Integrative Analysis of Transcriptome and Metabolome Reveals Molecular Responses in Eriocheir sinensis with Hepatopancreatic Necrosis Disease
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Samples
2.2. Histological Analysis
2.3. Metabolite Extraction and Profiling Analysis
2.4. Metabolomics Data Analysis
2.5. RNA Extraction and RNA-Sequencing
2.6. Transcriptome Profile Analysis
2.7. Integrative Analysis of Metabolomics and Transcriptomics
3. Results
3.1. Histopathology of Crab Tissues
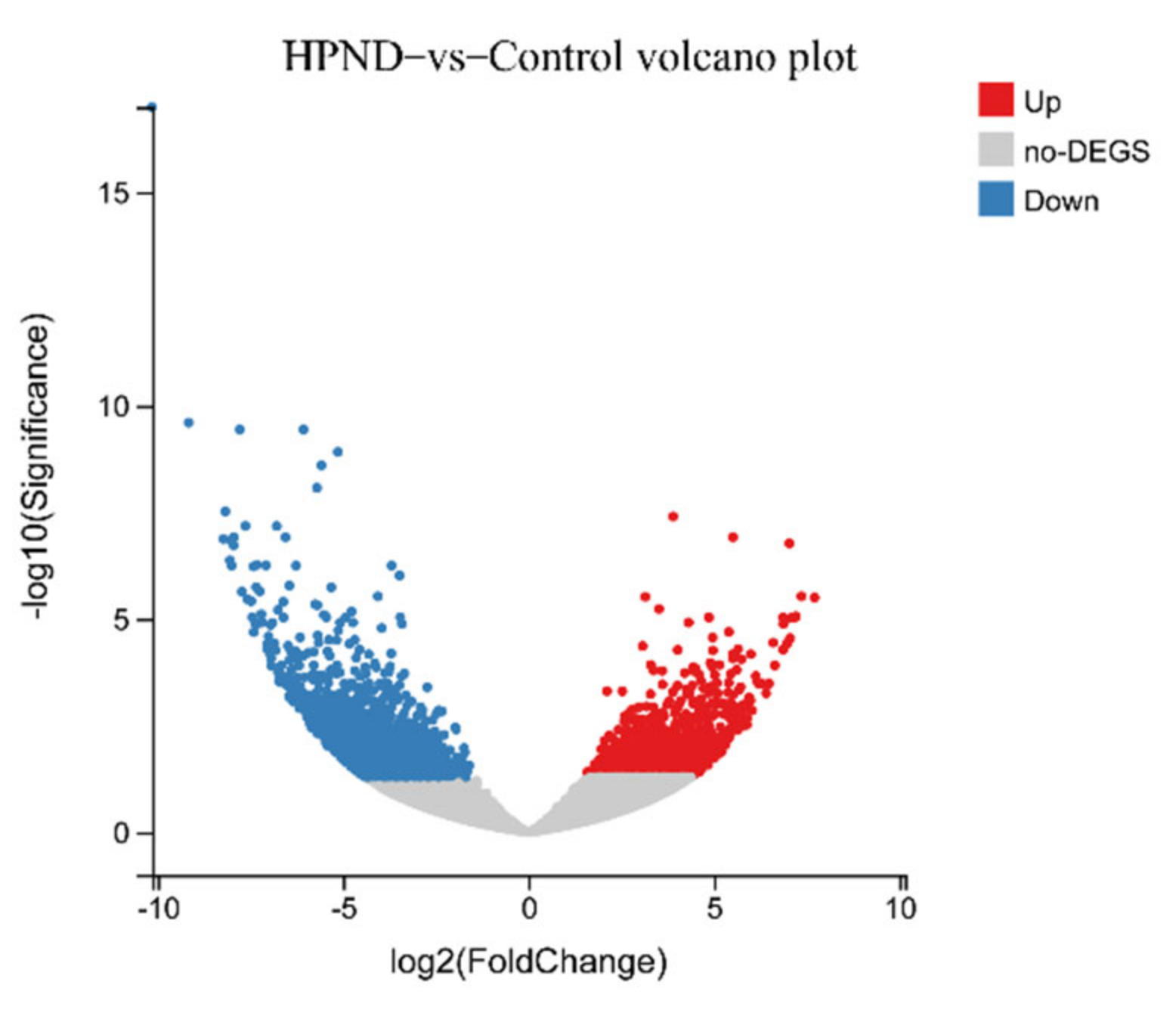
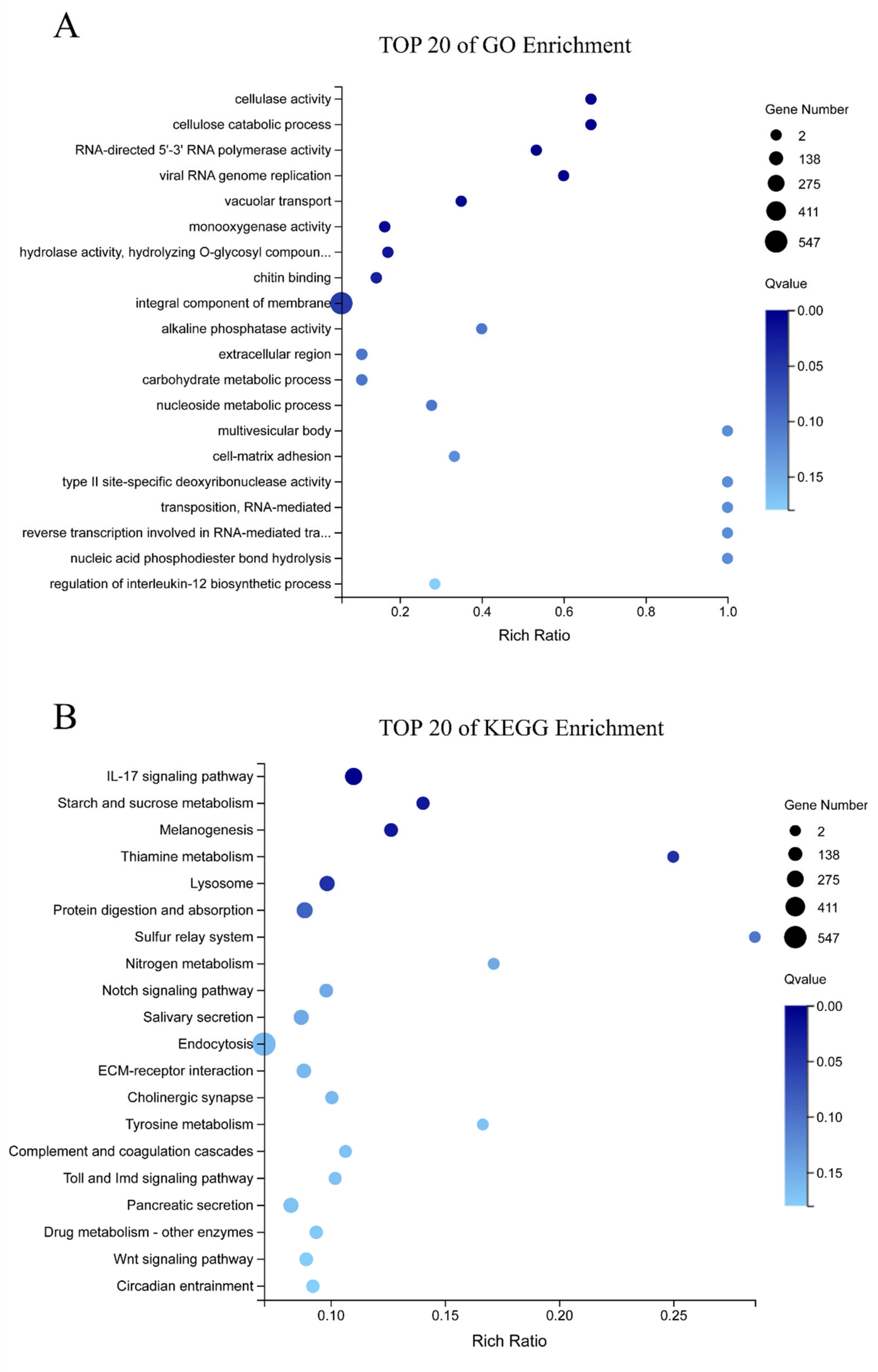
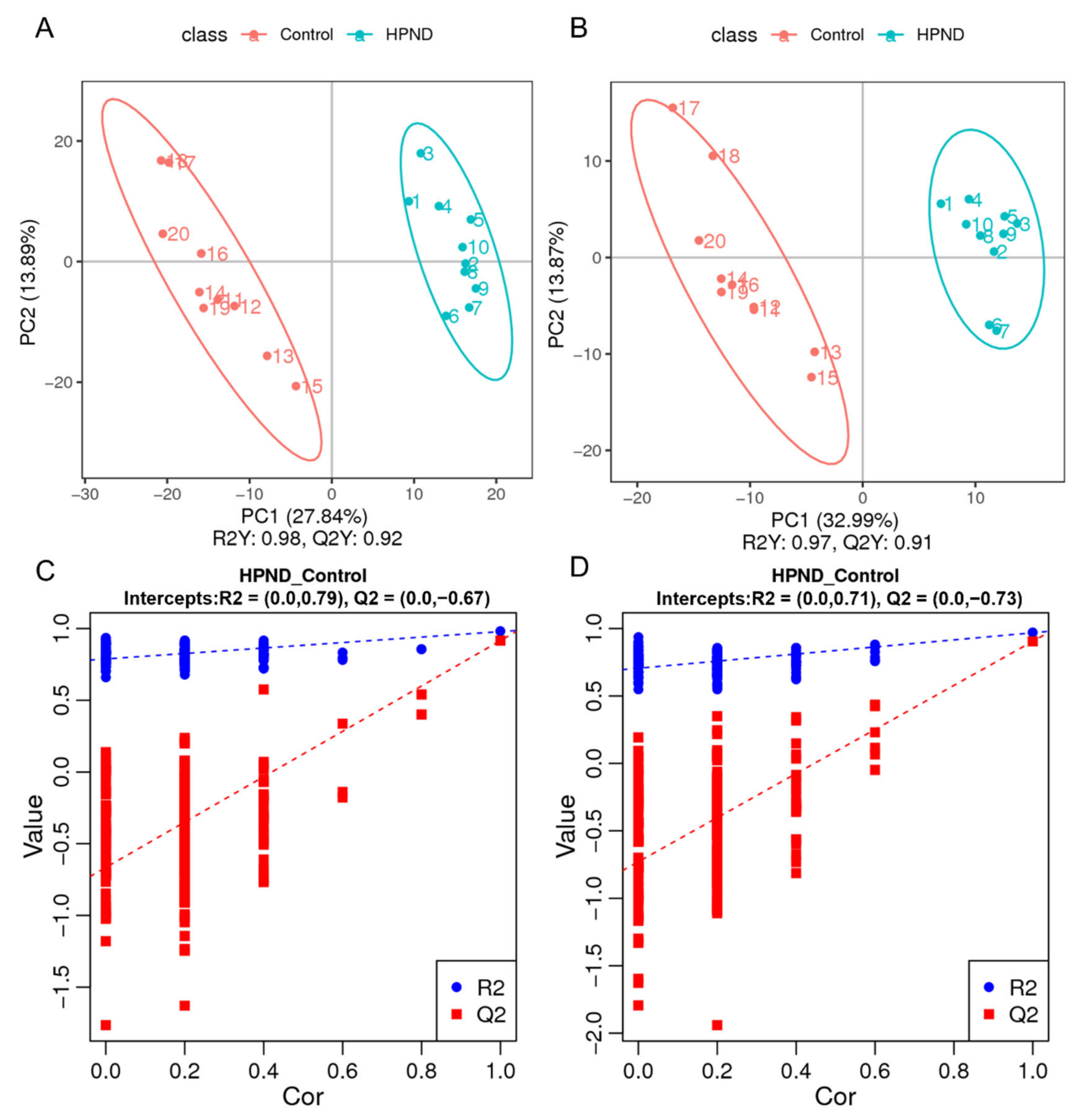
3.2. Transcriptomic Analysis of E. sinensis with HPND
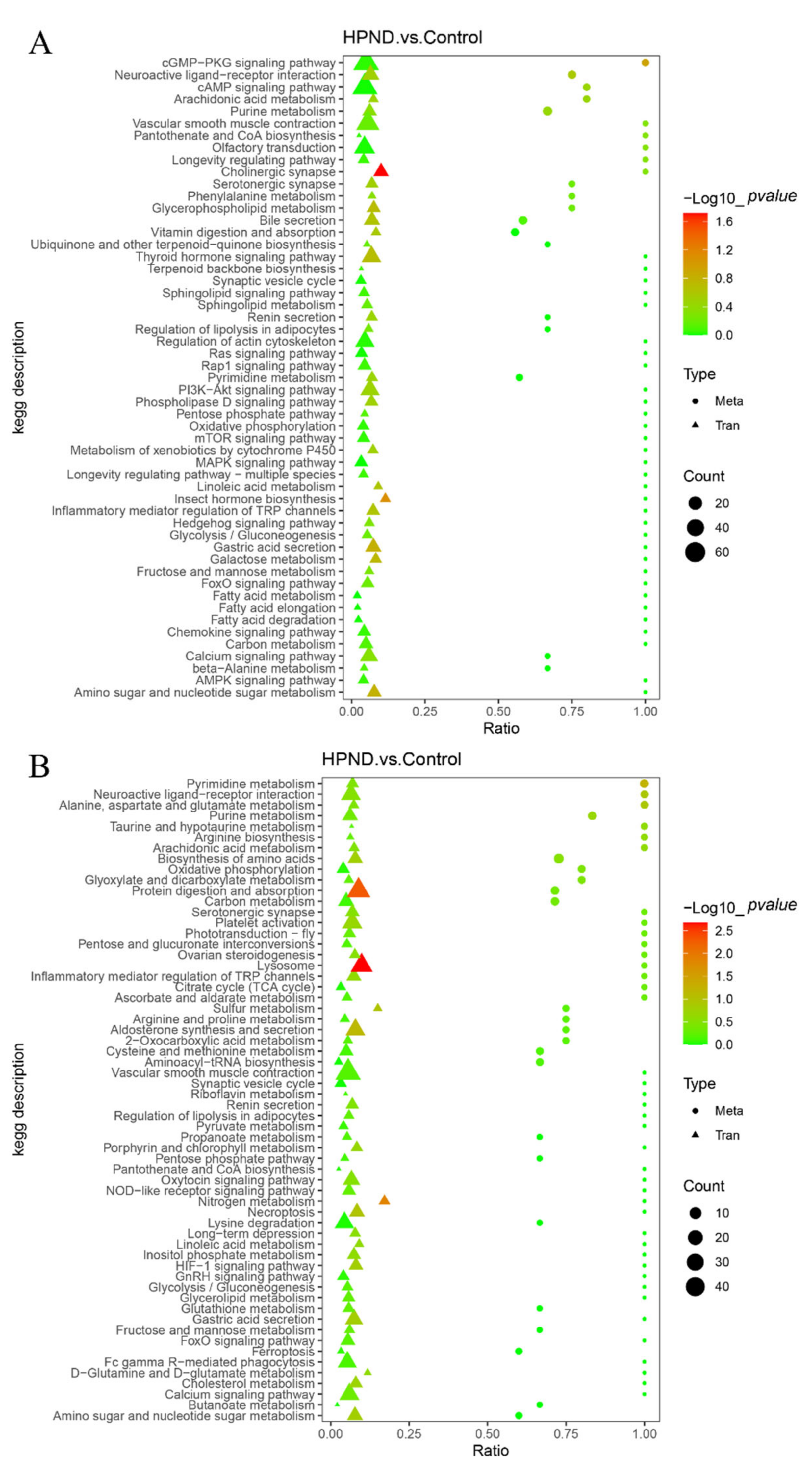
3.3. Metabolic Analysis of E. sinensis with HPND
3.4. Integrated Analysis of Metabolome and Transcriptome
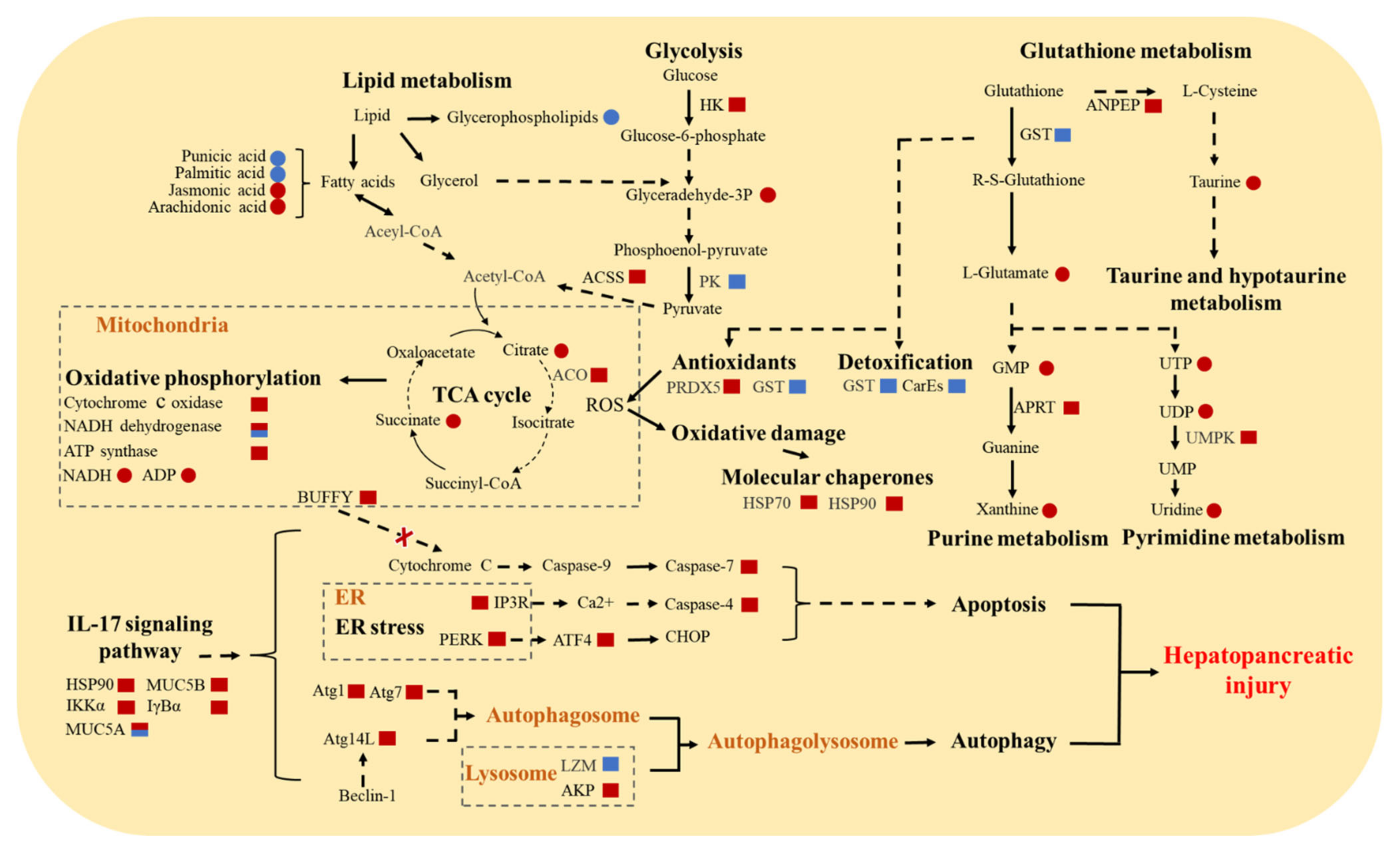
3.5. Molecular Responses of E. sinensis with HPND
4. Discussion
4.1. Abnormalities of the Nervous System
4.2. Oxidative Stress and Impaired Detoxification
4.3. Increase in Autophagy and Apoptosis
4.4. Impairment of Immune System
4.5. Energy and Substance Metabolism Disorders
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kong, L.; Cai, C.; Ye, Y.; Chen, D.; Wu, P.; Li, E.; Chen, L.; Song, L. Comparison of non-volatile compounds and sensory characteristics of Chinese mitten crabs (Eriocheir sinensis) reared in lakes and ponds: Potential environmental factors. Aquaculture 2012, 364, 96–102. [Google Scholar] [CrossRef]
- Yearbook CFS. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2020; Volume 2020, pp. 24–34. [Google Scholar]
- Shen, H.; Zang, Y.; Song, K.; Ma, Y.; Dai, T.; Serwadda, A. A meta-transcriptomics survey reveals changes in the microbiota of the Chinese mitten crab Eriocheir sinensis infected with Hepatopancreatic necrosis disease. Front. Microbiol. 2017, 8, 732. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Tang, S.; Qi, R.; Lei, Y.; Li, Y.; Wang, J. Pathological study on “Shuibiezi” disease of crab Eriocheir sinensis. J. Yantai Univ. 2017, 30, 313–316. [Google Scholar]
- Pan, Z.; Song, X.; Hu, X.; Xue, R.; Cao, G.; Zar, M.S.; Kumar, D.; Feng, Y.; Wei, Y.; Zhang, W. Pathological changes and risk factors of hepatopancreas necrosis disease of mitten crab, Eriocheir Sinensis. Fish. Aquac. J. 2017, 8, 3. [Google Scholar] [CrossRef]
- Zhan, M.; Xi, C.; Gong, J.; Zhu, M.; Sui, Y.; Xu, Z.; Xu, G.; Shen, H. 16S rRNA gene sequencing analysis reveals an imbalance in the intestinal flora of Eriocheir sinensis with hepatopancreatic necrosis disease. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100988. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Meng, Q.; Liu, H.; Yuan, S.; Zhang, F.; Sun, M.; Zhao, Y.; Shen, M.; Zhou, G.; Pan, J. First case of hepatopancreatic necrosis disease in pond-reared Chinese mitten crab, Eriocheir sinensis, associated with microsporidian. J. Fish Dis. 2016, 39, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Xu, Y.; Wang, K.; Deng, Y.; Yang, Y.; Lu, Q.; Pan, J.; Xu, Z. Comparative LC-MS based non-targeted metabolite profiling of the Chinese mitten crab Eriocheir sinensis suffering from hepatopancreatic necrosis disease (HPND). Aquaculture 2018, 491, 338–345. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Hu, K.; Liu, L.; Cai, H.; Zhang, F.; Yang, X. Etiological and histopathological study on hepatopancreatic necrosis syndrome in Eriocheir sinensis. Acta Hydrobiol. Sin. 2018, 42, 17–25. [Google Scholar]
- Zhu, J.; Wang, Z.; Cai, C.; Tang, X.; Shen, J.; Wu, D. Inducement and prevention technology of “Shuibiezi” disease of Chinese mitten crab Eriocheir sinensis. Sci. Fish Farming 2016, 6, 13–15. [Google Scholar]
- Gu, X.; Jiang, G.; Wei, B.; Niu, J.; Song, X. Investigation and analysis on the correlation of pesticides and hypoxia with “Shuibiezi” disease of Eriocheir sinensis. Sci. Fish Farming 2017, 4, 59–61. [Google Scholar]
- Zhu, M.; Gong, J.; Zhan, M.; Xi, C.; Shen, G.; Shen, H. Transcriptome analysis reveals the molecular mechanism of long-term exposure of Eriocheir sinensis to low concentration of trichlorfon.Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100916. [Google Scholar]
- Shen, G.; Shui, Y.; Zhang, X.; Song, K.; Wang, Y.; Xu, Z.; Shen, H. Hepatopancreatic necrosis disease (HPND) in Chinese mitten crab Eriocheir sinensis tightly linked to low concentration of two insecticides. Aquac. Res. 2021, 52, 2294–2304. [Google Scholar] [CrossRef]
- Shen, Z.; Kumar, D.; Liu, X.; Yan, B.; Fang, P.; Gu, Y.; Li, M.; Xie, M.; Yuan, R.; Feng, Y. Metatranscriptomic analysis reveals an imbalance of hepatopancreatic flora of Chinese mitten crab Eriocheir sinensis with hepatopancreatic necrosis disease. Biology 2021, 10, 462. [Google Scholar] [CrossRef]
- Wang, T.; Yang, C.; Zhang, S.; Rong, L.; Yang, X.; Wu, Z.; Sun, W. Metabolic changes and stress damage induced by ammonia exposure in juvenile Eriocheir sinensis. Ecotoxicol. Environ. Saf. 2021, 223, 112608. [Google Scholar] [CrossRef]
- Kong, T.; Ren, X.; Lin, S.; Li, S.; Gong, Y. Elucidation of metabolic responses in mud crab Scylla paramamosain challenged to WSSV infection by integration of metabolomics and transcriptomics. Dev. Comp. Immunol. 2020, 113, 103799. [Google Scholar] [CrossRef]
- Qiao, F.; Lei, K.; Li, Z.; Wei, Z.; Liu, Q.; Yang, L.; He, J.; An, L.; Qi, H.; Cui, S. Transcriptomic responses of the freshwater snail (Parafossarulus striatulus) following dietary exposure to cyanobacteria. Sci. Total Environ. 2018, 624, 153–161. [Google Scholar] [CrossRef]
- Mercuro, G.; Bassareo, P.P.; Deidda, M.; Cadeddu, C.; Barberini, L.; Atzori, L. Metabolomics: A new era in cardiology? J. Cardiovasc. Med. 2011, 12, 800–805. [Google Scholar] [CrossRef]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z. Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol. Cell. Proteom. 2016, 15, 154–163. [Google Scholar] [CrossRef]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef]
- Scott, R.C.; Juhász, G.; Neufeld, T.P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007, 17, 1–11. [Google Scholar] [CrossRef]
- Kothari, S.; Cizeau, J.; McMillan-Ward, E.; Israels, S.J.; Bailes, M.; Ens, K.; Kirshenbaum, L.A.; Gibson, S.B. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 2003, 22, 4734–4744. [Google Scholar] [CrossRef]
- Quinn, L.; Coombe, M.; Mills, K.; Daish, T.; Colussi, P.; Kumar, S.; Richardson, H. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 2003, 22, 3568–3579. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 2000, 5, 897–904. [Google Scholar] [CrossRef]
- Hitomi, J.; Katayama, T.; Eguchi, Y.; Kudo, T.; Taniguchi, M.; Koyama, Y.; Manabe, T.; Yamagishi, S.; Bando, Y.; Imaizumi, K. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J. Cell Biol. 2004, 165, 347–356. [Google Scholar] [CrossRef]
- Srinivasula, S.M.; Ahmad, M.; Fernandes-Alnemri, T.; Alnemri, E.S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1998, 1, 949–957. [Google Scholar] [CrossRef]
- Rajalakshmi, S.; Mohandas, A. Copper-induced changes in tissue enzyme activity in a freshwater mussel. Ecotoxicol. Environ. Saf. 2005, 62, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Blom, A.M. Complement in removal of the dead–balancing inflammation. Immunol. Rev. 2016, 274, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Walbrecq, G.; Wang, B.; Becker, S.; Hannotiau, A.; Fransen, M.; Knoops, B. Antioxidant cytoprotection by peroxisomal peroxiredoxin-5. Free Radic. Biol. Med. 2015, 84, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Shengming, S.; Jian, Z.; Xianping, G.; Chengfeng, Z.; Linghong, M.; Wuxiao, Z.; Qiong, Z. Molecular cloning, characterization and mRNA expression of Mu-typ glutathione S-transferases from Megalobrama amblycephala. Asian J. Ecotoxicol. 2016, 11, 295–305. [Google Scholar]
- Parsell, D.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Ann. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Hemingway, J.; Ranson, H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef]
- Wheelock, C.E.; Miller, J.L.; Miller, M.J.; Phillips, B.M.; Huntley, S.A.; Gee, S.J.; Tjeerdema, R.S.; Hammock, B.D. Use of carboxylesterase activity to remove pyrethroid-associated toxicity to Ceriodaphnia dubia and Hyalella azteca in toxicity identification evaluations. Environ. Toxicol. Chem. 2006, 25, 973–984. [Google Scholar] [CrossRef]
- Grieshaber, M.; Hardewig, I.; Kreutzer, U.; Pörtner, H.-O. Physiological and metabolic responses to hypoxia in invertebrates. Rev. Physiol. Biochem. Pharm. 1993, 125, 43–147. [Google Scholar]
- Calder, P.C. The relationship between the fatty acid composition of immune cells and their function. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 101–108. [Google Scholar] [CrossRef]
- Paone, A.; Marani, M.; Fiascarelli, A.; Rinaldo, S.; Giardina, G.; Contestabile, R.; Paiardini, A.; Cutruzzolà, F. SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 2014, 5, e1525. [Google Scholar] [CrossRef]
- Irving, A.; Collingridge, G.; Schofield, J. L-glutamate and acetylcholine mobilise Ca2+ from the same intracellular pool in cerebellar granule cells using transduction mechanisms with different Ca2+ sensitivities. Cell Calcium 1992, 13, 293–301. [Google Scholar] [CrossRef]
- Dvir, H.; Silman, I.; Harel, M.; Rosenberry, T.L.; Sussman, J.L. Acetylcholinesterase: From 3D structure to function. Chem. Interact. 2010, 187, 10–22. [Google Scholar] [CrossRef]
- Mileson, B.E.; Chambers, J.E.; Chen, W.; Dettbarn, W.; Ehrich, M.; Eldefrawi, A.T.; Gaylor, D.W.; Hamernik, K.; Hodgson, E.; Karczmar, A.G. Common mechanism of toxicity: A case study of organophosphorus pesticides. Toxicol. Sci. 1998, 41, 8–20. [Google Scholar]
- Kagias, K.; Nehammer, C.; Pocock, R. Neuronal responses to physiological stress. Front. Genet. 2012, 3, 222. [Google Scholar] [CrossRef]
- Kim, K.W.; Jin, Y. Neuronal responses to stress and injury in C. elegans. FEBS Lett. 2015, 589, 1644–1652. [Google Scholar] [CrossRef]
- Sun, X.; Tu, K.; Li, L.; Wu, B.; Wu, L.; Liu, Z.; Zhou, L.; Tian, J.; Yang, A. Integrated transcriptome and metabolome analysis reveals molecular responses of the clams to acute hypoxia. Mar. Environ. Res. 2021, 168, 105317. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Jiang, Y.; Zhu, F.; Zeng, L.; Wang, Y.; Lei, X.; Yao, Y.; Hou, Y.; Xu, L. Transcriptional responses in the hepatopancreas of Eriocheir sinensis exposed to deltamethrin. PLoS ONE 2017, 12, e0184581. [Google Scholar] [CrossRef]
- Tu, H.T.; Silvestre, F.; De Meulder, B.; Thome, J.-P.; Phuong, N.T.; Kestemont, P. Combined effects of deltamethrin, temperature and salinity on oxidative stress biomarkers and acetylcholinesterase activity in the black tiger shrimp (Penaeus monodon). Chemosphere 2012, 86, 83–91. [Google Scholar] [CrossRef]
- Slotkin, T.A.; Brown, K.K.; Seidler, F.J. Developmental exposure of rats to chlorpyrifos elicits sex-selective hyperlipidemia and hyperinsulinemia in adulthood. Environ. Health Perspect. 2005, 113, 1291–1294. [Google Scholar] [CrossRef][Green Version]
- Talesa, V.; Contenti, S.; Principato, G.; Pascolini, R.; Giovannini, E.; Rosi, G. Cholinesterases from Maia verrucosa and Palinurus vulgaris: A comparative study. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1992, 101, 499–503. [Google Scholar] [CrossRef]
- Redmond, L.; Ghosh, A. Regulation of dendritic development by calcium signaling. Cell Calcium 2005, 37, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wang, Y.-X.; Yu, J.; Yi, S. Critical signaling pathways during Wallerian degeneration of peripheral nerve. Neural Regen. Res. 2017, 12, 995. [Google Scholar] [PubMed]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef]
- Marklund, S.; Marklund, G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 1974, 47, 469–474. [Google Scholar] [CrossRef]
- Meng, X.; Jayasundara, N.; Zhang, J.; Ren, X.; Gao, B.; Li, J.; Liu, P. Integrated physiological, transcriptome and metabolome analyses of the hepatopancreas of the female swimming crab Portunus trituberculatus under ammonia exposure. Ecotoxicol. Environ. Saf. 2021, 228, 113026. [Google Scholar] [CrossRef]
- Hendrick, J.P.; Hartl, F.-U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef]
- Downs, C.A.; Fauth, J.E.; Woodley, C.M. Assessing the health of grass shrimp (Palaeomonetes pugio) exposed to natural and anthropogenic stressors: A molecular biomarker system. Mar. Biotechnol. 2001, 3, 380–397. [Google Scholar] [CrossRef]
- Kim, B.-M.; Rhee, J.-S.; Jeong, C.-B.; Seo, J.S.; Park, G.S.; Lee, Y.-M.; Lee, J.-S. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2014, 166, 65–74. [Google Scholar] [CrossRef]
- Rutherford, S.L.; Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 1998, 396, 336–342. [Google Scholar] [CrossRef]
- Hong, Y.; Huang, Y.; Huang, Z. Oxidative stress, immunological response, and heat shock proteins induction in the Chinese Mitten Crab, Eriocheir sinensis following avermectin exposure. Environ. Toxicol. 2020, 35, 213–222. [Google Scholar] [CrossRef]
- Hong, Y.; Huang, Y.; Wu, S.; Yang, X.; Dong, Y.; Xu, D.; Huang, Z. Effects of imidacloprid on the oxidative stress, detoxification and gut microbiota of Chinese mitten crab, Eriocheir sinensis. Sci. Total Environ. 2020, 729, 138276. [Google Scholar] [CrossRef]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Ann. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef]
- Song, P.; Ping, L.; Gao, J.; Li, X.; Zhu, M.; Wang, J. Ecotoxicological effects of fertilizers made from pulping waste liquor on earthworm Eisenia fetida. Ecotoxicol. Environ. Saf. 2018, 166, 237–241. [Google Scholar] [CrossRef]
- Hong, Y.; Huang, Y.; Yan, G.; Pan, C.; Zhang, J. Antioxidative status, immunological responses, and heat shock protein expression in hepatopancreas of Chinese mitten crab, Eriocheir sinensis under the exposure of glyphosate. Fish Shellfish Immunol. 2019, 86, 840–845. [Google Scholar] [CrossRef]
- Cui, Y.-Q.; Liu, Y.-J.; Zhang, F. The suppressive effects of Britannin (Bri) on human liver cancer through inducing apoptosis and autophagy via AMPK activation regulated by ROS. Biochem. Biophys. Res. Commun. 2018, 497, 916–923. [Google Scholar] [CrossRef]
- Lv, S.-X.; Qiao, X. Isovitexin (IV) induces apoptosis and autophagy in liver cancer cells through endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2018, 496, 1047–1054. [Google Scholar] [CrossRef]
- Kamada, Y.; Yoshino, K.-i.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhu, Q.; Dee, R.; Opheim, Z.; Mack, C.P.; Cyr, D.M.; Taylor, J.M. Focal Adhesion Kinase-mediated Phosphorylation of Beclin1 Protein Suppresses Cardiomyocyte Autophagy and Initiates Hypertrophic Growth. J. Biol. Chem. 2017, 292, 2065–2079. [Google Scholar] [CrossRef]
- Cohen, G.M. Caspases: The executioners of apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [CrossRef]
- Pinton, P.; Giorgi, C.; Siviero, R.; Zecchini, E.; Rizzuto, R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 2008, 27, 6407–6418. [Google Scholar] [CrossRef]
- Imaizumi, K.; Miyoshi, K.; Katayama, T.; Yoneda, T.; Taniguchi, M.; Kudo, T.; Tohyama, M. The unfolded protein response and Alzheimer’s disease. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2001, 1536, 85–96. [Google Scholar] [CrossRef]
- Li, J.; Lee, B.; Lee, A.S. Endoplasmic reticulum stress-induced apoptosis: Multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J. Biolog. Chem. 2006, 281, 7260–7270. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Patergnani, S.; Missiroli, S.; Morciano, G.; Rimessi, A.; Wieckowski, M.R.; Giorgi, C.; Pinton, P. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium 2018, 69, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Chukai, Y.; Ito, G.; Konno, M.; Sakata, Y.; Ozaki, T. Mitochondrial calpain-5 truncates caspase-4 during endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 2022, 608, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Slee, E.A.; Harte, M.T.; Kluck, R.M.; Wolf, B.B.; Casiano, C.A.; Newmeyer, D.D.; Wang, H.-G.; Reed, J.C.; Nicholson, D.W.; Alnemri, E.S. Ordering the cytochrome c–initiated caspase cascade: Hierarchical activation of caspases-2,-3,-6,-7,-8, and-10 in a caspase-9–dependent manner. J. Cell Biol. 1999, 144, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Panda, P.K.; Sinha, N.; Das, D.N.; Bhutia, S.K. Autophagy and apoptosis: Where do they meet? Apoptosis 2014, 19, 555–566. [Google Scholar] [CrossRef]
- Rikka, S.; Quinsay, M.N.; Thomas, R.L.; Kubli, D.A.; Zhang, X.; Murphy, A.N.; Gustafsson, Å.B. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011, 18, 721–731. [Google Scholar] [CrossRef]
- Igaki, T.; Miura, M. Role of Bcl-2 family members in invertebrates. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2004, 1644, 73–81. [Google Scholar] [CrossRef]
- Gooderham, M.; Posso-De Los Rios, C.J.; Rubio-Gomez, G.A.; Papp, K. Interleukin-17 (IL-17) inhibitors in the treatment of plaque psoriasis: A review. Ski. Ther. Lett. 2015, 20, 1–5. [Google Scholar]
- Chenuet, P.; Fauconnier, L.; Madouri, F.; Marchiol, T.; Rouxel, N.; Ledru, A.; Mauny, P.; Lory, R.; Uttenhove, C.; van Snick, J. Neutralization of either IL-17A or IL-17F is sufficient to inhibit house dust mite induced allergic asthma in mice. Clin. Sci. 2017, 131, 2533–2548. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, M.; Li, H.-H.; Long, Q.; Liang, W.; Wen, S.; Wang, M.; Guo, H.-P.; Cheng, X.; Liao, Y.-H. Autophagy contributes to IL-17-induced plasma cell differentiation in experimental autoimmune myocarditis. Int. Immunopharmacol. 2014, 18, 98–105. [Google Scholar] [CrossRef]
- Kim, E.K.; Kwon, J.-E.; Lee, S.-Y.; Lee, E.-J.; Kim, D.S.; Moon, S.-J.; Lee, J.; Kwok, S.-K.; Park, S.-H.; Cho, M.-L. IL-17-mediated mitochondrial dysfunction impairs apoptosis in rheumatoid arthritis synovial fibroblasts through activation of autophagy. Cell Death Dis. 2018, 8, e2565. [Google Scholar] [CrossRef]
- Qiu, A.-W.; Bian, Z.; Mao, P.-A.; Liu, Q.-H. IL-17A exacerbates diabetic retinopathy by impairing Müller cell function via Act1 signaling. Exp. Mol. Med. 2016, 48, e280. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Wang, J.; Manthari, R.K.; Wang, J. Fluoride induces apoptosis and autophagy through the IL-17 signaling pathway in mice hepatocytes. Arch. Toxicol. 2018, 92, 3277–3289. [Google Scholar] [CrossRef]
- Li, F.; Xiang, J. Recent advances in researches on the innate immunity of shrimp in China. Dev. Comp. Immunol. 2013, 39, 11–26. [Google Scholar] [CrossRef]
- Yu, F.; Chen, Z.; Wang, B.; Jin, Z.; Hou, Y.; Ma, S.; Liu, X. The role of lysosome in cell death regulation. Tumor Biol. 2016, 37, 1427–1436. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2002, 2, 647–656. [Google Scholar] [CrossRef]
- Shields, J.D. Climate change enhances disease processes in crustaceans: Case studies in lobsters, crabs, and shrimps. J. Crustac. Biol. 2019, 39, 673–683. [Google Scholar] [CrossRef]
- Yu, N.; Ding, Q.; Li, E.; Qin, J.G.; Chen, L.; Wang, X. Growth, energy metabolism and transcriptomic responses in Chinese mitten crab (Eriocheir sinensis) to benzo [α] pyrene (BaP) toxicity. Aquat. Toxicol. 2018, 203, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Nakamura, K. Metabolic characteristics of the Japanese clam Ruditapes philippinarum (Adams & Reeve) during aerial exposure. Aquac. Res. 2000, 31, 157–165. [Google Scholar]
- Shao, Y.; Li, C.; Xu, W.; Zhang, P.; Zhang, W.; Zhao, X. miR-31 links lipid metabolism and cell apoptosis in bacteria-challenged Apostichopus japonicus via targeting CTRP9. Front. Immunol. 2017, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Cengiz, E.I.; Kan, Y.; Kizmaz, V.; Başhan, M.; Yanar, M. The protective role of vitamin E on the fatty acid composition of phospholipid structure in gill and liver tissues of Oreochromis niloticus exposed to deltamethrin. Ecotoxicol. Environ. Saf. 2012, 80, 381–385. [Google Scholar] [CrossRef]
- Carter, N.S.; Yates, P.; Arendt, C.S.; Boitz, J.M.; Ullman, B. Purine and pyrimidine metabolism in Leishmania. Drug Targets Kinetoplastid Parasites 2008, 625, 141–154. [Google Scholar]
- Chandler, J.D.; Hu, X.; Ko, E.-J.; Park, S.; Lee, Y.-T.; Orr, M.; Fernandes, J.; Uppal, K.; Kang, S.-M.; Jones, D.P. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef]
- Kong, T.; Lin, S.; Ren, X.; Li, S.; Gong, Y. Transcriptome and metabolome integration analysis of mud crab Scylla paramamosain challenged to Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 2020, 103, 430–437. [Google Scholar] [CrossRef]
- Xiao, J.; Li, Q.-Y.; Tu, J.-P.; Chen, X.-L.; Chen, X.-H.; Liu, Q.-Y.; Liu, H.; Zhou, X.-Y.; Zhao, Y.-Z.; Wang, H.-L. Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotoxicol. Environ. Saf. 2019, 180, 491–500. [Google Scholar] [CrossRef]
- Nyhan, W.L. Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 2005, 86, 25–33. [Google Scholar] [CrossRef]


| Name | Gene/Metabolite | Up/Down | Function | Reference |
|---|---|---|---|---|
| Immune system | ||||
| beclin 1-associated autophagy-related key regulator (ATG14L) | gene | up | promote the formation of autophagosomes | [24] |
| ubiquitin-like modifier-activating enzyme ATG7 (ATG7) | gene | up | associated with starvation-induced autophagy | [25] |
| threonine-protein kinase ULK2 (ATG1) | gene | up | induce high levels of autophagy | [26] |
| BCL2/adenovirus E1B 19 kDa protein-interacting protein 3(BNIP3) | gene | up | cell death protein | [27] |
| run domain Beclin-1 interacting and cysteine-rich containing protein (RUBICON) | gene | up | inhibit autophagy | [24] |
| Bcl-2 family protein (BUFFY) | gene | up | inhibit apoptosis | [28] |
| ER protein kinase (PERK) | gene | up | promote apoptosis | [29] |
| Inositol 1,4,5-Trisphosphate Receptor (IP3R) | gene | up | release calcium ions | [30] |
| caspase 7(CASP7) | gene | up | molecular triggers of apoptosis | [31] |
| caspase 4 (CASP4) | gene | up | molecular triggers of apoptosis | [30] |
| Lysozyme (LZM) | gene | down | lysosomal hydrolase | [32] |
| alkaline phosphatase (AKP) | gene | up | lysosomal hydrolase | [32] |
| C-type lectin (CTL) | gene | down | aids in phagocytosis | [33] |
| Antioxidant system | ||||
| peroxiredoxin 5(PRDX5) | gene | up | thioredoxin peroxidase | [34] |
| glutathione S-transferase (GST) | gene | down | removes lipid peroxides and hydrogen peroxide | [35] |
| heat shock protein 70 (HSP70) | gene | up | maintain cellular homeostasis | [36] |
| heat shock protein 90 (HSP90) | gene | up | prevents irreversible protein aggregation | [37] |
| Detoxification | ||||
| glutathione S-transferase (GST) | gene | down | decomposition of pesticides | [38] |
| carboxylesterase (CarEs) | gene | down | enzyme for decomposing pyrethroids | [39] |
| metabolic system | ||||
| succinate | metabolite | up | alternative end product of anaerobic metabolism | [40] |
| arachidonic acid | metabolite | up | involved in immune response | [41] |
| uracil | metabolite | up | related to apoptosis | [42] |
| xanthine | metabolite | up | related to apoptosis | [42] |
| nervous system | ||||
| L-glutamate | metabolite | up | neurotransmitter | [43] |
| acetylcholinesterase (AChE) | gene | down | hydrolyzed acetylcholine | [44] |
| acetylcholine (ACh) | metabolite | up | neurotransmitters | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, M.; Wen, L.; Zhu, M.; Gong, J.; Xi, C.; Wen, H.; Xu, G.; Shen, H. Integrative Analysis of Transcriptome and Metabolome Reveals Molecular Responses in Eriocheir sinensis with Hepatopancreatic Necrosis Disease. Biology 2022, 11, 1267. https://doi.org/10.3390/biology11091267
Zhan M, Wen L, Zhu M, Gong J, Xi C, Wen H, Xu G, Shen H. Integrative Analysis of Transcriptome and Metabolome Reveals Molecular Responses in Eriocheir sinensis with Hepatopancreatic Necrosis Disease. Biology. 2022; 11(9):1267. https://doi.org/10.3390/biology11091267
Chicago/Turabian StyleZhan, Ming, Lujie Wen, Mengru Zhu, Jie Gong, Changjun Xi, Haibo Wen, Gangchun Xu, and Huaishun Shen. 2022. "Integrative Analysis of Transcriptome and Metabolome Reveals Molecular Responses in Eriocheir sinensis with Hepatopancreatic Necrosis Disease" Biology 11, no. 9: 1267. https://doi.org/10.3390/biology11091267
APA StyleZhan, M., Wen, L., Zhu, M., Gong, J., Xi, C., Wen, H., Xu, G., & Shen, H. (2022). Integrative Analysis of Transcriptome and Metabolome Reveals Molecular Responses in Eriocheir sinensis with Hepatopancreatic Necrosis Disease. Biology, 11(9), 1267. https://doi.org/10.3390/biology11091267





