Integrating Expression Data-Based Deep Neural Network Models with Biological Networks to Identify Regulatory Modules for Lung Adenocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
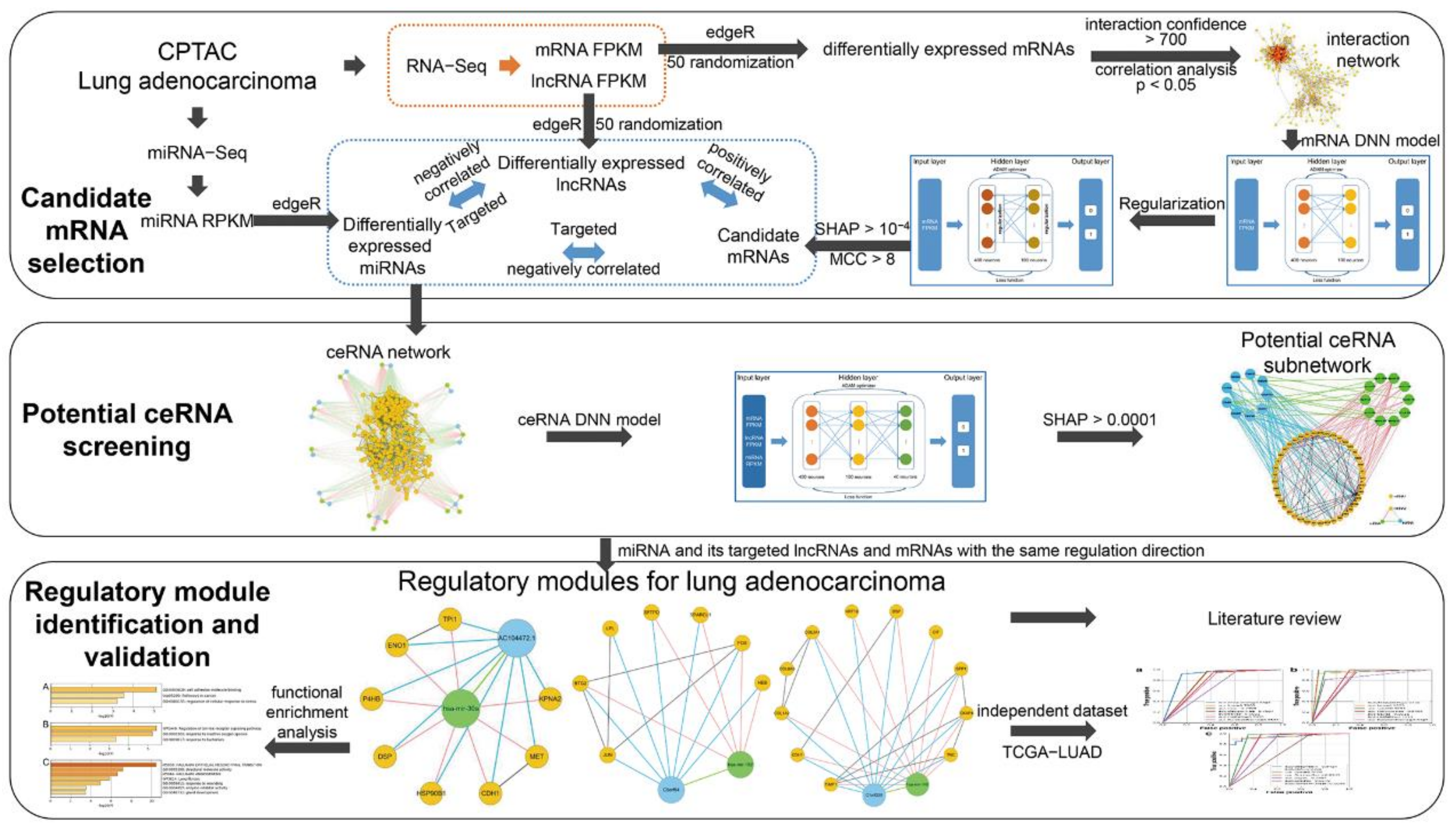
2. Materials and Methods
2.1. Data
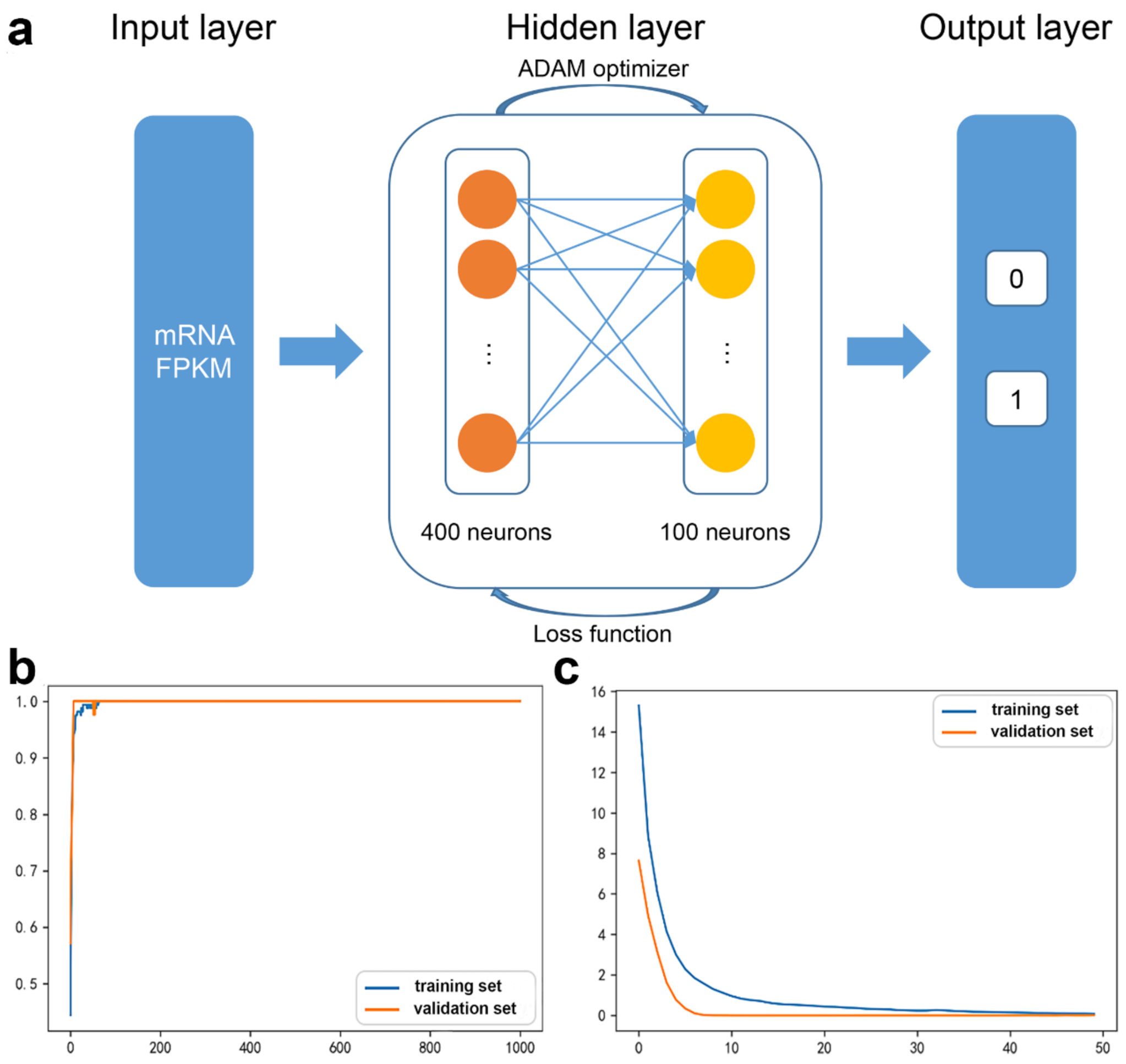
2.2. DNN Model
2.2.1. DNN Model Feature
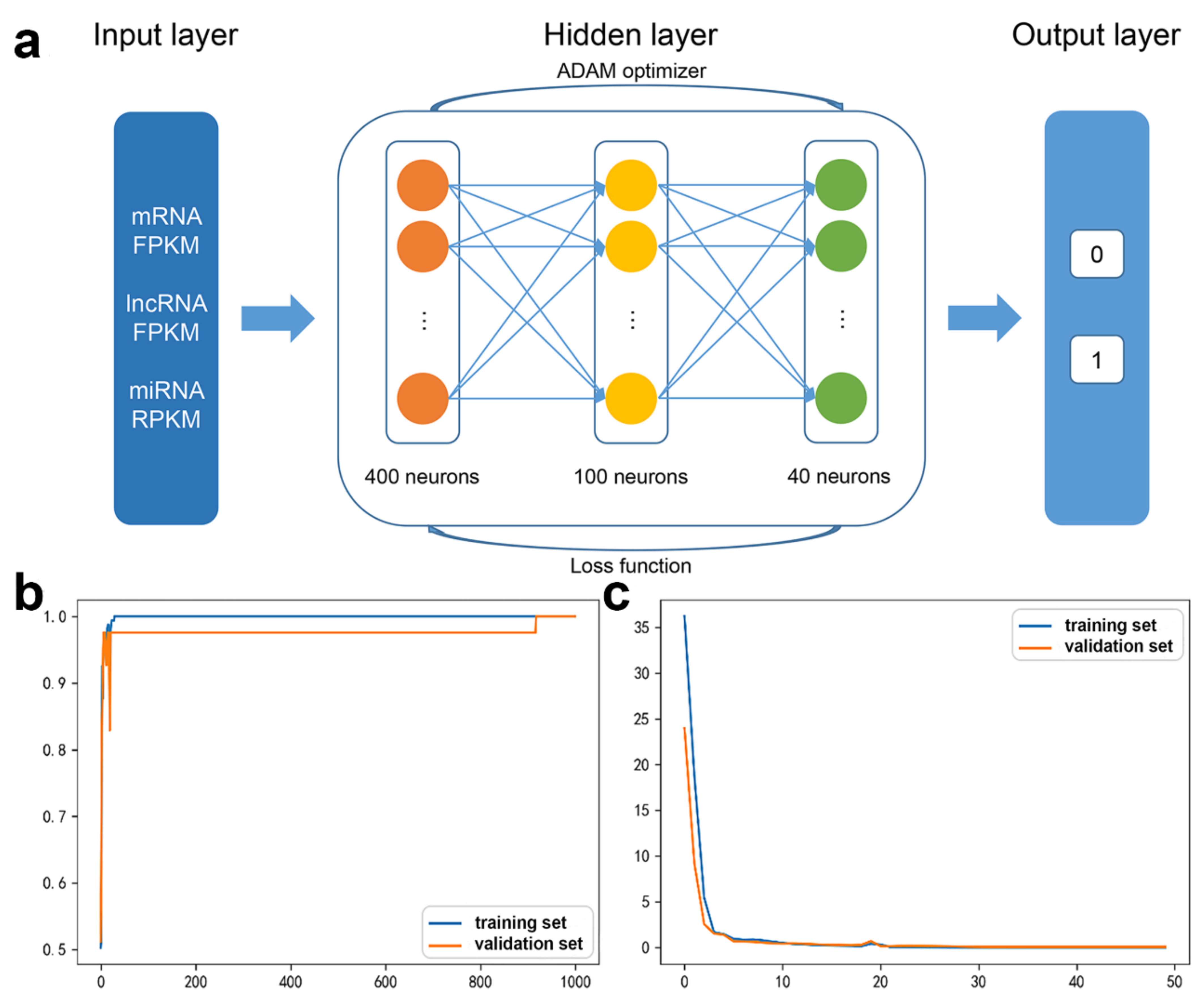
2.2.2. DNN Model Construction
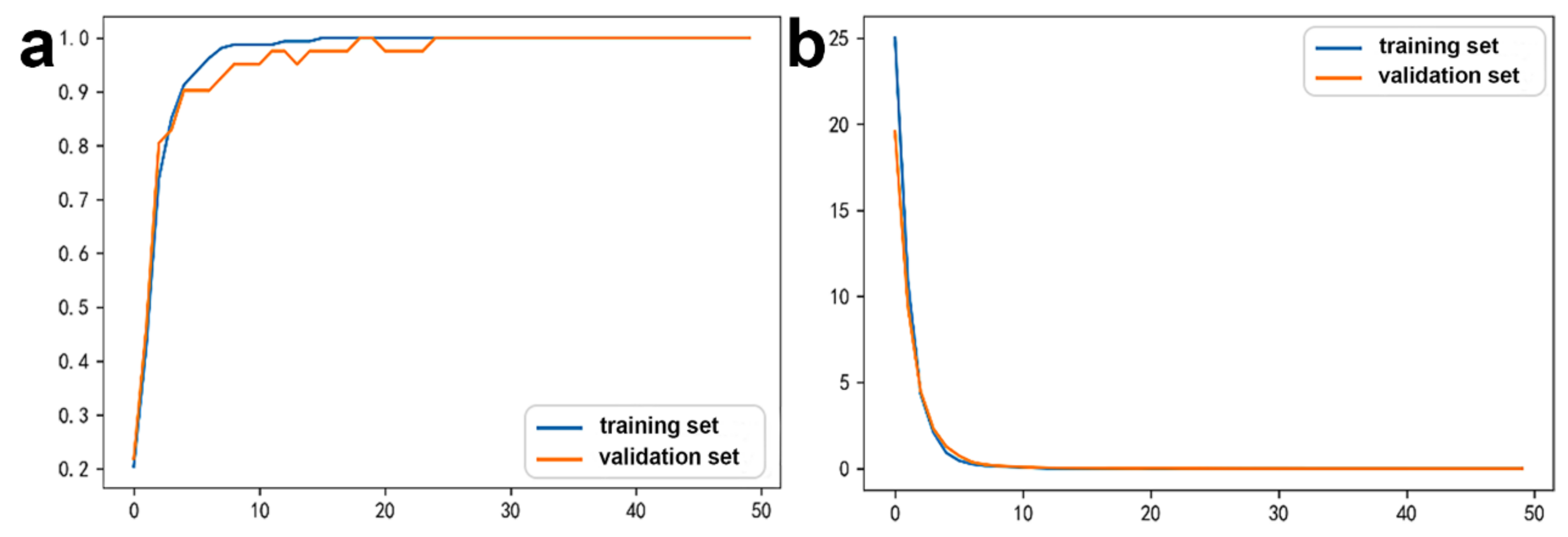
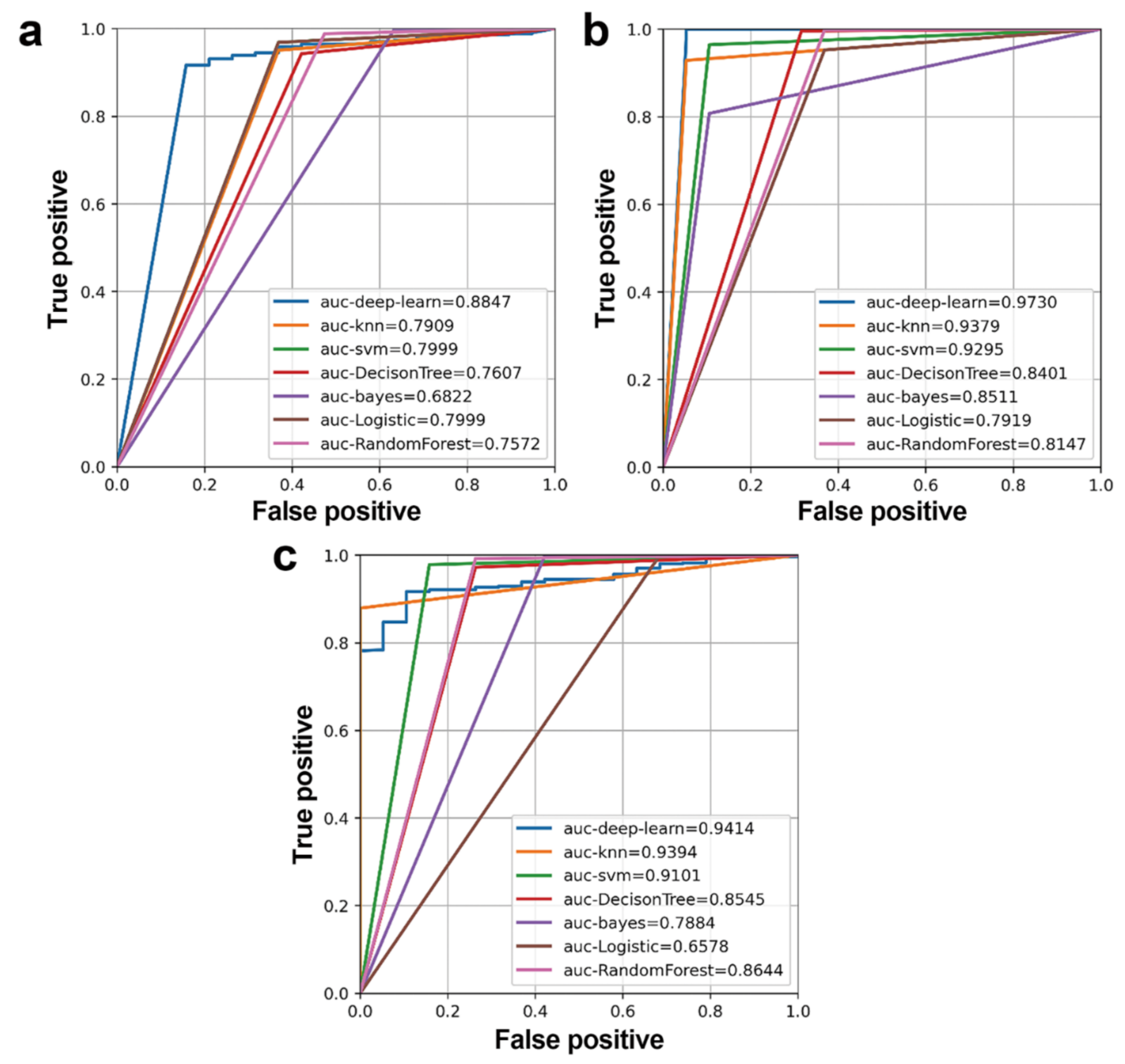
2.2.3. DNN Model Evaluation
2.3. Regulatory Modules for Lung Adenocarcinoma
2.3.1. Candidate mRNA Selection
2.3.2. Potential ceRNA Screening
2.3.3. Regulatory Module Identification and Validation
3. Results
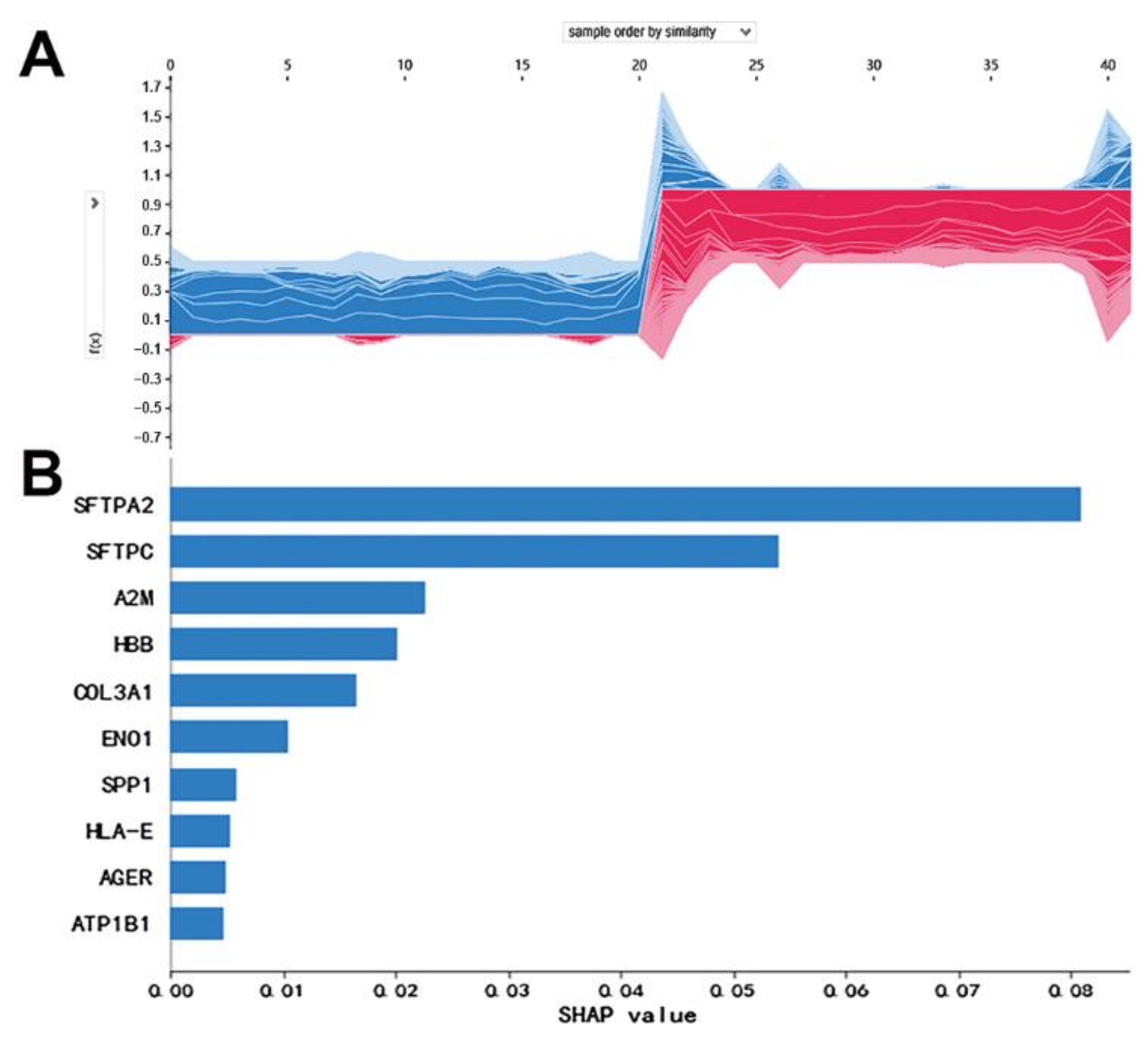
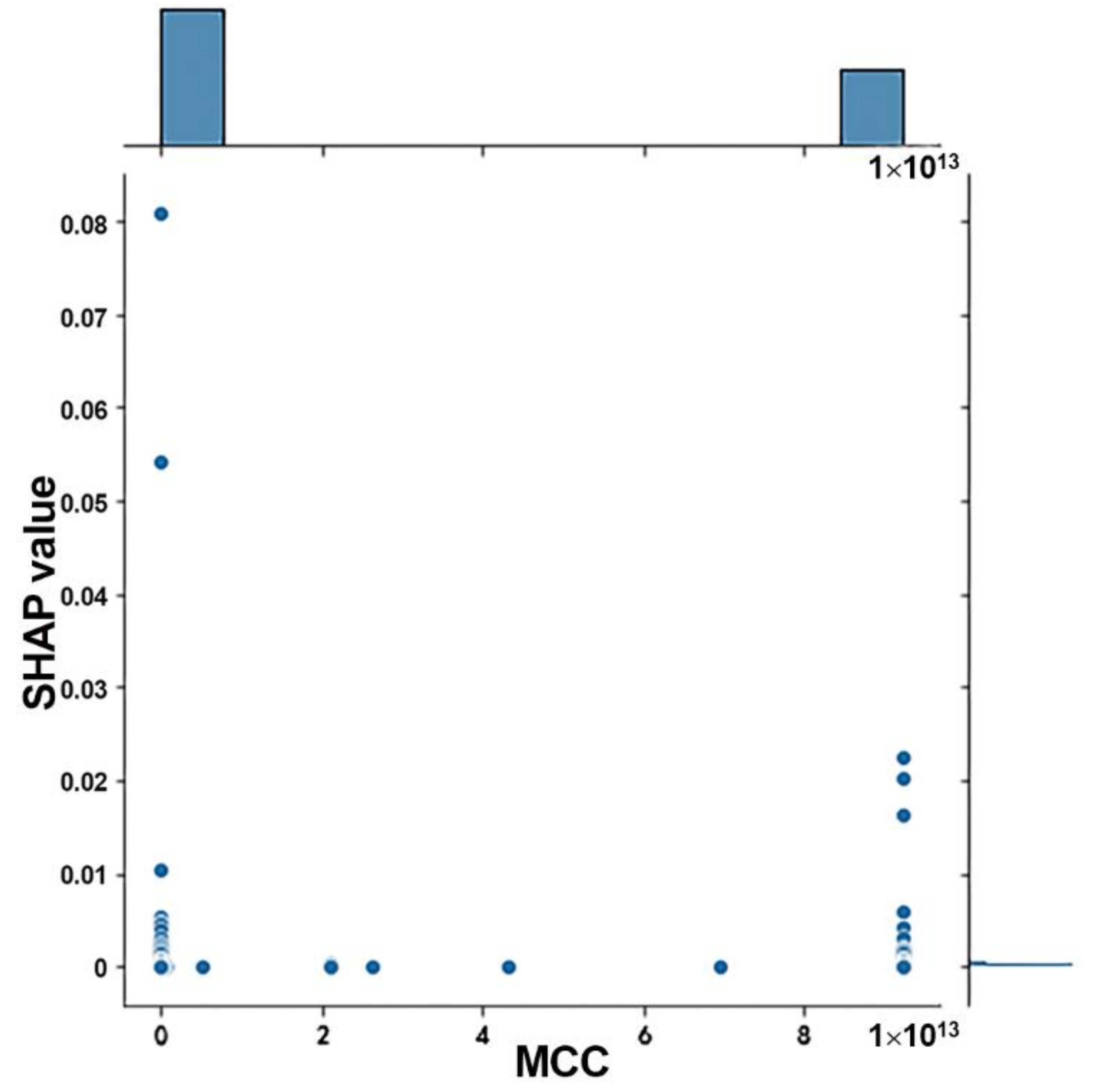
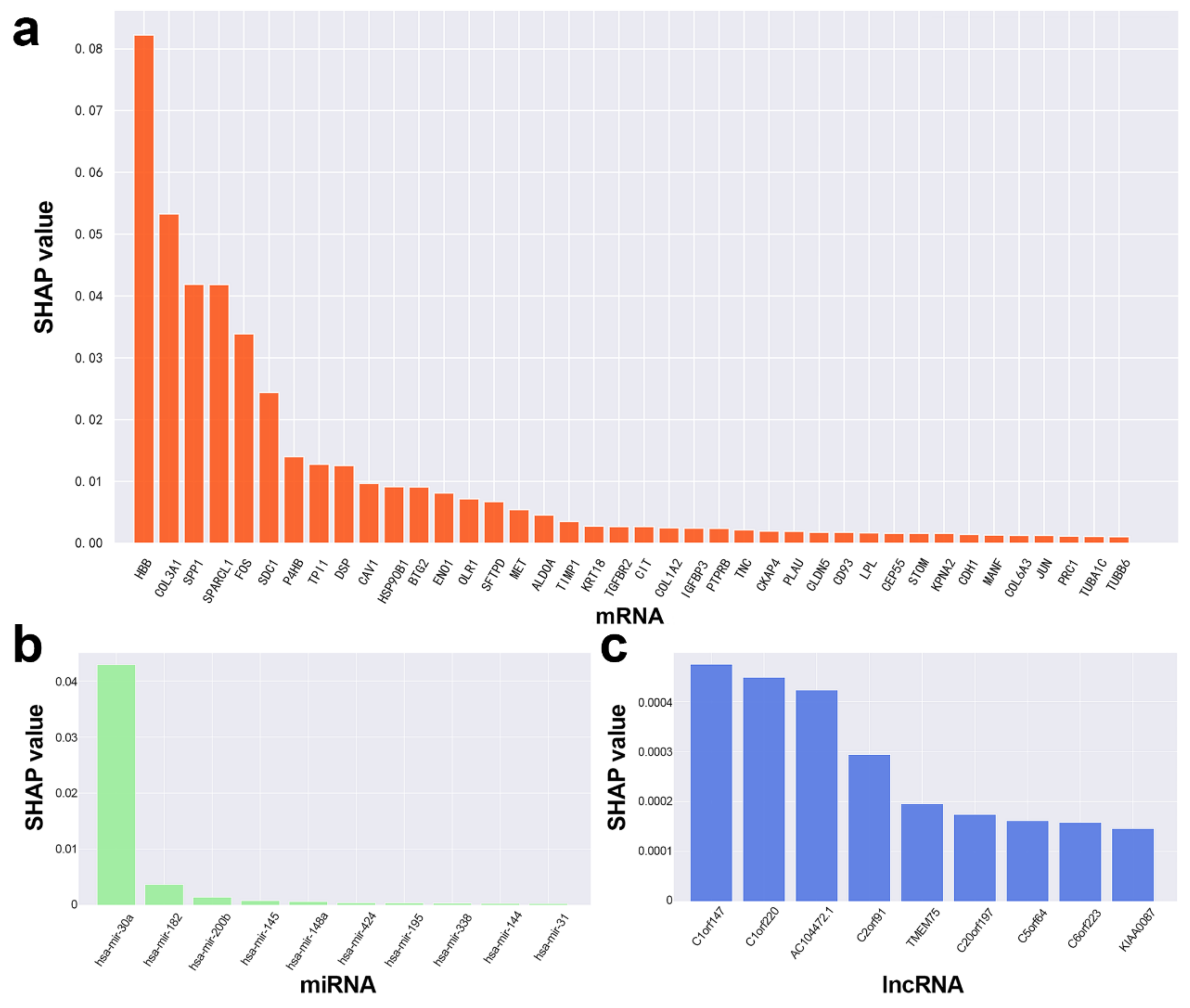
3.1. Candidate mRNAs
3.2. Potential ceRNAs
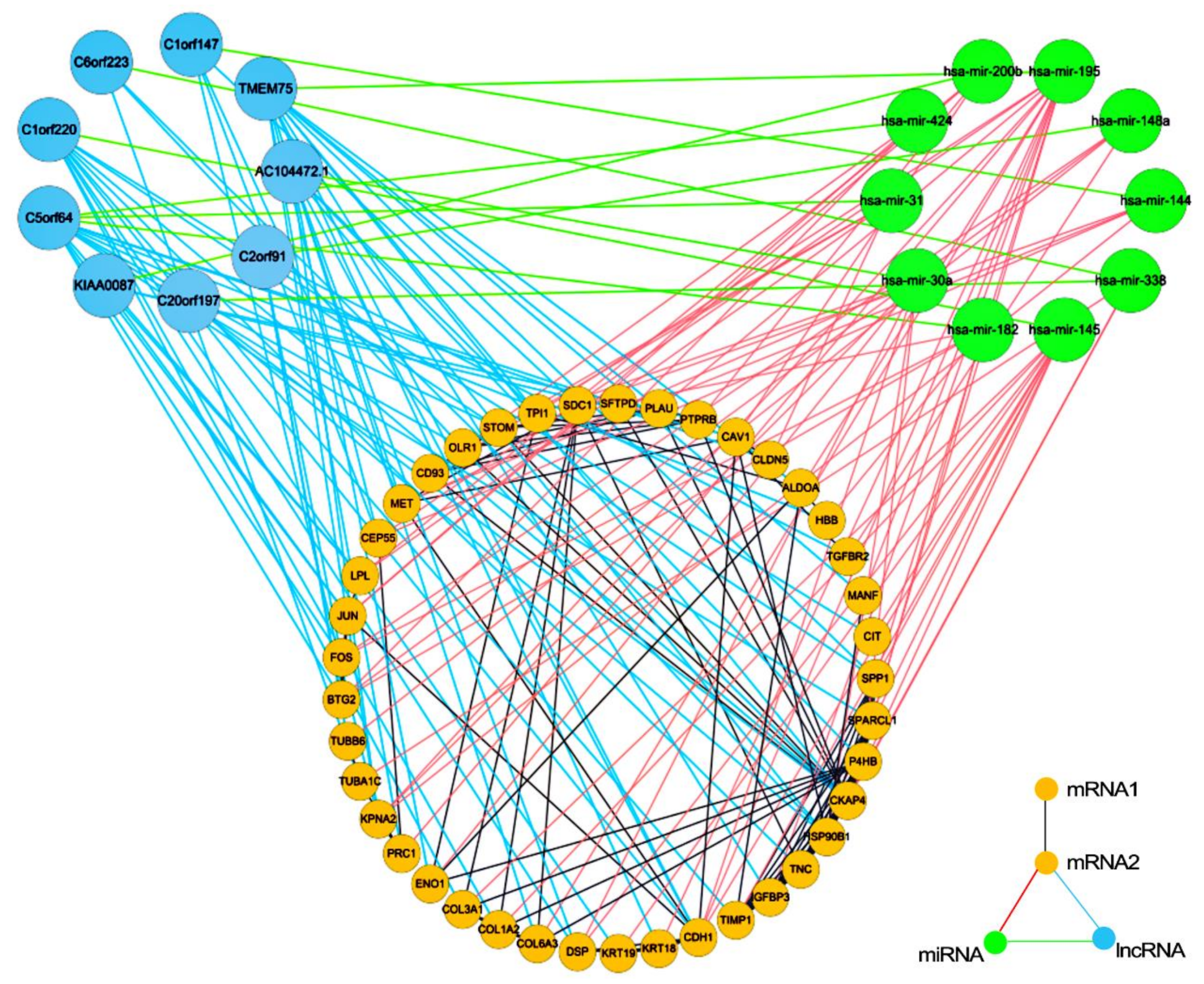
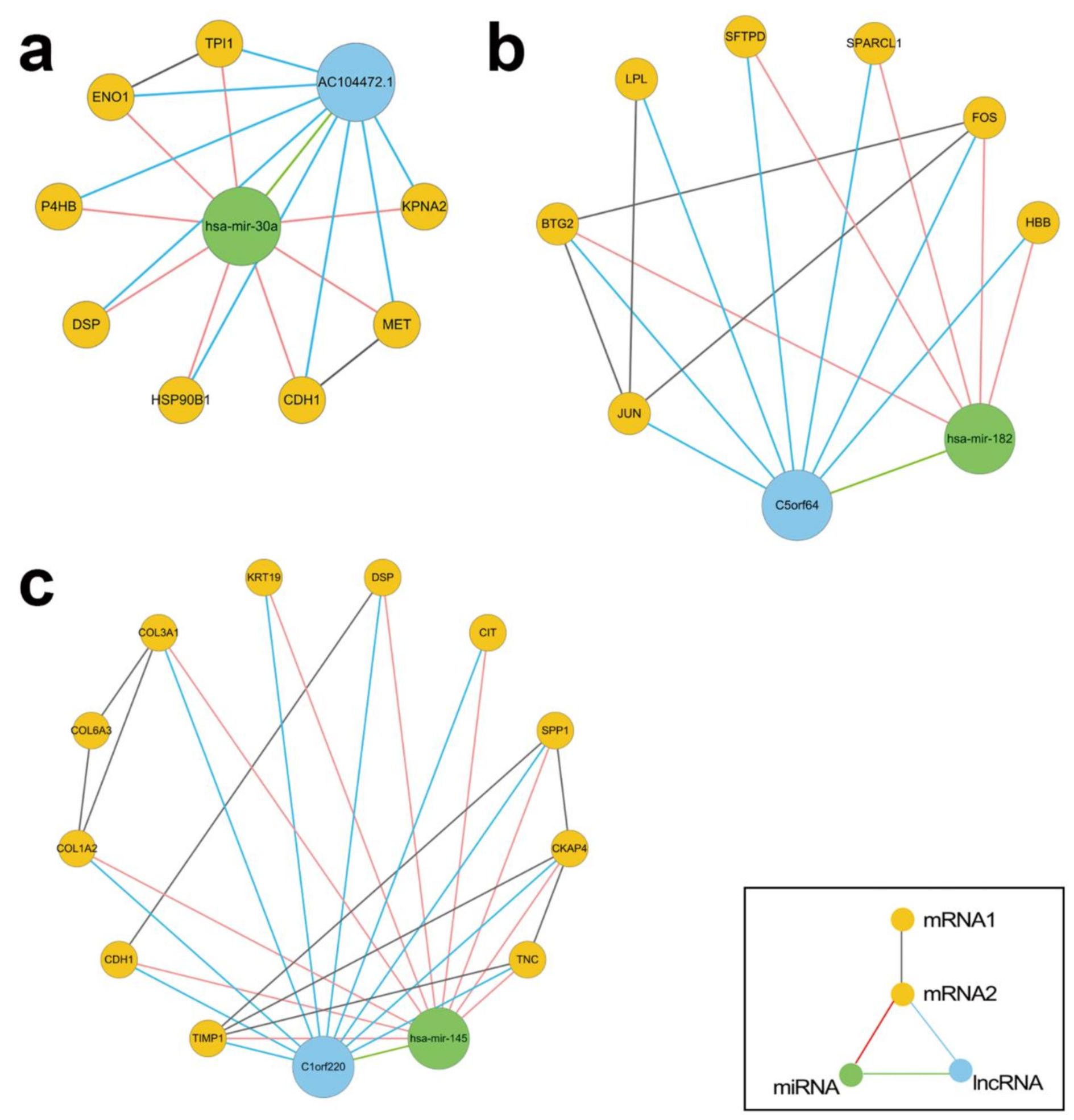
3.3. Regulatory Modules for Lung Adenocarcinoma
3.3.1. Literature Review
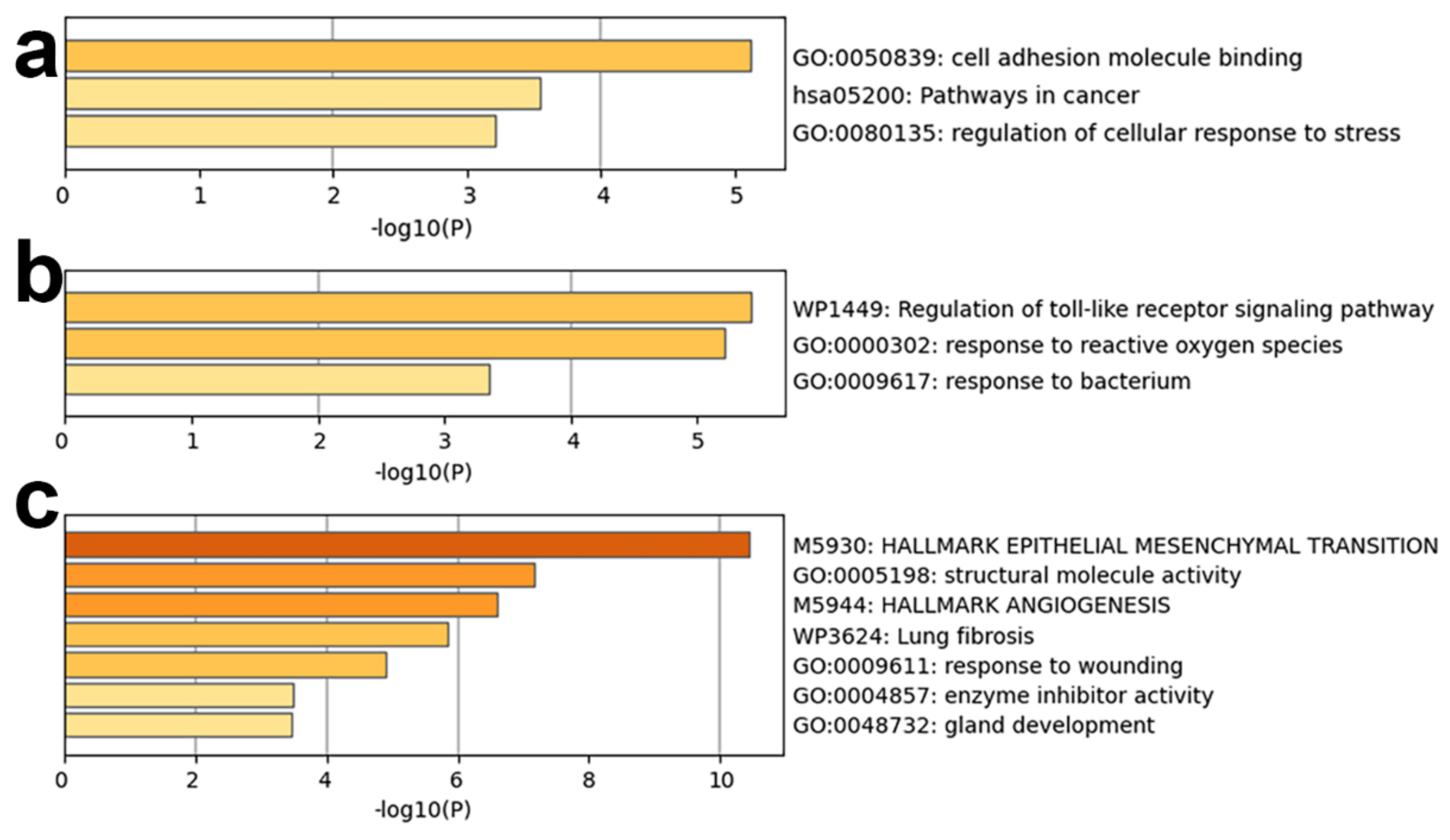
3.3.2. Functional Enrichment Analysis
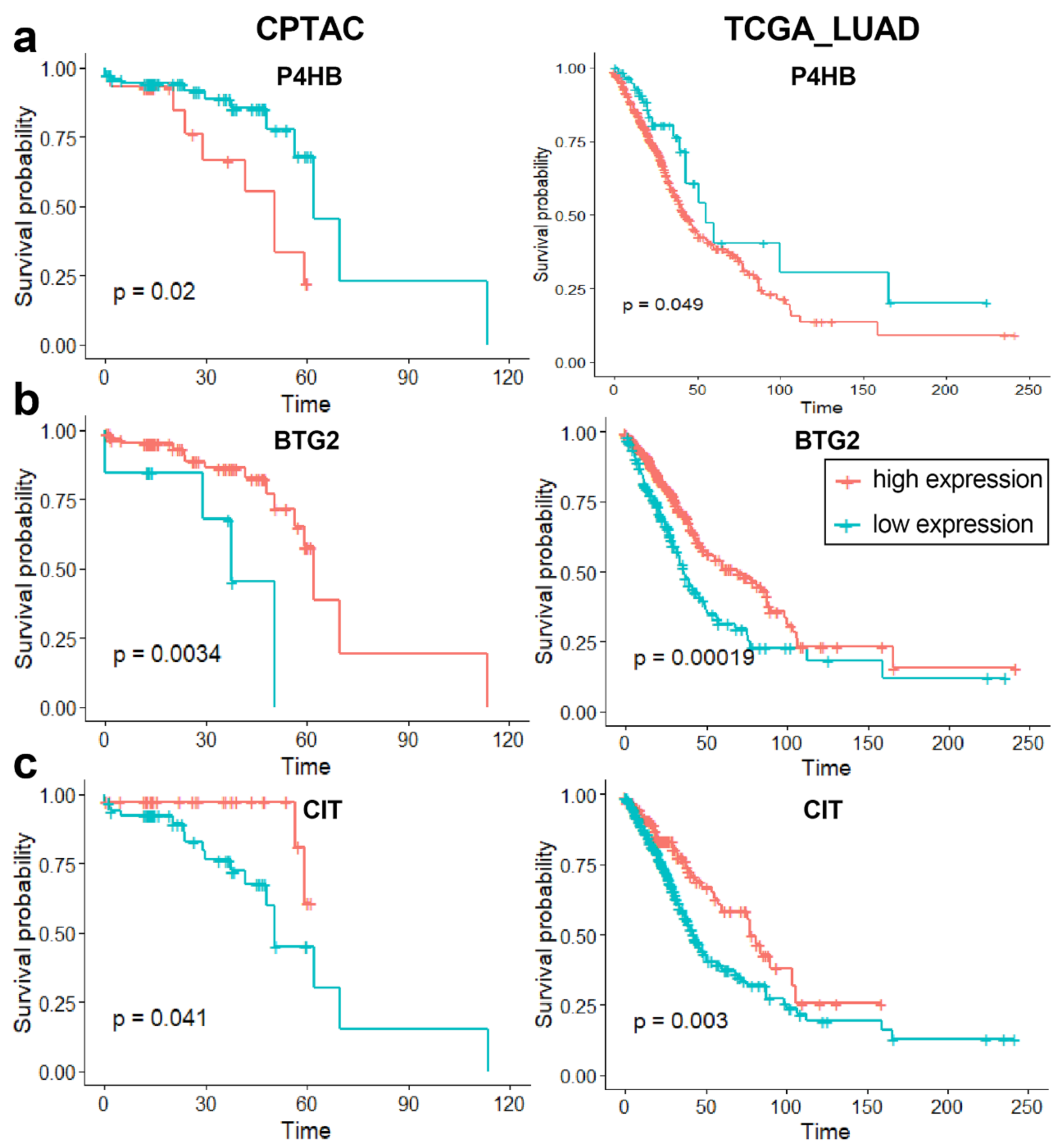
3.3.3. Independent Dataset Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hutchinson, B.D.; Shroff, G.S.; Truong, M.T.; Ko, J.P. Spectrum of Lung Adenocarcinoma. Semin. Ultrasound CT MR 2019, 40, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, E.; Morbini, P.; Cancellieri, A.; Damiani, S.; Cavazza, A.; Comin, C.E. Adenocarcinoma classification: Patterns and prognosis. Pathologica 2018, 110, 5–11. [Google Scholar] [PubMed]
- Zhang, Y.; Huang, Y.X.; Wang, D.L.; Yang, B.; Yan, H.Y.; Lin, L.H.; Li, Y.; Chen, J.; Xie, L.M.; Huang, Y.S.; et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 2020, 10, 10823–10837. [Google Scholar] [CrossRef] [PubMed]
- Athanasios, A.; Charalampos, V.; Vasileios, T.; Ashraf, G.M. Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Curr. Drug Metab. 2017, 18, 5–10. [Google Scholar] [CrossRef]
- Al-Harazi, O.; Kaya, I.H.; El Allali, A.; Colak, D. A Network-Based Methodology to Identify Subnetwork Markers for Diagnosis and Prognosis of Colorectal Cancer. Front. Genet. 2021, 12, 721949. [Google Scholar] [CrossRef]
- Khedkar, H.N.; Wang, Y.C.; Yadav, V.K.; Srivastava, P.; Lawal, B.; Mokgautsi, N.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.S. In-Silico Evaluation of Genetic Alterations in Ovarian Carcinoma and Therapeutic Efficacy of NSC777201, as a Novel Multi-Target Agent for TTK, NEK2, and CDK1. Int. J. Mol. Sci. 2021, 22, 5895. [Google Scholar] [CrossRef]
- Roudi, R.; Beikzadeh, B.; Roviello, G.; D'angelo, A.; Hadizadeh, M. Identification of hub genes, modules and biological pathways associated with lung adenocarcinoma: A system biology approach. Gene Rep. 2022, 27, 101638. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Jafarinejad-Farsangi, S.; Jazi, M.M.; Rostamzadeh, F.; Hadizadeh, M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Noncoding RNA Res. 2020, 5, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Jiang, L.H.; Sun, D.W.; Li, J.; Ji, Z.L. The role of miR-130a in cancer. Breast Cancer 2017, 24, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Huang, C.; Li, Q.; Wu, X. Construction and Comprehensive Analysis for Dysregulated Long Non-Coding RNA (lncRNA)-Associated Competing Endogenous RNA (ceRNA) Network in Gastric Cancer. Med. Sci. Monit. 2018, 24, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, J.; Tian, T.; Zhou, X.; Gu, H.; Xu, L.; Zeng, Y.; Miao, R.; Jin, G.; Ma, H.; et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Investig. 2008, 118, 2600–2608. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jager, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Huys, Q.J.; Maia, T.V.; Frank, M.J. Computational psychiatry as a bridge from neuroscience to clinical applications. Nat. Neurosci. 2016, 19, 404–413. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Gueguen, L.; Coppee, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Nguyen, L.C.; Nguyen-Xuan, H. Deep learning for computational structural optimization. ISA Trans. 2020, 103, 177–191. [Google Scholar] [CrossRef]
- Park, D.J.; Park, M.W.; Lee, H.; Kim, Y.J.; Kim, Y.; Park, Y.H. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci. Rep. 2021, 11, 7567. [Google Scholar] [CrossRef]
- Luan, H.; Zhang, C.; Zhang, T.; He, Y.; Su, Y.; Zhou, L. Identification of Key Prognostic Biomarker and Its Correlation with Immune Infiltrates in Pancreatic Ductal Adenocarcinoma. Dis. Markers 2020, 2020, 8825997. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.D.; Cheng, H.; Martin, S.E.; Si, H.; Ormanoglu, P.; Carlson, S.; Clavijo, P.E.; Yang, X.; Das, R.; Cornelius, S.; et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin. Cancer Res. 2019, 25, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mi, M.; Li, X.; Zheng, X.; Wu, G.; Zhang, L. lncRNA OSTN-AS1 May Represent a Novel Immune-Related Prognostic Marker for Triple-Negative Breast Cancer Based on Integrated Analysis of a ceRNA Network. Front. Genet. 2019, 10, 850. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, G.; Hu, M.; Huang, J.; Li, B.; Zhou, L.; Hong, L. Has-miR-30a regulates autophagic activity in cervical cancer upon hydroxycamptothecin exposure. Biomed. Pharm. 2015, 75, 67–74. [Google Scholar] [CrossRef]
- Xie, L.; Li, H.; Zhang, L.; Ma, X.; Dang, Y.; Guo, J.; Liu, J.; Ge, L.; Nan, F.; Dong, H.; et al. Autophagy-related gene P4HB: A novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging (Albany NY) 2020, 12, 1828–1842. [Google Scholar] [CrossRef]
- Wang, H.; Wu, M.; Lu, Y.; He, K.; Cai, X.; Yu, X.; Lu, J.; Teng, L. LncRNA MIR4435-2HG targets desmoplakin and promotes growth and metastasis of gastric cancer by activating Wnt/beta-catenin signaling. Aging (Albany NY) 2019, 11, 6657–6673. [Google Scholar] [CrossRef]
- Ye, T.; Li, J.; Sun, Z.; Liu, D.; Zeng, B.; Zhao, Q.; Wang, J.; Xing, H.R. Cdh1 functions as an oncogene by inducing self-renewal of lung cancer stem-like cells via oncogenic pathways. Int. J. Biol. Sci. 2020, 16, 447–459. [Google Scholar] [CrossRef]
- Yao, H.P.; Hudson, R.; Wang, M.H. Progress and challenge in development of biotherapeutics targeting MET receptor for treatment of advanced cancer. Biochim Biophys Acta. Rev. Cancer 2020, 1874, 188425. [Google Scholar] [CrossRef]
- Lin, G.; Li, J.; Cai, J.; Zhang, H.; Xin, Q.; Wang, N.; Xie, W.; Zhang, Y.; Xu, N. RNA-binding Protein MBNL2 regulates Cancer Cell Metastasis through MiR-182-MBNL2-AKT Pathway. J. Cancer 2021, 12, 6715–6726. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, X.; Wang, Y.; Wang, Y.; Yan, T.; Wan, J.; Wang, K.; Du, J. Long non-coding RNA C5orf64 is a potential indicator for tumor microenvironment and mutation pattern remodeling in lung adenocarcinoma. Genom. 2021, 113, 291–304. [Google Scholar] [CrossRef]
- Pirker, R.; Wiesenberger, K.; Pohl, G.; Minar, W. Anemia in lung cancer: Clinical impact and management. Clin. Lung Cancer 2003, 5, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, X.; Lu, B.; Gu, Y.; Chen, Q.; Lei, T.; Nie, F.; Gu, J.; Huang, J.; Wei, C.; et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J. Hematol Oncol. 2020, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, F.; Narayanan, A.; Gallotti, A.L.; Pieri, V.; Mazzoleni, S.; Cominelli, M.; Rezzola, S.; Corsini, M.; Brugnara, G.; Altabella, L.; et al. Enhanced SPARCL1 expression in cancer stem cells improves preclinical modeling of glioblastoma by promoting both tumor infiltration and angiogenesis. Neurobiol. Dis. 2020, 134, 104705. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, Y.; Yan, C.; Su, H.; Ying, K. Identification of key genes and evaluation of clinical outcomes in lung squamous cell carcinoma using integrated bioinformatics analysis. Oncol.Lett. 2019, 18, 5859–5870. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.H.; Ruan, X.L.; Huang, J.Y.; Liu, X.P.; Ma, H.L.; Chen, C.; Hu, W.D.; Li, S. Analysis of the Interaction Network of Hub miRNAs-Hub Genes, Being Involved in Idiopathic Pulmonary Fibers and Its Emerging Role in Non-small Cell Lung Cancer. Front. Genet. 2020, 11, 302. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, F.; Yang, X.; Xue, M.; Li, X.; Gao, Y.; Liu, L. Secreted Phosphoprotein 1 (SPP1) Contributes to Second-Generation EGFR Tyrosine Kinase Inhibitor Resistance in Non-Small Cell Lung Cancer. Oncol. Res. 2019, 27, 871–877. [Google Scholar] [CrossRef]
- Schoeps, B.; Eckfeld, C.; Prokopchuk, O.; Bottcher, J.; Haussler, D.; Steiger, K.; Demir, I.E.; Knolle, P.; Soehnlein, O.; Jenne, D.E.; et al. TIMP1 Triggers Neutrophil Extracellular Trap Formation in Pancreatic Cancer. Cancer Res. 2021, 81, 3568–3579. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Zhou, K.; Wang, C.; Jiang, L.; Zhang, L.; Yang, Y.; Luo, W.; Qiao, W.; Wang, G.; et al. Deciphering cell lineage specification of human lung adenocarcinoma with single-cell RNA sequencing. Nat. Commun. 2021, 12, 6500. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Matsubara, D.; Soda, M.; Ueno, T.; Amano, Y.; Kihara, A.; Sakatani, T.; Nakano, T.; Shibano, T.; Endo, S.; et al. Mucin 21 is a key molecule involved in the incohesive growth pattern in lung adenocarcinoma. Cancer Sci. 2019, 110, 3006–3011. [Google Scholar] [CrossRef]
- Kasiri, S.; Chen, B.; Wilson, A.N.; Reczek, A.; Mazambani, S.; Gadhvi, J.; Noel, E.; Marriam, U.; Mino, B.; Lu, W.; et al. Stromal Hedgehog pathway activation by IHH suppresses lung adenocarcinoma growth and metastasis by limiting reactive oxygen species. Oncogene 2020, 39, 3258–3275. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.L.; Lee, C.H.; Chen, C.H.; Zhan, J.F.; Wu, S.Y. Angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers might be associated with lung adenocarcinoma risk: A nationwide population-based nested case-control study. Am. J. Transl. Res. 2020, 12, 6615–6625. [Google Scholar] [CrossRef] [PubMed]
- Gergen, A.K.; Kohtz, P.D.; Halpern, A.L.; Li, A.; Meng, X.; Reece, T.B.; Fullerton, D.A.; Weyant, M.J. Activation of Toll-Like Receptor 2 Promotes Proliferation of Human Lung Adenocarcinoma Cells. Anticancer Res. 2020, 40, 5361–5369. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, W.; Wu, B.; Liu, Y.; Li, D.; Guo, Y.; Fu, H.; Li, Y. KDM3A promotes inhibitory cytokines secretion by participating in TLR4 regulation of Foxp3 transcription in lung adenocarcinoma cells. Oncol. Lett. 2017, 13, 3529–3537. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karampitsakos, T.; Tzilas, V.; Tringidou, R.; Steiropoulos, P.; Aidinis, V.; Papiris, S.A.; Bouros, D.; Tzouvelekis, A. Lung cancer in patients with idiopathic pulmonary fibrosis. Pulm. Pharmacol. Ther. 2017, 45, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sarode, P.; Mansouri, S.; Karger, A.; Schaefer, M.B.; Grimminger, F.; Seeger, W.; Savai, R. Epithelial cell plasticity defines heterogeneity in lung cancer. Cell Signal. 2020, 65, 109463. [Google Scholar] [CrossRef]
- Cai, S.; Guo, X.; Huang, C.; Deng, Y.; Du, L.; Liu, W.; Yang, C.; Zhao, H.; Ma, K.; Wang, L.; et al. Integrative analysis and experiments to explore angiogenesis regulators correlated with poor prognosis, immune infiltration and cancer progression in lung adenocarcinoma. J. Transl. Med. 2021, 19, 361. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef]

| CPTAC | |
|---|---|
| Patient (n) | 100 |
| Age, years | |
| median | 63.5 |
| range | 35–81 |
| Sex (%) | |
| male | 63 (63%) |
| female | 37 (37%) |
| Tumor_grade (%) | |
| G1 | 7 (7%) |
| G2 | 55 (55%) |
| G3 | 37 (37%) |
| GX | 1 (1%) |
| Ajcc_pathologic_stage (%) | |
| Stage I | 54 (54%) |
| Stage II | 29 (29%) |
| Stage III | 17 (17%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Luo, K.; Lv, J.; Wang, X.; Qin, S.; Zhang, Z.; Sun, S.; Wang, X.; Yun, B.; He, Y.; et al. Integrating Expression Data-Based Deep Neural Network Models with Biological Networks to Identify Regulatory Modules for Lung Adenocarcinoma. Biology 2022, 11, 1291. https://doi.org/10.3390/biology11091291
Fu L, Luo K, Lv J, Wang X, Qin S, Zhang Z, Sun S, Wang X, Yun B, He Y, et al. Integrating Expression Data-Based Deep Neural Network Models with Biological Networks to Identify Regulatory Modules for Lung Adenocarcinoma. Biology. 2022; 11(9):1291. https://doi.org/10.3390/biology11091291
Chicago/Turabian StyleFu, Lei, Kai Luo, Junjie Lv, Xinyan Wang, Shimei Qin, Zihan Zhang, Shibin Sun, Xu Wang, Bei Yun, Yuehan He, and et al. 2022. "Integrating Expression Data-Based Deep Neural Network Models with Biological Networks to Identify Regulatory Modules for Lung Adenocarcinoma" Biology 11, no. 9: 1291. https://doi.org/10.3390/biology11091291
APA StyleFu, L., Luo, K., Lv, J., Wang, X., Qin, S., Zhang, Z., Sun, S., Wang, X., Yun, B., He, Y., He, W., Li, W., & Chen, L. (2022). Integrating Expression Data-Based Deep Neural Network Models with Biological Networks to Identify Regulatory Modules for Lung Adenocarcinoma. Biology, 11(9), 1291. https://doi.org/10.3390/biology11091291






