | 46,XY[20] | K700E | I290T | No | 28 | Died |
| AML | ||||||||||||
| 10 | 70 | F | 6.9 | 6.9 | 5 | 135 | 46,XX[20] | K700E | c.585+1G>A p.? | No | 11 | Died |
| 11 | 82 | M | 2.5 | 9.1 | 53 | 130 | 46,XY[20] | I704N | H302R | No | 90 | Died |
| 12 | 82 | M | 1.2 | 11.3 | 111 | 232 | –Y,+1, der(1;13)(q10;q10) [4/11] | K666N | R274Q | No | 51 | Died |
| 13 | 75 | M | 1.0 | 8.0 | 52 | 189 | 46,XY[20] | K700E | K130fs | No | 13 | Died |
| 14 | 58 | F | 5.8 | 8.9 | 199 | 126 | 46,XX[20] | K700E | H239R | No | 9 | CR |
| MPN | ||||||||||||
| 15 | 67 | F | 2.9 | 8.6 | 39 | 247 | del(20)(q11.2q13.3) [20] | K666N | H239Y | No | 57 | Died |
| 16 | 59 | M | 7.1 | 9.0 | 56 | 375 | del(20)(q11.2q13.3) [20] | K666T | I314T | No | 37 | Died |
| 17 | 76 | M | 3.6 | 9.3 | 196 | 266 | 46,XY[20] | K666Q | R274Q | No | 41 | Died |
| 18 | 58 | M | NA | NA | NA | NA | NA | K700E | E27fs, R225*, Y240C | No | 72 | Died |
| MDS/MPN | ||||||||||||
| 19 | 73 | F | 11.5 | 7.2 | 25 | 987 | 46,XX[20] | G740E | C215S | No | 16 | Died |
| 20 | 69 | F | 17.2 | 5.2 | 595 | 666 | 46,XX[20] | K700E | F263* | No | 87 | Died |
| 21 | 66 | F | 60.4 | 9.1 | 26 | 570 | del(20)(q11.2q13.1) [6/20] | K700E | I314T | No | 20 | Died |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Z.; Medeiros, L.J.; Garces, S.; Routbort, M.J.; Ok, C.Y.; Loghavi, S.; Kanagal-Shamanna, R.; Jelloul, F.Z.; Garcia-Manero, G.; Chien, K.S.; et al. Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms. Biology 2023, 12, 13. https://doi.org/10.3390/biology12010013
Zuo Z, Medeiros LJ, Garces S, Routbort MJ, Ok CY, Loghavi S, Kanagal-Shamanna R, Jelloul FZ, Garcia-Manero G, Chien KS, et al. Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms. Biology. 2023; 12(1):13. https://doi.org/10.3390/biology12010013
Chicago/Turabian StyleZuo, Zhuang, L. Jeffrey Medeiros, Sofia Garces, Mark J. Routbort, Chi Young Ok, Sanam Loghavi, Rashmi Kanagal-Shamanna, Fatima Zahra Jelloul, Guillermo Garcia-Manero, Kelly S. Chien, and et al. 2023. "Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms" Biology 12, no. 1: 13. https://doi.org/10.3390/biology12010013
APA StyleZuo, Z., Medeiros, L. J., Garces, S., Routbort, M. J., Ok, C. Y., Loghavi, S., Kanagal-Shamanna, R., Jelloul, F. Z., Garcia-Manero, G., Chien, K. S., Patel, K. P., Luthra, R., & Yin, C. C. (2023). Concurrent Mutations in SF3B1 and PHF6 in Myeloid Neoplasms. Biology, 12(1), 13. https://doi.org/10.3390/biology12010013







