Optimization of Transcription Factor Genetic Circuits
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Number of Parameters
2.2. Biological Bounds on Parameters
2.3. Optimization of Bounded Parameters
2.4. Initial Parameters and Hyperparameters
2.5. Stochastic Fluctuations Vary with Abundance
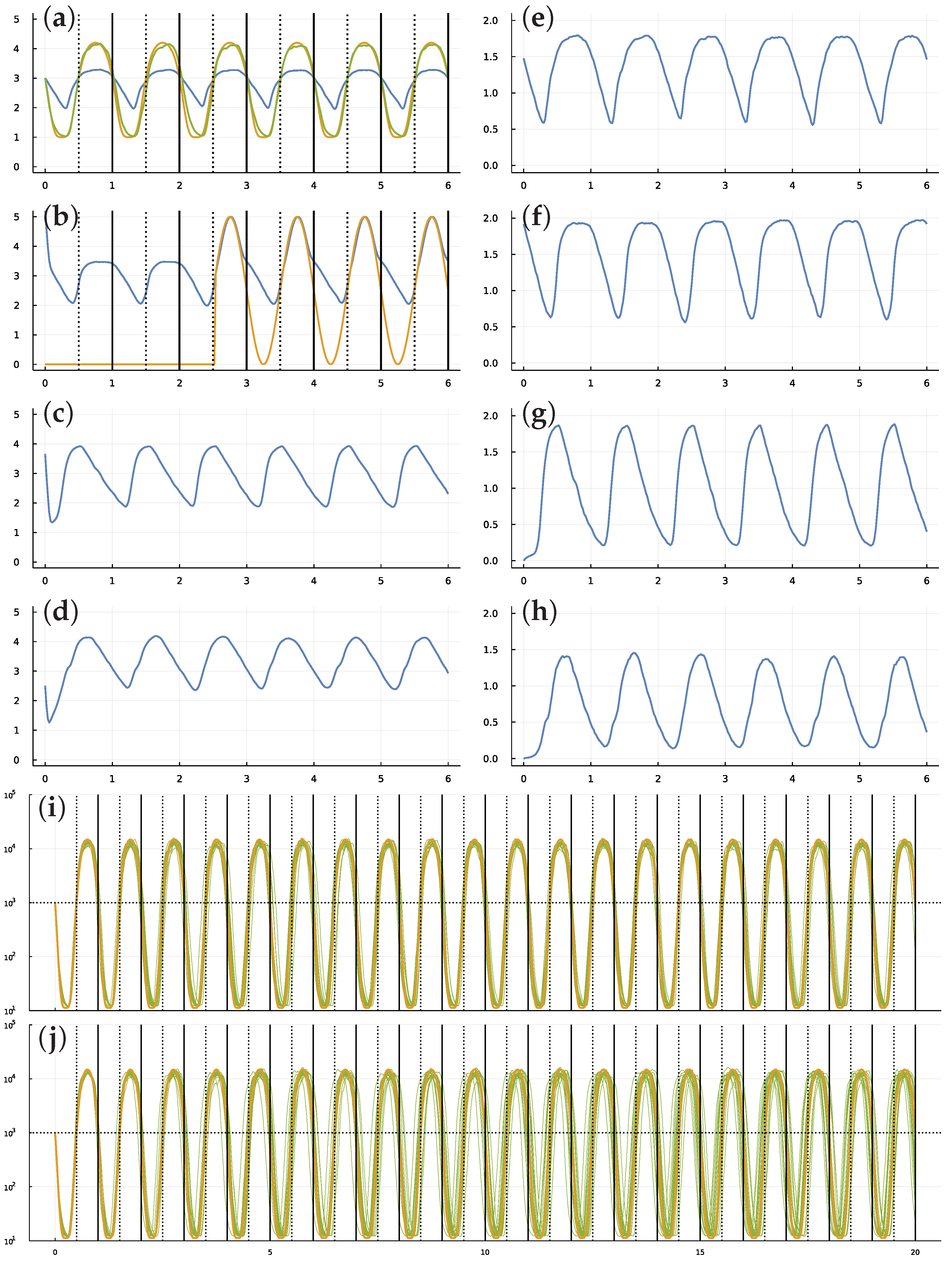
3. Results
3.1. Dynamics of TF Networks
3.2. TF Network as Input-Output Function
3.3. Maintaining Circadian Rhythm as a Design Challenge
3.4. Stochastic Molecular Dynamics
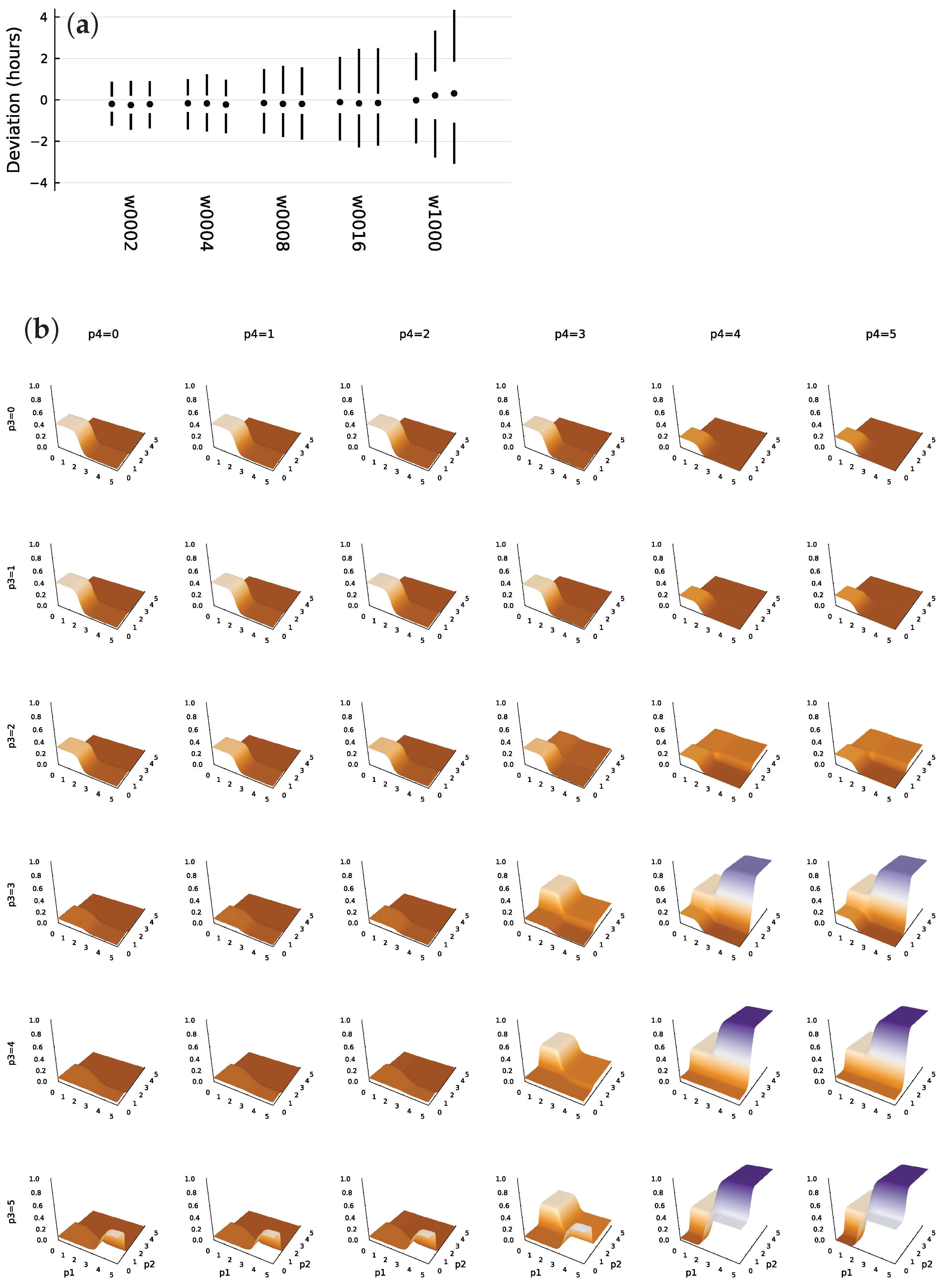
3.5. Random External Light Signal for Entrainment
3.6. Dynamics of an Optimized System
3.7. TF Logic of an Optimized System
4. Discussion
4.1. Optimize a Neural Network and Fit a TF Network
4.2. Large Networks, Flat Fitness Surfaces, and Genetic Variation
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| SciML | scientific machine learning (Julia language packages) |
| TF | transcription factor |
References
- Baydin, A.G.; Pearlmutter, B.A.; Radul, A.A.; Siskind, J.M. Automatic differentiation in machine learning: A survey. J. Mach. Learn. Res. 2018, 18, 1–43. [Google Scholar]
- Margossian, C.C. A review of automatic differentiation and its efficient implementation. WIREs Data Min. Knowl. Discov. 2019, 9, e1305. [Google Scholar] [CrossRef]
- Frank, S.A. Automatic differentiation and the optimization of differential equation models in biology. arXiv 2022, arXiv:2207.04487. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Fröhlich, F.; Kessler, T.; Weindl, D.; Shadrin, A.; Schmiester, L.; Hache, H.; Muradyan, A.; Schütte, M.; Lim, J.H.; Heinig, M.; et al. Efficient parameter estimation enables the prediction of drug response using a mechanistic pan-cancer pathway model. Cell Syst. 2018, 7, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Hiscock, T.W. Adapting machine-learning algorithms to design gene circuits. BMC Bioinform. 2019, 20, 214. [Google Scholar] [CrossRef]
- Liu, Y.; Barr, K.; Reinitz, J. Fully interpretable deep learning model of transcriptional control. Bioinformatics 2020, 36, i499–i507. [Google Scholar] [CrossRef]
- Nilsson, A.; Peters, J.M.; Meimetis, N.; Bryson, B.; Lauffenburger, D.A. Artificial neural networks enable genome-scale simulations of intracellular signaling. Nat. Commun. 2022, 13, 3069. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Collins, J.J. Predictive biology: Modelling, understanding and harnessing microbial complexity. Nat. Rev. Microbiol. 2020, 18, 507–520. [Google Scholar] [CrossRef]
- Rackauckas, C.; Ma, Y.; Martensen, J.; Warner, C.; Zubov, K.; Supekar, R.; Skinner, D.; Ramadhan, A. Universal differential equations for scientific machine learning. arXiv 2020, arXiv:2001.04385. [Google Scholar]
- Bezanson, J.; Edelman, A.; Karpinski, S.; Shah, V.B. Julia: A fresh approach to numerical computing. SIAM Rev. 2017, 59, 65–98. [Google Scholar] [CrossRef]
- Frank, S.A. Optimization of Transcription Factor Genetic Circuits: Julia Software Code. Zenodo 2022. Available online: https://doi.org/10.5281/zenodo.6798420 (accessed on 7 August 2022).
- Milo, R.; Phillips, R. Cell Biology by the Numbers; Garland Science: New York, NY, USA, 2015. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Frank, S.A. Optimizing differential equations to fit data and predict outcomes. arXiv 2022, arXiv:2204.07833. [Google Scholar] [CrossRef]
- Marbach, D.; Prill, R.J.; Schaffter, T.; Mattiussi, C.; Floreano, D.; Stolovitzky, G. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. USA 2010, 107, 6286–6291. [Google Scholar] [CrossRef] [PubMed]
- Bintu, L.; Buchler, N.E.; Garcia, H.G.; Gerland, U.; Hwa, T.; Kondev, J.; Phillips, R. Transcriptional regulation by the numbers: Models. Curr. Opin. Genet. Dev. 2005, 15, 116–124. [Google Scholar] [CrossRef]
- Bintu, L.; Buchler, N.E.; Garcia, H.G.; Gerland, U.; Hwa, T.; Kondev, J.; Kuhlman, T.; Phillips, R. Transcriptional regulation by the numbers: Applications. Curr. Opin. Genet. Dev. 2005, 15, 125–135. [Google Scholar] [CrossRef]
- Stadler, P.F. Fitness landscapes. In Biological Evolution and Statistical Physics; Lässig, M., Valleriani, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 183–204. [Google Scholar] [CrossRef]
- Malan, K.M. A survey of advances in landscape analysis for optimisation. Algorithms 2021, 14, 40. [Google Scholar] [CrossRef]
- De Jong, H.; Gouzé, J.L.; Hernandez, C.; Page, M.; Sari, T.; Geiselmann, J. Qualitative simulation of genetic regulatory networks using piecewise-linear models. Bull. Math. Biol. 2004, 66, 301–340. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Belkin, M.; Uhler, C. Overparameterized neural networks implement associative memory. Proc. Natl. Acad. Sci. USA 2020, 117, 27162–27170. [Google Scholar] [CrossRef]
- Elad, M.; Simon, D.; Aberdam, A. Another step toward demystifying deep neural networks. Proc. Natl. Acad. Sci. USA 2020, 117, 27070–27072. [Google Scholar] [CrossRef]
- Baraniuk, R.; Donoho, D.; Gavish, M. The science of deep learning. Proc. Natl. Acad. Sci. USA 2020, 117, 30029–230032. [Google Scholar] [CrossRef]
- Poggio, T.; Banburski, A.; Liao, Q. Theoretical issues in deep networks. Proc. Natl. Acad. Sci. USA 2020, 117, 30039–30045. [Google Scholar] [CrossRef]
- Ciliberti, S.; Martin, O.C.; Wagner, A. Innovation and robustness in complex regulatory gene networks. Proc. Natl. Acad. Sci. USA 2007, 104, 13591–13596. [Google Scholar] [CrossRef]
- Frank, S.A. Genetic variation in cancer predisposition: Mutational decay of a robust genetic control network. Proc. Natl. Acad. Sci. USA 2004, 101, 8061–8065. [Google Scholar] [CrossRef]
- Frank, S.A. Evolutionary design of regulatory control. II. Robust error-correcting feedback increases genetic and phenotypic variability. J. Theor. Biol. 2019, 468, 72–81. [Google Scholar] [CrossRef]
- Frank, S.A. Robustness increases heritability: Implications for familial disease. bioRxiv 2022. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frank, S.A. Optimization of Transcription Factor Genetic Circuits. Biology 2022, 11, 1294. https://doi.org/10.3390/biology11091294
Frank SA. Optimization of Transcription Factor Genetic Circuits. Biology. 2022; 11(9):1294. https://doi.org/10.3390/biology11091294
Chicago/Turabian StyleFrank, Steven A. 2022. "Optimization of Transcription Factor Genetic Circuits" Biology 11, no. 9: 1294. https://doi.org/10.3390/biology11091294
APA StyleFrank, S. A. (2022). Optimization of Transcription Factor Genetic Circuits. Biology, 11(9), 1294. https://doi.org/10.3390/biology11091294






