Myosin Transducer Inter-Strand Communication Is Critical for Normal ATPase Activity and Myofibril Structure
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Structure Modeling
2.2. DNA Constructs
2.3. P Element Transformation of Mhc Genes
2.4. Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Determination of Myosin Expression Levels
2.6. Flight Testing
2.7. Confocal Imaging of Hemithoraces and IFM Myofibrils
2.8. Transmission Electron Microscopy
2.9. ATPase and In Vitro Motility Assays
3. Results
3.1. Molecular Modeling of R249 Interactions
3.2. Production and Verification of R249D, D262R and R249D-D262R Transgenic Lines
3.3. Effects of the R249D, D262R and R249D-D262R Mutations on Flight Ability
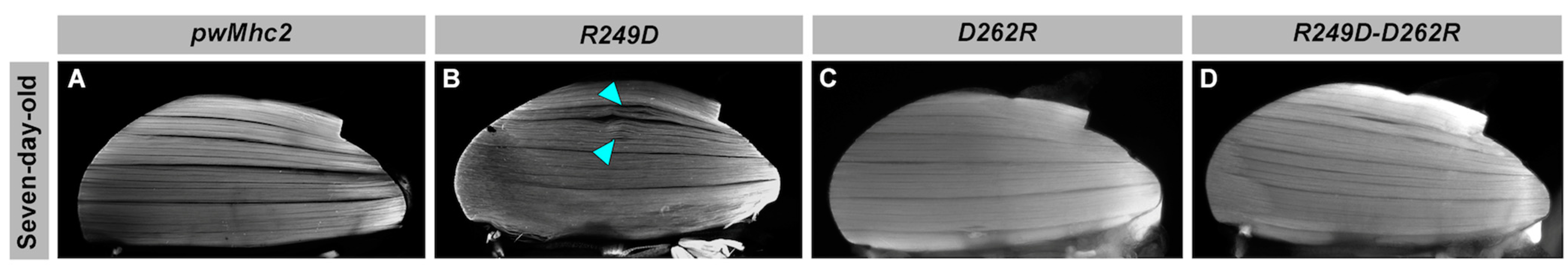
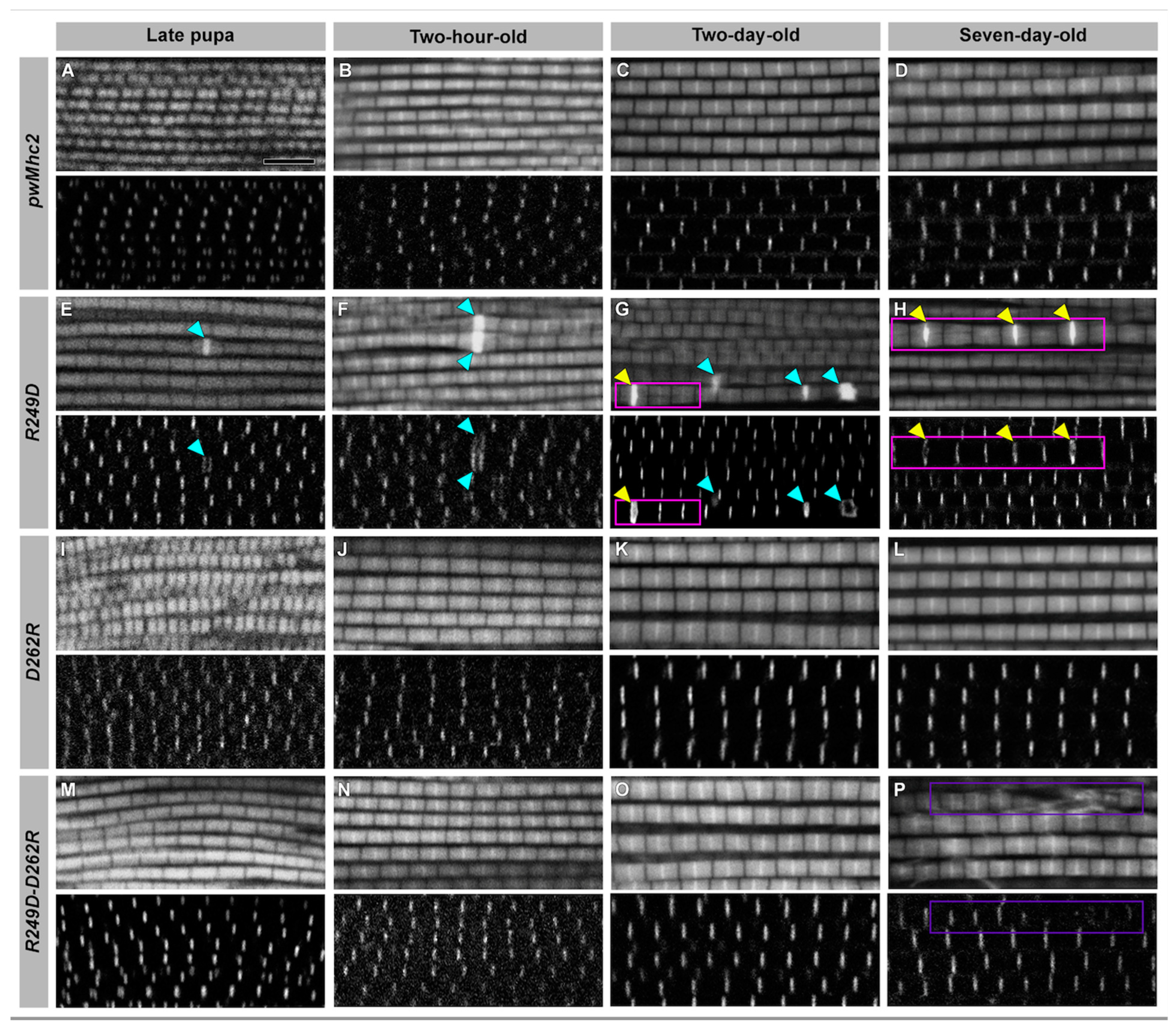
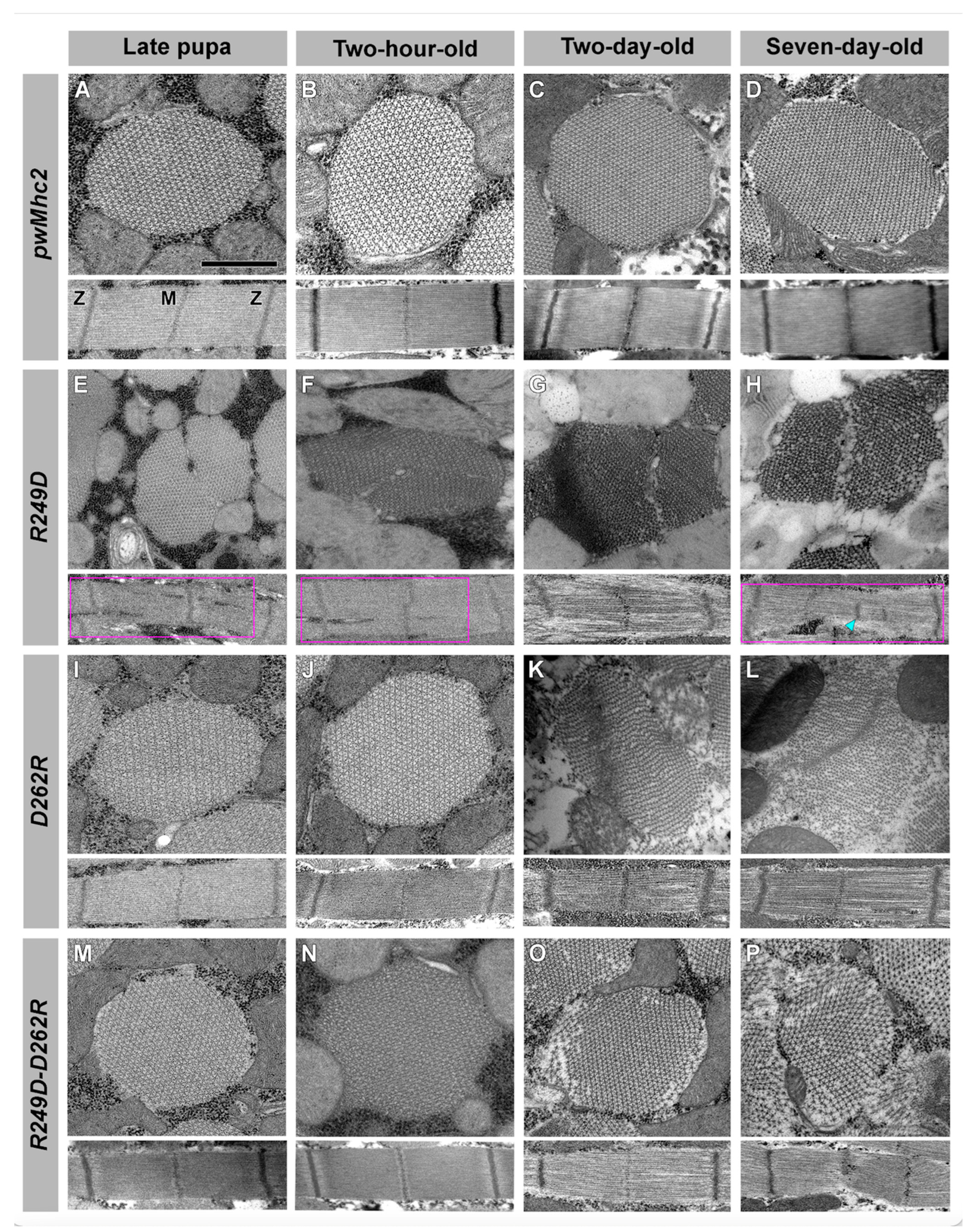
3.4. Effects of the R249D, D262R and R249D-D262R Mutations on Muscle Ultrastructure
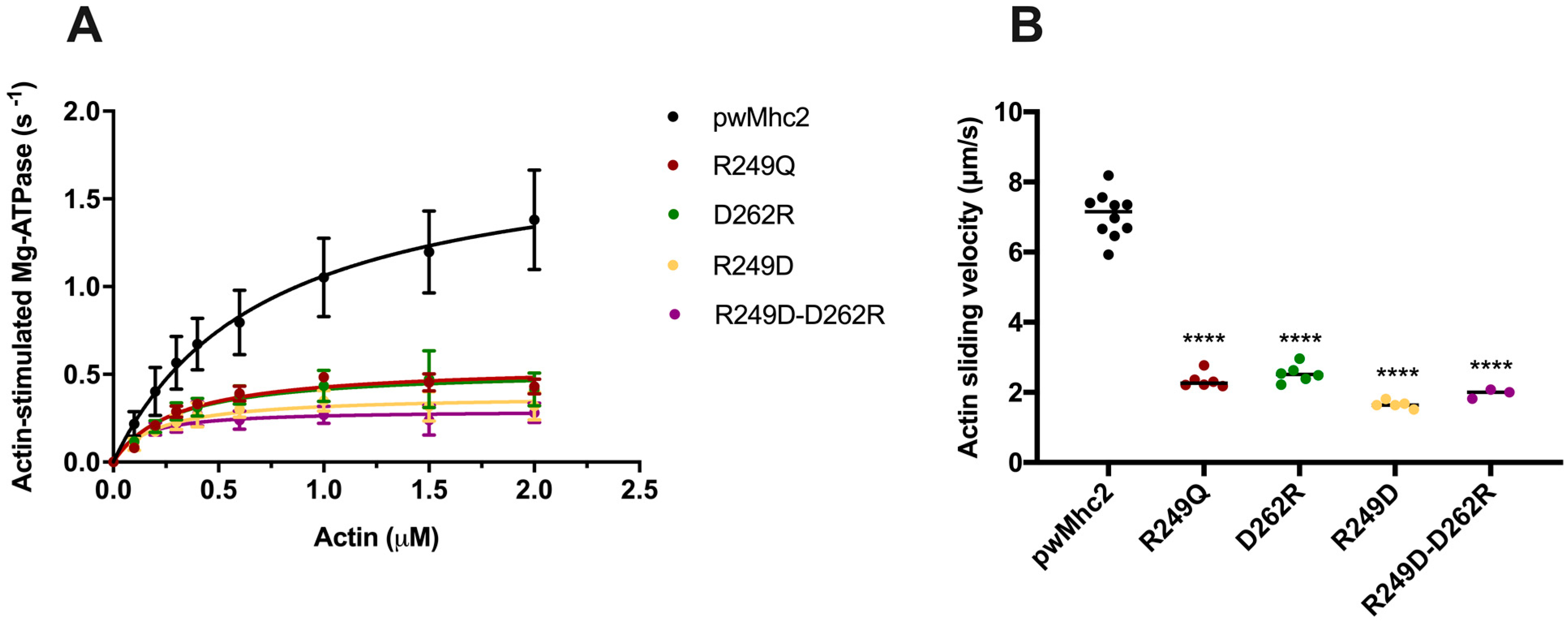
3.5. Effects of the R249Q, R249D, D262R and R249D-D262R Mutations on Myosin ATPase and Actin Sliding Velocity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sweeney, H.L.; Houdusse, A. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 2010, 39, 539–557. [Google Scholar] [CrossRef]
- Coureux, P.D.; Sweeney, H.L.; Houdusse, A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004, 23, 4527–4537. [Google Scholar] [CrossRef] [PubMed]
- Wulf, S.F.; Ropars, V.; Fujita-Becker, S.; Oster, M.; Hofhaus, G.; Trabuco, L.G.; Pylypenko, O.; Sweeney, H.L.; Houdusse, A.M.; Schroder, R.R. Force-producing ADP state of myosin bound to actin. Proc. Natl. Acad. Sci. USA 2016, 113, E1844–E1852. [Google Scholar] [CrossRef] [PubMed]
- Pospich, S.; Sweeney, H.L.; Houdusse, A.; Raunser, S. High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism. Elife 2021, 10, e73724. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.; Watkins, H.; Hwang, D.S.; Miri, M.; McKenna, W.; Traill, T.A.; Seidman, J.G.; Seidman, C.E. Preclinical diagnosis of familial hypertrophic cardiomyopathy by genetic analysis of blood lymphocytes. N. Engl. J. Med. 1991, 325, 1753–1760. [Google Scholar] [CrossRef]
- Greber-Platzer, S.; Marx, M.; Fleischmann, C.; Suppan, C.; Dobner, M.; Wimmer, M. Beta-myosin heavy chain gene mutations and hypertrophic cardiomyopathy in Austrian children. J. Mol. Cell. Cardiol. 2001, 33, 141–148. [Google Scholar] [CrossRef]
- Woo, A.; Rakowski, H.; Liew, J.C.; Zhao, M.S.; Liew, C.C.; Parker, T.G.; Zeller, M.; Wigle, E.D.; Sole, M.J. Mutations of the beta myosin heavy chain gene in hypertrophic cardiomyopathy: Critical functional sites determine prognosis. Heart 2003, 89, 1179–1185. [Google Scholar] [CrossRef]
- Bell, K.M.; Kronert, W.A.; Huang, A.; Bernstein, S.I.; Swank, D.M. The R249Q hypertrophic cardiomyopathy myosin mutation decreases contractility in Drosophila by impeding force production. J. Physiol. 2019, 597, 2403–2420. [Google Scholar] [CrossRef]
- Adhikari, A.S.; Trivedi, D.V.; Sarkar, S.S.; Song, D.; Kooiker, K.B.; Bernstein, D.; Spudich, J.A.; Ruppel, K.M. Beta-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 2019, 10, 2685. [Google Scholar] [CrossRef]
- Robert-Paganin, J.; Auguin, D.; Houdusse, A. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 2018, 9, 4019. [Google Scholar] [CrossRef]
- Woodhead, J.L.; Craig, R. The mesa trail and the interacting heads motif of myosin II. Arch. Biochem. Biophys. 2020, 680, 108228. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Swank, D.M.; Wells, L.; Kronert, W.A.; Morrill, G.E.; Bernstein, S.I. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc. Res. Tech. 2000, 50, 430–442. [Google Scholar] [CrossRef]
- Thummel, C.; Pirrotta, V. Technical notes: New pCasper P-element vectors. Dros. Inf. Serv. 1992, 71, 150. [Google Scholar]
- Rubin, G.M.; Spradling, A.C. Genetic transformation of Drosophila with transposable element vectors. Science 1982, 218, 348–353. [Google Scholar] [CrossRef]
- Collier, V.L.; Kronert, W.A.; O’Donnell, P.T.; Edwards, K.A.; Bernstein, S.I. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990, 4, 885–895. [Google Scholar] [CrossRef]
- Becker, K.D.; O’Donnell, P.T.; Heitz, J.M.; Vito, M.; Bernstein, S.I. Analysis of Drosophila paramyosin: Identification of a novel isoform which is restricted to a subset of adult muscles. J. Cell Biol. 1992, 116, 669–681. [Google Scholar] [CrossRef]
- O’Donnell, P.T.; Collier, V.L.; Mogami, K.; Bernstein, S.I. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989, 3, 1233–1246. [Google Scholar] [CrossRef]
- Drummond, D.R.; Hennessey, E.S.; Sparrow, J.C. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol. Gen. Genet. 1991, 226, 70–80. [Google Scholar] [CrossRef]
- Tohtong, R.; Yamashita, H.; Graham, M.; Haeberle, J.; Simcox, A.; Maughan, D. Impairment of muscle function caused by mutations of phosphorylation sites in myosin regulatory light chain. Nature 1995, 374, 650–653. [Google Scholar] [CrossRef]
- O’Donnell, P.T.; Bernstein, S.I. Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: Differential effects on muscle function produced by similar thick filament abnormalities. J. Cell Biol. 1988, 107, 2601–2612. [Google Scholar] [CrossRef] [PubMed]
- Swank, D.M.; Bartoo, M.L.; Knowles, A.F.; Iliffe, C.; Bernstein, S.I.; Molloy, J.E.; Sparrow, J.C. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J. Biol. Chem. 2001, 276, 15117–15124. [Google Scholar] [CrossRef] [PubMed]
- Kronert, W.A.; Dambacher, C.M.; Knowles, A.F.; Swank, D.M.; Bernstein, S.I. Alternative relay domains of Drosophila melanogaster myosin differentially affect ATPase activity, in vitro motility, myofibril structure and muscle function. J. Mol. Biol. 2008, 379, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.S.; Kronert, W.A.; Guo, Y.; Hsu, K.H.; Sarsoza, F.; Bernstein, S.I. Reductions in ATPase activity, actin sliding velocity, and myofibril stability yield muscle dysfunction in Drosophila models of myosin-based Freeman-Sheldon syndrome. Mol. Biol. Cell 2019, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Pardee, J.D.; Spudich, J.A. Purification of muscle actin. Methods Enzym. 1982, 85 Pt B, 164–181. [Google Scholar] [CrossRef]
- Caldwell, J.T.; Mermelstein, D.J.; Walker, R.C.; Bernstein, S.I.; Huxford, T. X-ray crystallographic and molecular dynamic analyses of Drosophila melanogaster embryonic muscle myosin define domains responsible for isoform-specific properties. J. Mol. Biol. 2020, 432, 427–447. [Google Scholar] [CrossRef] [PubMed]
- Katti, P.; Thimmaya, D.; Madan, A.; Nongthomba, U. Overexpression of miRNA-9 generates muscle hypercontraction through translational repression of troponin-T in Drosophila melanogaster indirect flight muscles. G3 Genes Genomes Genet. 2017, 7, 3521–3531. [Google Scholar] [CrossRef][Green Version]
- Gonzalez-Morales, N.; Holenka, T.K.; Schock, F. Filamin actin-binding and titin-binding fulfill distinct functions in Z-disc cohesion. PLoS Genet. 2017, 13, e1006880. [Google Scholar] [CrossRef]
- Loison, O.; Weitkunat, M.; Kaya-Copur, A.; Nascimento Alves, C.; Matzat, T.; Spletter, M.L.; Luschnig, S.; Brasselet, S.; Lenne, P.F.; Schnorrer, F. Polarization-resolved microscopy reveals a muscle myosin motor-independent mechanism of molecular actin ordering during sarcomere maturation. PLoS Biol. 2018, 16, e2004718. [Google Scholar] [CrossRef]
- Spletter, M.L.; Barz, C.; Yeroslaviz, A.; Zhang, X.; Lemke, S.B.; Bonnard, A.; Brunner, E.; Cardone, G.; Basler, K.; Habermann, B.H.; et al. A transcriptomics resource reveals a transcriptional transition during ordered sarcomere morphogenesis in flight muscle. Elife 2018, 7, e34058. [Google Scholar] [CrossRef]
- Kao, S.Y.; Nikonova, E.; Chaabane, S.; Sabani, A.; Martitz, A.; Wittner, A.; Heemken, J.; Straub, T.; Spletter, M.L. A candidate RNAi screen reveals diverse RNA-binding protein phenotypes in Drosophila flight muscle. Cells 2021, 10, 2505. [Google Scholar] [CrossRef]
- Vander Roest, A.S.; Liu, C.; Morck, M.M.; Kooiker, K.B.; Jung, G.; Song, D.; Dawood, A.; Jhingran, A.; Pardon, G.; Ranjbarvaziri, S.; et al. Hypertrophic cardiomyopathy beta-cardiac myosin mutation (P710R) leads to hypercontractility by disrupting super relaxed state. Proc. Natl. Acad. Sci. USA 2021, 118, e2025030118. [Google Scholar] [CrossRef]
- Spudich, J.A. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflug. Arch. 2019, 471, 701–717. [Google Scholar] [CrossRef]
- Nag, S.; Trivedi, D.V.; Sarkar, S.S.; Adhikari, A.S.; Sunitha, M.S.; Sutton, S.; Ruppel, K.M.; Spudich, J.A. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 2017, 24, 525–533. [Google Scholar] [CrossRef]
- Alamo, L.; Pinto, A.; Sulbaran, G.; Mavarez, J.; Padron, R. Lessons from a tarantula: New insights into myosin interacting-heads motif evolution and its implications on disease. Biophys. Rev. 2018, 10, 1465–1477. [Google Scholar] [CrossRef]
- Chu, S.; Muretta, J.M.; Thomas, D.D. Direct detection of the myosin super-relaxed state and interacting-heads motif in solution. J. Biol. Chem. 2021, 297, 101157. [Google Scholar] [CrossRef]
- Hooijman, P.; Stewart, M.A.; Cooke, R. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 2011, 100, 1969–1976. [Google Scholar] [CrossRef]
- Lee, K.H.; Sulbaran, G.; Yang, S.; Mun, J.Y.; Alamo, L.; Pinto, A.; Sato, O.; Ikebe, M.; Liu, X.; Korn, E.D.; et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc. Natl. Acad. Sci. USA 2018, 115, E1991–E2000. [Google Scholar] [CrossRef]
- Menetret, J.F.; Schroder, R.R.; Hofmann, W. Cryo-electron microscopic studies of relaxed striated muscle thick filaments. J. Muscle Res. Cell Motil. 1990, 11, 1–11. [Google Scholar] [CrossRef]
- Daneshparvar, N.; Taylor, D.W.; O’Leary, T.S.; Rahmani, H.; Abbasiyeganeh, F.; Previs, M.J.; Taylor, K.A. CryoEM structure of Drosophila flight muscle thick filaments at 7 Å resolution. Life Sci. Alliance 2020, 3, e202000823. [Google Scholar] [CrossRef]
- Alamo, L.; Qi, D.; Wriggers, W.; Pinto, A.; Zhu, J.; Bilbao, A.; Gillilan, R.E.; Hu, S.; Padron, R. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J. Mol. Biol. 2016, 428, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Phung, L.A.; Foster, A.D.; Miller, M.S.; Lowe, D.A.; Thomas, D.D. Super-relaxed state of myosin in human skeletal muscle is fiber-type dependent. Am. J. Physiol. Cell Physiol. 2020, 319, C1158–C1162. [Google Scholar] [CrossRef] [PubMed]
- Maughan, D.; Moore, J.; Vigoreaux, J.; Barnes, B.; Mulieri, L.A. Work production and work absorption in muscle strips from vertebrate cardiac and insect flight muscle fibers. Adv. Exp. Med. Biol. 1998, 453, 471–480. [Google Scholar] [CrossRef] [PubMed]

| Line Name | Chromosomal Location a | Protein Accumulation ± SEM b |
|---|---|---|
| pwMhc2 | X | 1.00 ± 0.04 |
| pwMhcR249D-6 | 3 | 0.98 ± 0.07 |
| pwMhcR249D-12 | 3 | 0.95 ± 0.06 |
| pwMhcD262R-1 | 3 | 0.96 ± 0.09 |
| pwMhcD262R-6 | 3 | 0.95 ± 0.02 |
| pwMhcD262R-7 | 3 | 0.97 ± 0.01 |
| pwMhcR249D-D262R-2 | 3 | 0.95 ± 0.02 |
| pwMhcR249D-D262R-4 | 4 | 0.95 ± 0.01 |
| pwMhcR249D-D262R-5 | 3 | 0.91 ± 0.02 |
| Line Name | Fly Age (Days) | Number Tested | Up (%) a | Horizontal (%) | Down (%) | Not at All (%) | Flight Index ± SEM b |
|---|---|---|---|---|---|---|---|
| pwMhc2 | 2 | 148 | 54.1 | 24.3 | 17.6 | 4.1 | 4.6 ± 0.01 |
| pwMhcR249D-6 | 2 | 119 | 0 | 0 | 0 | 100 | 0 |
| pwMhcR249D-12 | 2 | 127 | 0 | 0 | 0 | 100 | 0 |
| pwMhcD262R-1 | 2 | 130 | 0.8 | 18.5 | 40.8 | 40.0 | 1.6 ± 0.02 |
| pwMhcD262R-6 | 2 | 121 | 1.7 | 19.0 | 40.5 | 38.8 | 1.7 ± 0.03 |
| pwMhcD262R-7 | 2 | 121 | 0.8 | 18.2 | 41.3 | 40.0 | 1.6 ± 0.03 |
| pwMhcR249D-D262R-2 | 2 | 128 | 0 | 0 | 15.6 | 84.4 | 0.3 ± 0.01 |
| pwMhcR249D-D262R-4 | 2 | 132 | 0 | 0 | 15.2 | 84.8 | 0.3 ± 0.01 |
| pwMhcR249D-D262R-5 | 2 | 130 | 0 | 0 | 15.4 | 84.6 | 0.3 ± 0.01 |
| pwMhc2 | 7 | 116 | 37.1 | 36.2 | 21.6 | 5.2 | 4.1 ± 0.02 |
| pwMhcD262R-1 | 7 | 131 | 0 | 0 | 44.3 | 55.7 | 0.88 ± 0.02 |
| pwMhcD262R-6 | 7 | 131 | 0 | 0 | 42.7 | 57.3 | 0.85 ± 0.02 |
| pwMhcD262R-7 | 7 | 117 | 0 | 0 | 35.9 | 64.1 | 0.72 ± 0.01 |
| pwMhcR249D-D262R-2 | 7 | 118 | 0 | 0 | 0 | 100 | 0 |
| pwMhcR249D-D262R-4 | 7 | 116 | 0 | 0 | 0 | 100 | 0 |
| pwMhcR249D-D262R-5 | 7 | 119 | 0 | 0 | 0 | 100 | 0 |
| Myosin Type (n for ATPase/n for Motility) | Basal Mg-ATPase (s−1) | Actin-Stimulated Vmax (s−1) | Actin-Stimulated Km (µM) | Actin Velocity (µm/s) |
|---|---|---|---|---|
| IFM control- pwMhc2 (18/10) | 0.30 ± 0.11 a | 1.79 ± 0.41 d | 0.71 ± 0.23 e | 7.06 ± 0.65 f |
| R249Q (6/6) | 0.25 ± 0.04 b | 0.56 ± 0.11 | 0.31 ± 0.09 | 2.34 ± 0.22 g |
| R249D (5/5) | 0.12 ± 0.05 c | 0.40 ± 0.22 | 0.22 ± 0.17 | 1.66 ± 0.11 h |
| D262R (4/6) | 0.11 ± 0.04 c | 0.58 ± 0.33 | 0.34 ± 0.30 | 2.54 ± 0.25 i |
| R249D-D262R (4/3) | 0.24 ± 0.03 | 0.30 ± 0.15 | 0.13 ± 0.09 | 1.97 ± 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kronert, W.A.; Hsu, K.H.; Madan, A.; Sarsoza, F.; Cammarato, A.; Bernstein, S.I. Myosin Transducer Inter-Strand Communication Is Critical for Normal ATPase Activity and Myofibril Structure. Biology 2022, 11, 1137. https://doi.org/10.3390/biology11081137
Kronert WA, Hsu KH, Madan A, Sarsoza F, Cammarato A, Bernstein SI. Myosin Transducer Inter-Strand Communication Is Critical for Normal ATPase Activity and Myofibril Structure. Biology. 2022; 11(8):1137. https://doi.org/10.3390/biology11081137
Chicago/Turabian StyleKronert, William A., Karen H. Hsu, Aditi Madan, Floyd Sarsoza, Anthony Cammarato, and Sanford I. Bernstein. 2022. "Myosin Transducer Inter-Strand Communication Is Critical for Normal ATPase Activity and Myofibril Structure" Biology 11, no. 8: 1137. https://doi.org/10.3390/biology11081137
APA StyleKronert, W. A., Hsu, K. H., Madan, A., Sarsoza, F., Cammarato, A., & Bernstein, S. I. (2022). Myosin Transducer Inter-Strand Communication Is Critical for Normal ATPase Activity and Myofibril Structure. Biology, 11(8), 1137. https://doi.org/10.3390/biology11081137







