CRISPRi-Mediated Down-Regulation of the Cinnamate-4-Hydroxylase (C4H) Gene Enhances the Flavonoid Biosynthesis in Nicotiana tabacum
Abstract
Simple Summary
Abstract
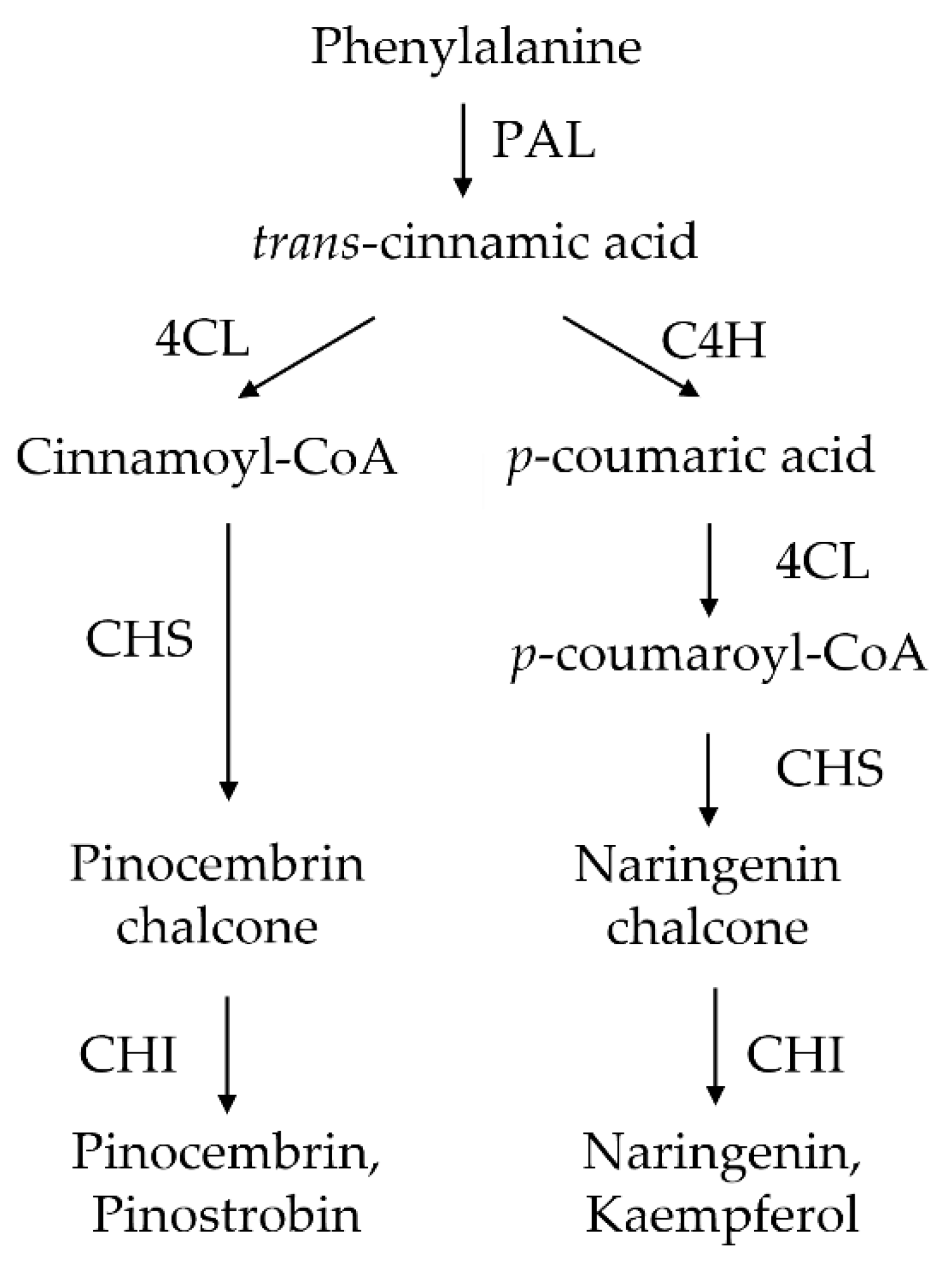
1. Introduction
2. Materials and Methods
2.1. Plant Material and Establishment of Cell Suspension Culture
2.2. sgRNA Design
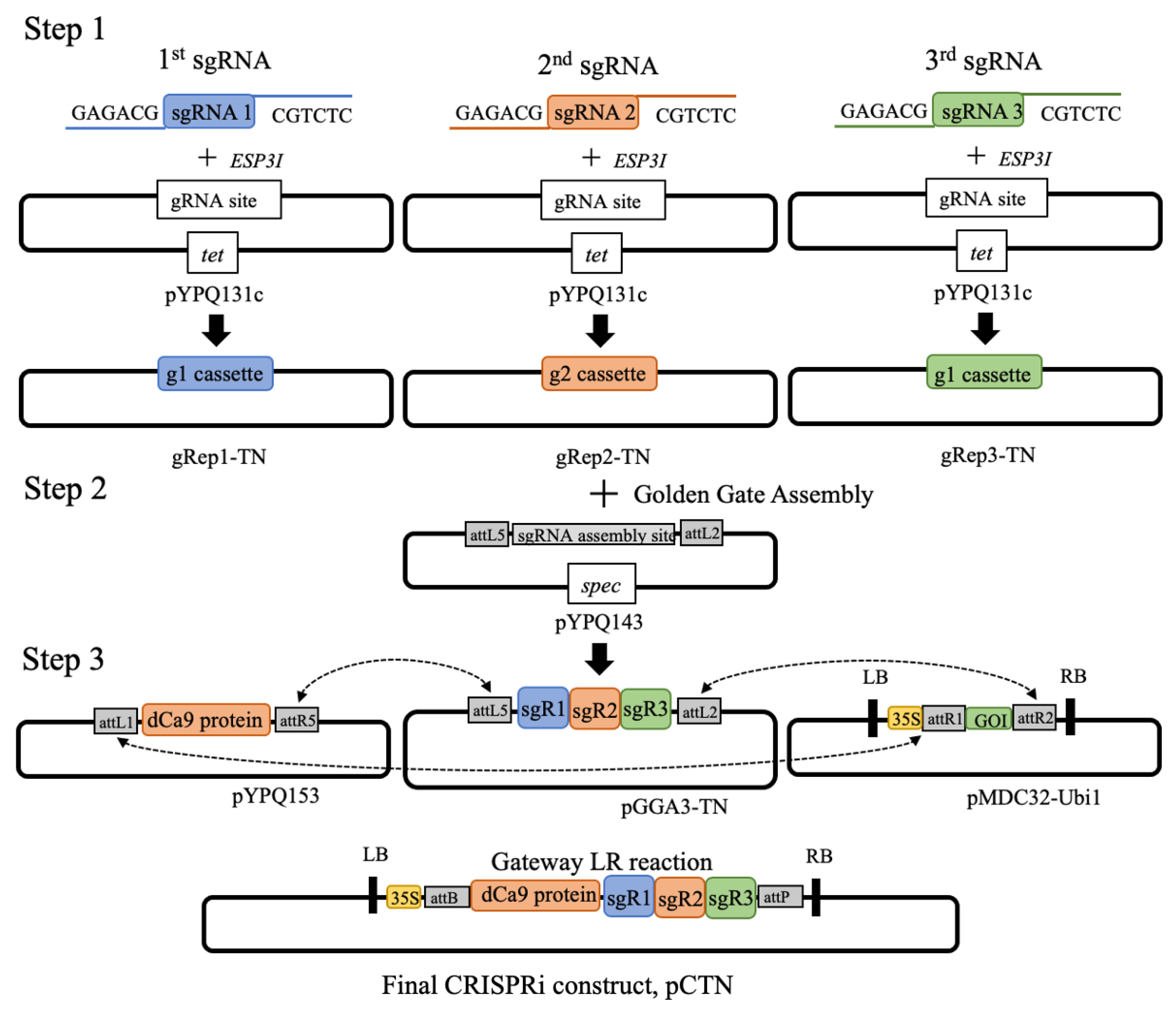
2.3. Cloning of sgRNAs into Expression Vectors
2.4. Golden Gate Assembly of Multiple sgRNAs
2.5. CRISPRi Vector Construction
2.6. Plant Transformation
2.7. PCR Amplification
2.8. Quantitative Real-Time PCR (qPCR)
2.9. Extraction of Metabolites
2.10. LC-MS Analysis
2.11. Statistical Analysis
3. Results
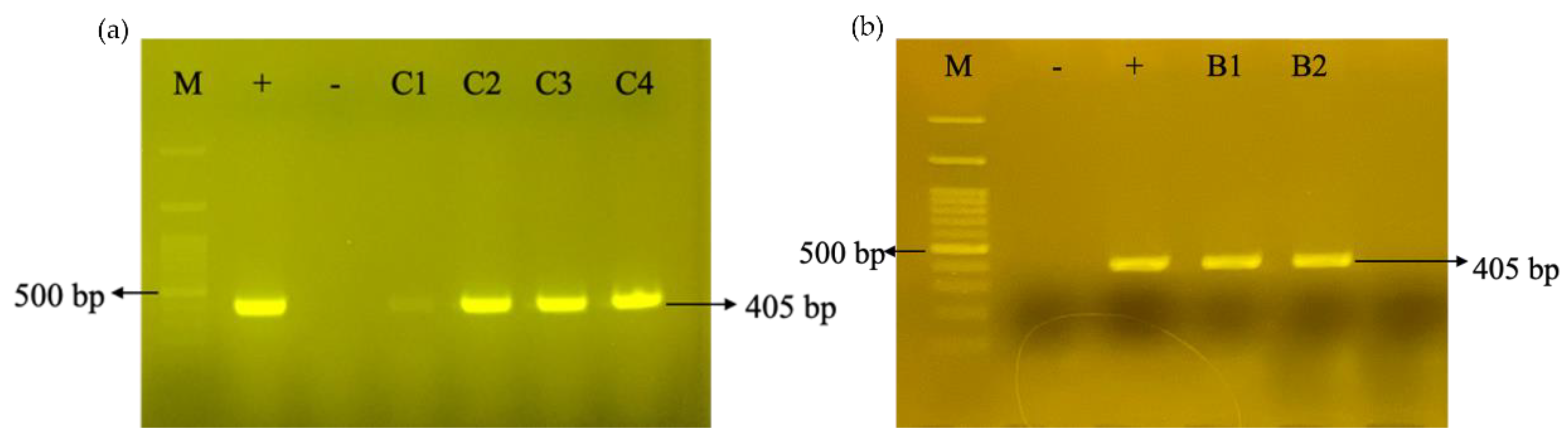
3.1. Introducing the CRISPRi Silencing Vector into N. tabacum
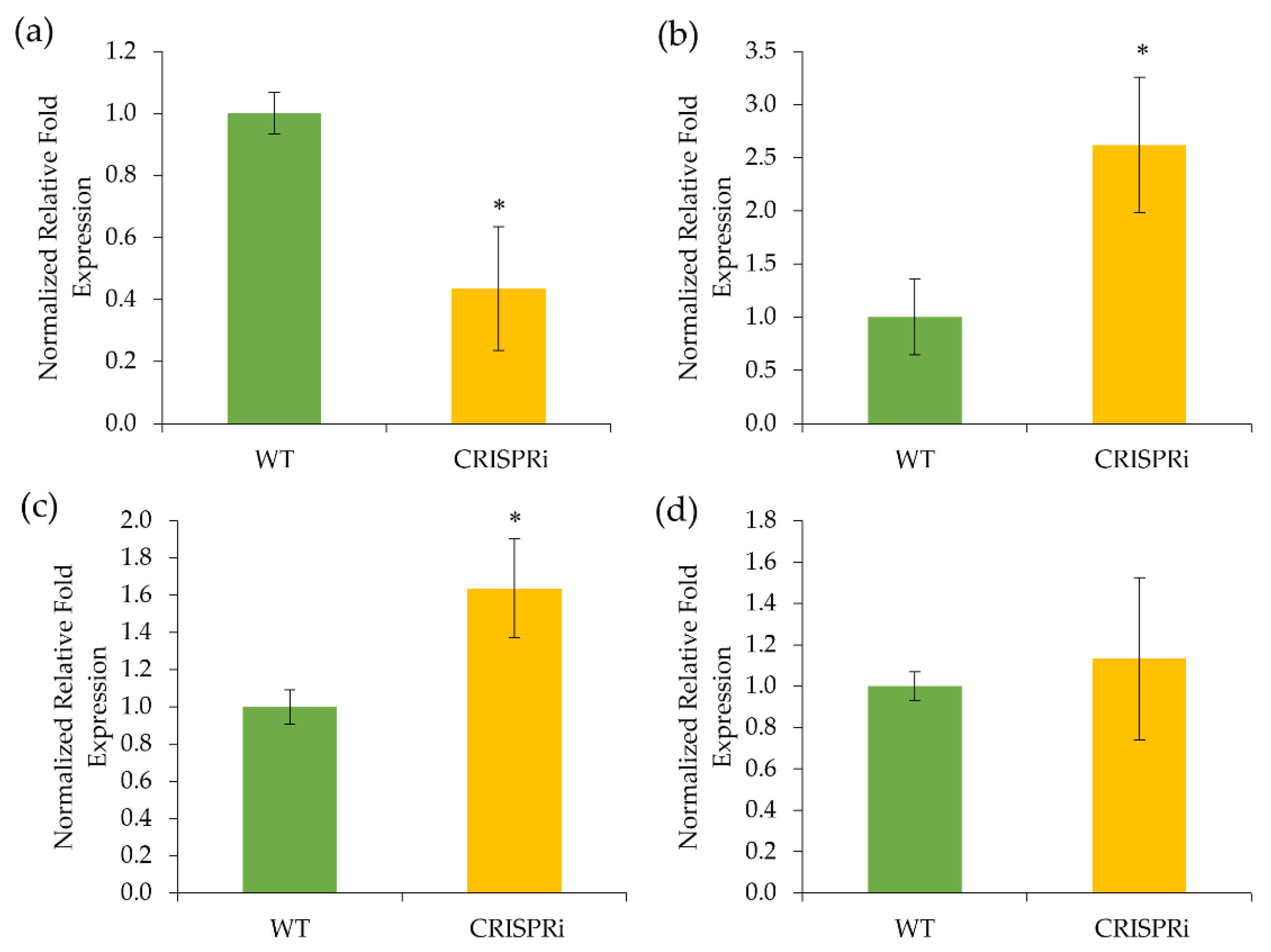
3.2. Gene Expression of the C4H-Silenced Cells
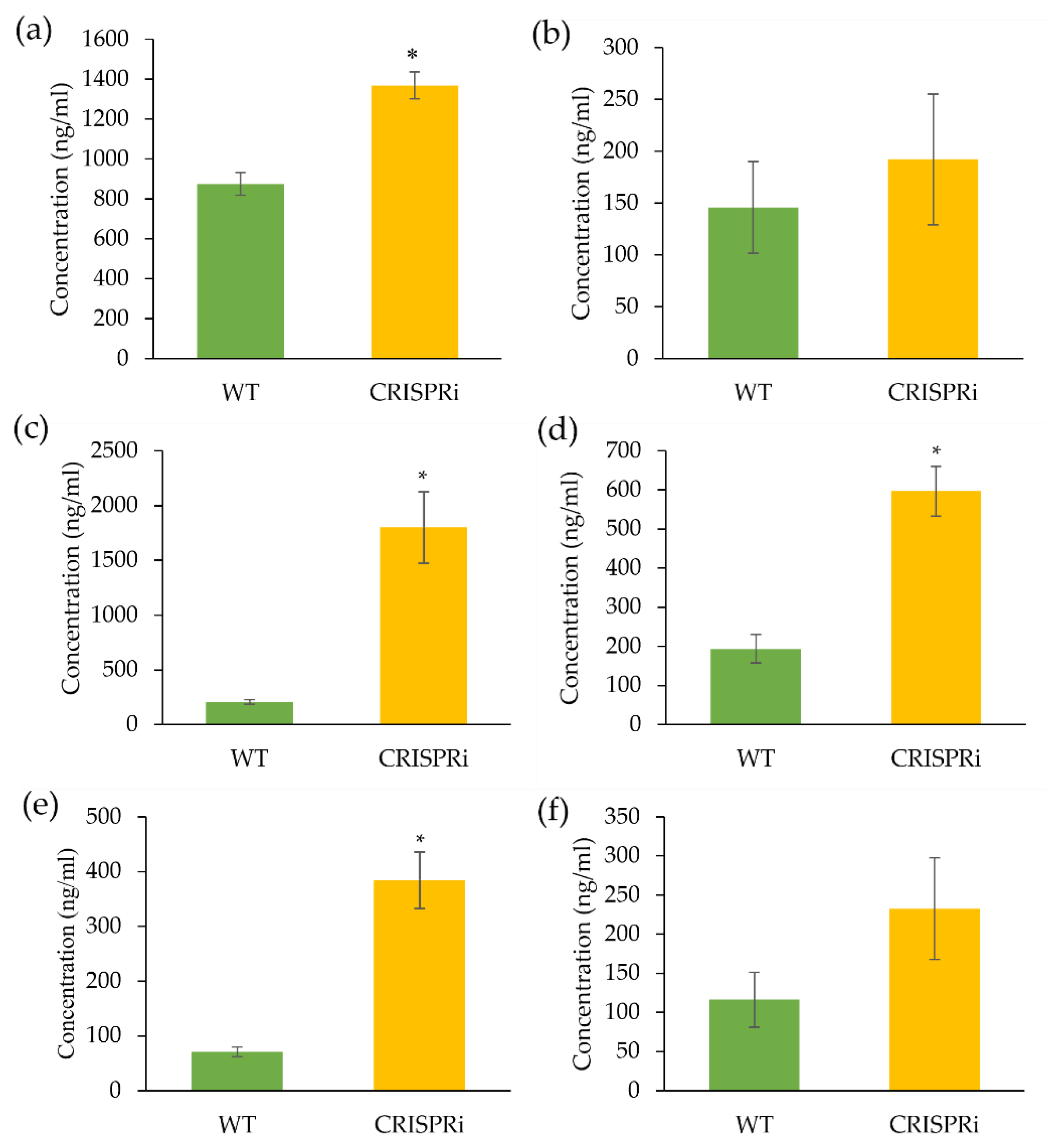
3.3. LC-MS Analysis of Flavonoids in the C4H-Silenced Cells
4. Discussion
4.1. The CRISPRi System Is a Powerful Tool for Silencing C4H
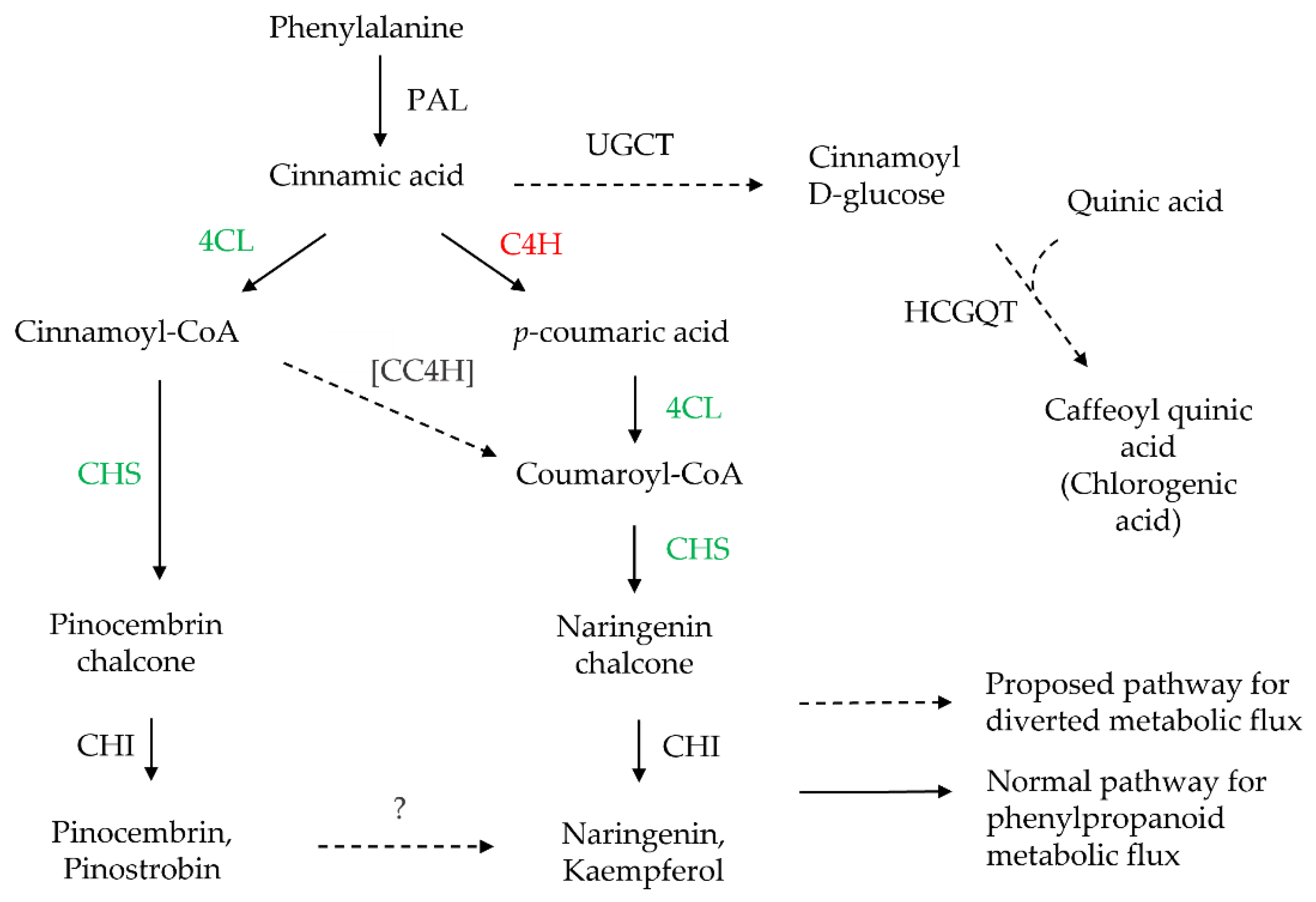
4.2. The Flavonoid-Related Genes Were Upregulated in the C4H-Silenced Cells
4.3. The Flavonoid Production Was Altered in the C4H-Silenced Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mutha, R.E.; Tatiya, A.U.; Surana, S.J. Flavonoids as natural phenolic compounds and their role in therapeutics: An overview. Futur. J. Pharm. Sci. 2021, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Liew, Y.J.M.; Lee, Y.K.; Khalid, N.; Rahman, N.A.; Tan, B.C. Enhancing flavonoid production by promiscuous activity of prenyltransferase, BrPT2 from Boesenbergia Rotunda. PeerJ 2020, 8, e9094. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Hu, L.; Lu, H.; Wei, T.; Chen, Q. Metabolic engineering of microbial cell factories for biosynthesis of flavonoids: A review. Molecules 2021, 26, 4522. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, J.; Liu, X.; Meng, M.; Wang, J.; Lin, J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics 2020, 112, 1781–1794. [Google Scholar] [CrossRef]
- Tan, B.C.; Tan, S.K.; Wong, S.M.; Ata, N.; Rahman, N.A.; Khalid, N. Distribution of flavonoids and cyclohexenyl chalcone derivatives in conventional propagated and in vitro-derived field-grown Boesenbergia rotunda(L.) Mansf. Evid. Based Complement. Alternat. Med. 2015, 2015, 451870. [Google Scholar] [CrossRef]
- Dudek, B.; Warskulat, A.-C.; Schneider, B. The occurrence of flavonoids and related compounds in flower sections of Papaver nudicaule. Plants 2016, 5, 28. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Wang, Y.; Wu, Z.; Fu, C.; Lin, H.; et al. NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef]
- Shojaie, B.; Mostajeran, A.; Ghannadian, M. Flavonoid dynamic responses to different drought conditions: Amount, type, and localization of flavonols in roots and shoots of Arabidopsis thaliana L. Turk. J. Biol. 2016, 40, 612–622. [Google Scholar] [CrossRef]
- Shah, A.; Smith, D.L. Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Udomthanadech, K.; Vajrodaya, S.; Paisooksantivatana, Y. Antibacterial properties of the extracts from some Zingibereous species in Thailand against bacteria causing diarrhea and food poisoning in human. Int. Transact. J. Eng. Manag. Appl. Sci. Technol. 2015, 6, 203–213. [Google Scholar] [CrossRef]
- Wu, N.; Kong, Y.; Zu, Y.; Fu, Y.; Liu, Z.; Meng, R.; Liu, X.; Efferth, T. Activity investigation of pinostrobin towards herpes simplex virus-1 as determined by atomic force microscopy. Phytomedicine 2011, 18, 110–118. [Google Scholar] [CrossRef]
- Chiang, M.; Kurmoo, Y.; Khoo, T.-J. Chemical and cell-based antioxidant capacity of methanolic extracts of three commonly edible plants from Zingiberaceae Family. Free Radic. 2016, 7, 57–62. [Google Scholar] [CrossRef]
- Isa, N.M.; Abdelwahab, S.I.; Mohan, S.; Abdul, A.B.; Sukari, M.A.; Taha, M.M.E.; Syam, S.; Narrima, P.; Cheah, S.C.; Ahmad, S.; et al. In Vitro anti-inflammatory, cytotoxic and antioxidant activities of boesenbergin A, a chalcone isolated from Boesenbergia rotunda (L.) (fingerroot). Braz. J. Med. Biol. Res. 2012, 45, 524–530. [Google Scholar] [CrossRef]
- Abdelwahab, S.I.; Mohan, S.; Abdulla, M.A.; Sukari, M.A.; Abdul, A.B.; Taha, M.M.; Syam, S.; Ahmad, S.; Lee, K.H. The methanolic extract of Boesenbergia rotunda (L.) Mansf. and its major compound pinostrobin induces anti-ulcerogenic property In Vivo: Possible involvement of indirect antioxidant action. J. Ethnopharmacol. 2011, 137, 963–970. [Google Scholar] [CrossRef]
- Cheah, S.-C.; Appleton, D.R.; Lee, S.-T.; Lam, M.-L.; Hadi, A.H.A.; Mustafa, M.R. Panduratin A inhibits the growth of A549 cells through induction of apoptosis and inhibition of NF-KappaB translocation. Molecules 2011, 16, 2583–2598. [Google Scholar] [CrossRef]
- Kim, D.U.; Chung, H.C.; Kim, C.; Hwang, J.K. Oral intake of Boesenbergia pandurata extract improves skin hydration, gloss, and wrinkling: A randomized, double-blind, and placebo-controlled study. J. Cosmet. Dermatol. 2017, 16, 512–519. [Google Scholar] [CrossRef]
- Vinayagam, R.; Xu, B. Antidiabetic properties of dietary flavonoids: A cellular mechanism review. Nutr. Metab. 2015, 12, 1–20. [Google Scholar] [CrossRef]
- Amelia, F.; Iryani; Sari, P.Y.; Parikesit, A.A.; Bakri, R.; Toepak, E.P.; Tambunan, U.S.F. Assessment of drug binding potential of pockets in the NS2B/NS3 Dengue virus protein. IOP Conf. Ser. Mater. Sci. Eng. 2018, 349, 012021. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Wang, L.; Shi, H.; Wu, J.; Cao, F. Alternative partial root-zone irrigation enhances leaf flavonoid accumulation and water use efficiency of Ginkgo biloba. New For. 2015, 47, 377–391. [Google Scholar] [CrossRef]
- Hernandez, I.; Alegre, L.; Munne-Bosch, S. Drought-induced changes in flavonoids and other low molecular weight antioxidants in Cistus clusii grown under Mediterranean field conditions. Tree Physiol. 2004, 24, 1303–1311. [Google Scholar] [CrossRef]
- Farré, G.; Blancquaert, D.; Capell, T.; Van Der Straeten, D.; Christou, P.; Zhu, C. Engineering complex metabolic pathways in plants. Annu. Rev. Plant Biol. 2014, 65, 187–223. [Google Scholar] [CrossRef]
- Yin, Y.-C.; Hou, J.-M.; Tian, S.-K.; Yang, L.; Zhang, Z.-X.; Li, W.-D.; Liu, Y. Overexpressing chalcone synthase (CHS) gene enhanced flavonoids accumulation in Glycyrrhiza uralensis hairy roots. Bot. Lett. 2019, 167, 219–231. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Xin, Y.; Wang, G.; Xu, L.A. Overexpression of the GbF3’H1 gene enhanced the epigallocatechin, gallocatechin, and catechin contents in transgenic Populus. J. Agric. Food Chem. 2020, 68, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/dCas9-based systems: Mechanisms and applications in plant sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Murahige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Shahmuradov, I.A.; Umarov, R.K.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 2017, 45, e65. [Google Scholar] [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Cloning and transformation with plasmid vectors. In Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; pp. 157–260. [Google Scholar]
- Shumin, Z.; Yanxia, C.; Bang, Z.; Wei, Z. Optimization of BY2 cell suspension as a stable transformable system. Not. Bot. Horti Agrobot. Cluj-Napoca 2014, 42, 472–477. [Google Scholar] [CrossRef][Green Version]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Toni, L.S.; Garcia, A.M.; Jeffrey, D.A.; Jiang, X.; Stauffer, B.L.; Miyamoto, S.D.; Sucharov, C.C. Optimization of phenol-chloroform RNA extraction. MethodsX 2018, 5, 599–608. [Google Scholar] [CrossRef]
- Rostami, Z.; Fazeli, A.; Hojati, Z. The isolation and expression analysis of cinnamate 4-hydroxylase and chalcone synthase genes of Scrophularia striata under different abiotic elicitors. Sci. Rep. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef]
- Cho, S.; Shin, J.; Cho, B.-K. Applications of CRISPR/Cas system to bacterial metabolic engineering. Int. J. Mol. Sci. 2018, 19, 1089. [Google Scholar] [CrossRef]
- Zhang, M.M.; Wong, F.T.; Wang, Y.; Luo, S.; Lim, Y.H.; Heng, E.; Yeo, W.L.; Cobb, R.E.; Enghiad, B.; Ang, E.L.; et al. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017, 13, 607–609. [Google Scholar] [CrossRef]
- Moscatiello, R.; Baldan, B.; Navazio, L. Plant cell suspension cultures. Methods Mol. Biol. 2013, 953, 77–93. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef]
- Sykes, R.W.; Gjersing, E.L.; Foutz, K.; Rottmann, W.H.; Kuhn, S.A.; Foster, C.E.; Ziebell, A.; Turner, G.B.; Decker, S.R.; Hinchee, M.A.W.; et al. Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol. Biofuels 2015, 8, 128. [Google Scholar] [CrossRef]
- Kumar, S.; Omer, S.; Chitransh, S.; Khan, B.M. Cinnamate 4-hydroxylase downregulation in transgenic tobacco alters transcript level of other phenylpropanoid pathway genes. Int. J. Adv. Biotechnol. Res. 2012, 3, 545–557. [Google Scholar]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Zha, J.; Wu, X.; Gong, G.; Koffas, M.A.G. Pathway enzyme engineering for flavonoid production in recombinant microbes. Metab. Eng. Commun. 2019, 9, e00104. [Google Scholar] [CrossRef] [PubMed]
- Biala, W.; Jasinski, M. The phenylpropanoid case-It is transport that matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef]
- Yang, Y.H.; Yang, M.R.; Chen, J.Y.; Liu, Z.Y.; Zhang, Y.X.; Zhang, Z.Y.; Li, R.F. Two 4-coumarate: Coenzyme A ligase genes involved in acteoside and flavonoids biosynthesis in Rehmannia glutinosa. Ind. Crops Prod. 2022, 185, 115117. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Feng, S.; Zou, W.; Guo, K.; Fan, C.; Si, S.; Peng, L. Analysis of five rice 4-coumarate: Coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 2013, 430, 1151–1156. [Google Scholar] [CrossRef]
- Zhang, C.; Zang, Y.; Liu, P.; Zheng, Z.; Ouyang, J. Characterization, functional analysis and application of 4-Coumarate: CoA ligase genes from Populus trichocarpa. J. Biotechnol. 2019, 302, 92–100. [Google Scholar] [CrossRef]
- Wang, M.; Ding, Y.; Wang, Q.; Wang, P.; Han, Y.; Gu, Z.; Yang, R. NaCl treatment on physio-biochemical metabolism and phenolics accumulation in barley seedlings. Food Chem. 2020, 331, 127282. [Google Scholar] [CrossRef]
- Chen, X.; Fang, X.; Zhang, Y.; Wang, X.; Zhang, C.; Yan, X.; Zhao, Y.; Wu, J.; Xu, P.; Zhang, S. Overexpression of a soybean 4-coumaric acid: Coenzyme A ligase (GmPI4L) enhances resistance to Phytophthora sojae in soybean. Funct. Plant Biol. 2019, 46, 304–313. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Samec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Yin, Y.C.; Zhang, X.D.; Gao, Z.Q.; Hu, T.; Liu, Y. The research progress of chalcone isomerase (CHI) in plants. Mol. Biotechnol. 2019, 61, 32–52. [Google Scholar] [CrossRef]
- Kumar, R.; Vashisth, D.; Misra, A.; Akhtar, M.Q.; Jalil, S.U.; Shanker, K.; Gupta, M.M.; Rout, P.K.; Gupta, A.K.; Shasany, A.K. RNAi down-regulation of cinnamate-4-hydroxylase increases artemisinin biosynthesis in Artemisia annua. Sci. Rep. 2016, 6, 26458. [Google Scholar] [CrossRef]
- Gifford, I.; Battenberg, K.; Vaniya, A.; Wilson, A.; Tian, L.; Fiehn, O.; Berry, A.M. Distinctive patterns of flavonoid biosynthesis in roots and nodules of Datisca glomerata and Medicago spp. revealed by metabolomic and gene expression profiles. Front. Plant Sci. 2018, 9, 1463. [Google Scholar] [CrossRef]
- Rastogi, S.; Kumar, R.; Chanotiya, C.S.; Shanker, K.; Gupta, M.M.; Nagegowda, D.A.; Shasany, A.K. 4-coumarate: CoA ligase partitions metabolites for eugenol biosynthesis. Plant Cell Physiol. 2013, 54, 1238–1252. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, L.; Deng, Z.; Yu, Y. Two putative parallel pathways for naringenin biosynthesis in Epimedium wushanense. RSC Adv. 2021, 11, 13919–13927. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Shakya, R.; Sengoda, V.G.; Munyaneza, J.E.; Swamy, P.; Navarre, D.A. Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux inHQT-silenced lines. Plant Biotechnol. J. 2015, 13, 551–564. [Google Scholar] [CrossRef]
- Volpi, E.S.N.; Mazzafera, P.; Cesarino, I. Should I stay or should I go: Are chlorogenic acids mobilized towards lignin biosynthesis? Phytochemistry 2019, 166, 112063. [Google Scholar] [CrossRef]
- Kundu, A.; Vadassery, J. Chlorogenic acid-mediated chemical defence of plants against insect herbivores. Plant Biol. 2019, 21, 185–189. [Google Scholar] [CrossRef]
- Petrulova, V.; Ducaiova, Z.; Repcak, M. Short-term UV-B dose stimulates production of protective metabolites in Matricaria chamomilla leaves. Photochem. Photobiol. 2014, 90, 1061–1068. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, J.; Lin, Y.; Zhu, H.; Tang, L.; Wang, X.; Wang, X. Comparative transcriptome and weighted correlation network analyses reveal candidate genes involved in chlorogenic acid biosynthesis in sweet potato. Sci. Rep. 2022, 12, 2770. [Google Scholar] [CrossRef]
| Plasmid | Backbone | Bacterial Antibiotic Resistance | Concentration of Antibiotics (mg/L) | Remarks |
|---|---|---|---|---|
| gRep1-TN | pYPQ131c | Tetracycline | 5 | 1st gRNA expression vector for tobacco |
| gRep2-TN | pYPQ132c | Tetracycline | 5 | 2nd gRNA expression vector for tobacco |
| gRep3-TN | pYPQ133c | Tetracycline | 5 | 3rd gRNA expression vector for tobacco |
| pGGA3-TN | pYPQ143 | Spectinomycin | 100 | gRNA entry vector for tobacco |
| pdCas9 | pYPQ153 | Spectinomycin | 100 | dCas9 entry vector |
| pCRi-0 | pMDC32-Ubi1 | Kanamycin | 50 | Plant destination vector/ CRISPRi empty vector |
| pCTN | pMDC32-Ubi1 | Kanamycin | 50 | CRISPRi vector |
| gRNA | gRNA Sequence (5′-3′) | Expression Vector | Plasmid |
|---|---|---|---|
| G1F-TN | GTG TGC GTT AAT ATT AAC GGA GAG TTG G | pYPQ131c | gRep1-TN |
| G1R-TN | AAA CCC AAC TCT CCG TTA ATA TTA ACG C | ||
| G2F-TN | GTG TGC CTC ACA CTT TCT TAT CTT ATG G | pYPQ132c | gRep2-TN |
| G2R-TN | AAA CCC ATA AGA TAA GAA AGT GTG AGG C | ||
| G3F-TN | GTG TGG AGA AAA GAA ACT TGG GAG TTG G | pYPQ133c | gRep3-TN |
| G3R-TN | AAA CCC AAC TCC CAA GTT TCT TTT CTC C |
| Gene | Primer Name | Primer Sequence (5′-3′) |
|---|---|---|
| EF1α | K-EF1aF K-Efa1R | CCCTTGGTGTCAAGCAAATG GGTAGGAAGAAACCTCCTTCAC |
| LF25 | K-L25F K-L25R | AAAGCTGATCCGTCCAAAAA GACAGCCTTGGCAACCTTAG |
| C4H | K-C4H F K-C4H R | GGAAGAAGCCCGAAGAGTTTAG CTCCTCCTACCAACACCAAATG |
| 4CL | 4CL-F 4CL-R | GGTTACACACTGGCGACATTGG GGAACTTCTCCTGCTTGCTCATC |
| CHS | K-CHS_TN F K-CHS_TN R | CCTTTGTTCGAGCTTGTCTCTG GCCCAGGAACATCTTTGAGTAAG |
| CHI | K-CHI_TN F K-CHI_TN R | ATCCAGTGATTGAGGAGAAACC TCAGGCTCAGTTGACAAAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karlson, C.K.S.; Mohd Noor, S.N.; Khalid, N.; Tan, B.C. CRISPRi-Mediated Down-Regulation of the Cinnamate-4-Hydroxylase (C4H) Gene Enhances the Flavonoid Biosynthesis in Nicotiana tabacum. Biology 2022, 11, 1127. https://doi.org/10.3390/biology11081127
Karlson CKS, Mohd Noor SN, Khalid N, Tan BC. CRISPRi-Mediated Down-Regulation of the Cinnamate-4-Hydroxylase (C4H) Gene Enhances the Flavonoid Biosynthesis in Nicotiana tabacum. Biology. 2022; 11(8):1127. https://doi.org/10.3390/biology11081127
Chicago/Turabian StyleKarlson, Chou Khai Soong, Siti Nurfadhlina Mohd Noor, Norzulaani Khalid, and Boon Chin Tan. 2022. "CRISPRi-Mediated Down-Regulation of the Cinnamate-4-Hydroxylase (C4H) Gene Enhances the Flavonoid Biosynthesis in Nicotiana tabacum" Biology 11, no. 8: 1127. https://doi.org/10.3390/biology11081127
APA StyleKarlson, C. K. S., Mohd Noor, S. N., Khalid, N., & Tan, B. C. (2022). CRISPRi-Mediated Down-Regulation of the Cinnamate-4-Hydroxylase (C4H) Gene Enhances the Flavonoid Biosynthesis in Nicotiana tabacum. Biology, 11(8), 1127. https://doi.org/10.3390/biology11081127






