Does the Actin Network Architecture Leverage Myosin-I Functions?
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Molecular Domains of Myosin-I
1.2. Myosin-I Members Have Diverse Functions
2. Challenges to Leverage the Molecular Conformational Changes of Myosin-I through Scales
2.1. Membrane Remodeling
2.2. Membrane and Cortical Tension
2.3. Role in Actin Remodeling
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sebé-Pedrós, A.; Grau-Bové, X.; Richards, T.A.; Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 2014, 6, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Kollmar, M.; Mühlhausen, S. Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 2017, 17, 211. [Google Scholar] [CrossRef] [PubMed]
- Burnette, D.T.; Manley, S.; Sengupta, P.; Sougrat, R.; Davidson, M.W.; Kachar, B.; Lippincott-Schwartz, J. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 2011, 13, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Maître, J.-L.; Niwayama, R.; Turlier, H.; Nédélec, F.; Hiiragi, T. Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol. 2015, 17, 849–855. [Google Scholar] [CrossRef]
- Maître, J.-L.; Turlier, H.; Illukkumbura, R.; Eismann, B.; Niwayama, R.; Nédélec, F.; Hiiragi, T. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 2016, 536, 344–348. [Google Scholar] [CrossRef]
- Schliffka, M.F.; Tortorelli, A.F.; Özgüç, Ö.; de Plater, L.; Polzer, O.; Pelzer, D.; Maître, J.-L. Multiscale analysis of single and double maternal-zygotic Myh9 and Myh10 mutants during mouse preimplantation development. eLife 2021, 10, e68536. [Google Scholar] [CrossRef]
- Vigouroux, C.; Henriot, V.; Le Clainche, C. Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force. Nat. Commun. 2020, 11, 3116. [Google Scholar] [CrossRef]
- Peckham, M. Coiled coils and SAH domains in cytoskeletal molecular motors. Biochem. Soc. Trans. 2011, 39, 1142–1148. [Google Scholar] [CrossRef]
- McIntosh, B.; Ostap, E.M. Myosin-I molecular motors at a glance. J. Cell Sci. 2016, 129, 2689–2695. [Google Scholar] [CrossRef]
- McConnell, R.E.; Tyska, M.J. Leveraging the membrane—Cytoskeleton interface with myosin-1. Trends Cell Biol. 2010, 20, 418–426. [Google Scholar] [CrossRef]
- Ruppert, C.; Godel, J.; Muller, R.; Kroschewski, R.; Reinhard, J.; Bahler, M. Localization of the rat myosin I molecules myr 1 and myr 2 and in vivo targeting of their tail domains. J. Cell Sci. 1995, 108, 3775–3786. [Google Scholar] [CrossRef]
- Mentes, A.; Huehn, A.; Liu, X.; Zwolak, A.; Dominguez, R.; Shuman, H.; Ostap, E.M.; Sindelar, C.V. High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc. Natl. Acad. Sci. USA 2018, 115, 1292–1297. [Google Scholar] [CrossRef]
- Brown, S. Myosins in yeast. Curr. Opin. Cell Biol. 1997, 9, 44–48. [Google Scholar] [CrossRef]
- Berg, J.S.; Powell, B.C.; Cheney, R.E. A millennial myosin census. Mol. Biol. Cell 2001, 12, 780–794. [Google Scholar] [CrossRef]
- Kim, S.V.; Flavell, R.A. Myosin I: From yeast to human. Cell. Mol. Life Sci. 2008, 65, 2128–2137. [Google Scholar] [CrossRef]
- Benesh, A.E.; Nambiar, R.; McConnell, R.E.; Mao, S.; Tabb, D.L.; Tyska, M.J. Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol. Biol. Cell 2010, 21, 970–978. [Google Scholar] [CrossRef]
- Prospéri, M.-T.; Lépine, P.; Dingli, F.; Paul-Gilloteaux, P.; Martin, R.; Loew, D.; Knölker, H.-J.; Coudrier, E. Myosin 1b functions as an effector of EphB signaling to control cell repulsion. J. Cell Biol. 2015, 210, 347–361. [Google Scholar] [CrossRef]
- Prospéri, M.-T.; Pernier, J.; Lachuer, H.; Coudrier, E. Plekhh1, a partner of myosin 1 and an effector of EphB2 controls the cortical actin network for cell repulsion. J. Cell Sci. 2021, 134, jcs258802. [Google Scholar] [CrossRef]
- Iuliano, O.; Yoshimura, A.; Prospéri, M.-T.; Martin, R.; Knölker, H.-J.; Coudrier, E. Myosin 1b promotes axon formation by regulating actin wave propagation and growth cone dynamics. J. Cell Biol. 2018, 217, 2033–2046. [Google Scholar] [CrossRef]
- Almeida, C.G.; Yamada, A.; Tenza, D.; Louvard, D.; Raposo, G.; Coudrier, E. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat. Cell Biol. 2011, 13, 779–789. [Google Scholar] [CrossRef]
- Yamada, A.; Mamane, A.; Lee-Tin-Wah, J.; Di Cicco, A.; Prévost, C.; Levy, D.; Joanny, J.-F.; Coudrier, E.; Bassereau, P. Catch-bond behaviour facilitates membrane tubulation by non-processive myosin 1b. Nat. Commun. 2014, 5, 3624. [Google Scholar] [CrossRef]
- Pernier, J.; Kusters, R.; Bousquet, H.; Lagny, T.; Morchain, A.; Joanny, J.-F.; Bassereau, P.; Coudrier, E. Myosin 1b is an actin depolymerase. Nat. Commun. 2019, 10, 5200. [Google Scholar] [CrossRef]
- Pernier, J.; Morchain, A.; Caorsi, V.; Bertin, A.; Bousquet, H.; Bassereau, P.; Coudrier, E. Myosin 1b flattens and prunes branched actin filaments. J. Cell Sci. 2020, 133, jcs247403. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Greenberg, M.J.; Moore, J.R.; Ostap, E.M. A hearing loss-associated myo1c mutation (R156W) decreases the myosin duty ratio and force sensitivity. Biochemistry 2011, 50, 1831–1838. [Google Scholar] [CrossRef]
- Dai, J.; Ting-Beall, H.P.; Hochmuth, R.M.; Sheetz, M.P.; Titus, M.A. Myosin I contributes to the generation of resting cortical tension. Biophys. J. 1999, 77, 1168–1176. [Google Scholar] [CrossRef]
- Brandstaetter, H.; Kendrick-Jones, J.; Buss, F. Molecular roles of Myo1c function in lipid raft exocytosis. Commun. Integr. Biol. 2012, 5, 508–510. [Google Scholar] [CrossRef]
- Boguslavsky, S.; Chiu, T.; Foley, K.P.; Osorio-Fuentealba, C.; Antonescu, C.N.; Bayer, K.U.; Bilan, P.J.; Klip, A. Myo1c binding to submembrane actin mediates insulin-induced tethering of GLUT4 vesicles. Mol. Biol. Cell 2012, 23, 4065–4078. [Google Scholar] [CrossRef]
- Capmany, A.; Yoshimura, A.; Kerdous, R.; Caorsi, V.; Lescure, A.; Del Nery, E.; Coudrier, E.; Goud, B.; Schauer, K. MYO1C stabilizes actin and facilitates arrival of transport carriers at the Golgi apparatus. J. Cell Sci. 2019, 132, jcs225029. [Google Scholar] [CrossRef]
- Brandstaetter, H.; Kishi-Itakura, C.; A Tumbarello, D.; Manstein, D.J.; Buss, F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy 2014, 10, 2310–2323. [Google Scholar] [CrossRef]
- Lebreton, G.; Géminard, C.; Lapraz, F.; Pyrpassopoulos, S.; Cerezo, D.; Spéder, P.; Ostap, E.M.; Noselli, S. Molecular to organismal chirality is induced by the conserved myosin 1D. Science 2018, 362, 949–952. [Google Scholar] [CrossRef]
- Pyrpassopoulos, S.; Feeser, E.A.; Mazerik, J.N.; Tyska, M.J.; Ostap, E.M. Membrane-bound Myo1c powers asymmetric motility of actin filaments. Curr. Biol. 2012, 22, 1688–1692. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Guilherme, A.; Robida, S.I.; Nicoloro, S.M.C.; Zhou, Q.L.; Jiang, Z.Y.; Pomerleau, D.P.; Czech, M.P. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature 2002, 420, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Robida, S.; Furcinitti, P.S.; Chawla, A.; Fogarty, K.; Corvera, S.; Czech, M.P. Unconventional myosin Myo1c promotes membrane fusion in a regulated exocytic pathway. Mol. Cell. Biol. 2004, 24, 5447–5458. [Google Scholar] [CrossRef]
- Yip, M.F.; Ramm, G.; Larance, M.; Hoehn, K.L.; Wagner, M.C.; Guilhaus, M.; James, D.E. CaMKII-Mediated phosphorylation of the myosin motor Myo1c Is required for insulin-stimulated GLUT4 translocation in adipocytes. Cell Metab. 2008, 8, 384–398. [Google Scholar] [CrossRef]
- Chen, X.-W.; Leto, D.; Chiang, S.-H.; Wang, Q.; Saltiel, A.R. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell 2007, 13, 391–404. [Google Scholar] [CrossRef]
- Huang, S.; Czech, M.P. The GLUT4 glucose transporter. Cell Metab. 2007, 5, 237–252. [Google Scholar] [CrossRef]
- Tiwari, A.; Jung, J.-J.; Inamdar, S.M.; Nihalani, D.; Choudhury, A. The myosin motor Myo1c is required for VEGFR2 delivery to the cell surface and for angiogenic signaling. Am. J. Physiol. Circ. Physiol. 2013, 304, H687–H696. [Google Scholar] [CrossRef]
- Kittelberger, N.; Breunig, M.; Martin, R.; Knölker, H.-J.; Miklavc, P. The role of myosin 1c and myosin 1b for surfactant exocytosis. J. Cell Sci. 2016, 129, 1685–1696. [Google Scholar] [CrossRef]
- Sarshad, A.; Sadeghifar, F.; Louvet, E.; Mori, R.; Böhm, S.; Al-Muzzaini, B.; Vintermist, A.; Fomproix, N.; Östlund, A.-K.; Percipalle, P. Nuclear myosin 1c facilitates the chromatin modifications required to activate rRNA gene transcription and cell cycle progression. PLoS Genet. 2013, 9, e1003397. [Google Scholar] [CrossRef]
- Hegan, P.S.; Ostertag, E.; Geurts, A.M.; Mooseker, M.S. Myosin Id is required for planar cell polarity in ciliated tracheal and ependymal epithelial cells: Myo1d is a determinant of planar cell polarity. Cytoskeleton 2015, 72, 503–516. [Google Scholar] [CrossRef]
- Lechler, T.; Shevchenko, A.; Shevchenko, A.; Li, R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 2000, 148, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Grassart, A.; Drubin, D.G. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol. Biol. Cell 2012, 23, 2891–2904. [Google Scholar] [CrossRef]
- Maravillas-Montero, J.L.; López-Ortega, O.; Patiño-López, G.; Santos-Argumedo, L. Myosin 1g regulates cytoskeleton plasticity, cell migration, exocytosis, and endocytosis in B lymphocytes: Molecular immunology. Eur. J. Immunol. 2013, 44, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Arif, E.; Wagner, M.C.; Johnstone, D.B.; Wong, H.N.; George, B.; Pruthi, P.A.; Lazzara, M.J.; Nihalani, D. Motor protein Myo1c is a podocyte protein that facilitates the transport of slit diaphragm protein Neph1 to the podocyte membrane. Mol. Cell. Biol. 2011, 31, 2134–2150. [Google Scholar] [CrossRef]
- Krendel, M.; Kim, S.V.; Willinger, T.; Wang, T.; Kashgarian, M.; Flavell, R.A.; Mooseker, M.S. Disruption of myosin 1e promotes podocyte injury. J. Am. Soc. Nephrol. 2008, 20, 86–94. [Google Scholar] [CrossRef]
- Mele, C.; Iatropoulos, P.; Donadelli, R.; Calabria, A.; Maranta, R.; Cassis, P.; Buelli, S.; Tomasoni, S.; Piras, R.; Krendel, M.; et al. MYO1E Mutations and Childhood Familial Focal Segmental Glomerulosclerosis. N. Engl. J. Med. 2011, 365, 295–306. [Google Scholar] [CrossRef]
- Wagner, M.C.; Blazer-Yost, B.L.; Boyd-White, J.; Srirangam, A.; Pennington, J.; Bennett, S. Expression of the unconventional myosin Myo1c alters sodium transport in M1 collecting duct cells. Am. J. Physiol. Cell Physiol. 2005, 289, C120–C129. [Google Scholar] [CrossRef]
- Bi, J.; Chase, S.E.; Pellenz, C.D.; Kurihara, H.; Fanning, A.S.; Krendel, M. Myosin 1e is a component of the glomerular slit diaphragm complex that regulates actin reorganization during cell-cell contact formation in podocytes. Am. J. Physiol. Ren. Physiol. 2013, 305, F532–F544. [Google Scholar] [CrossRef]
- Batters, C.; Arthur, C.P.; Lin, A.; Porter, J.; A Geeves, M.; A Milligan, R.; E Molloy, J.; Coluccio, L.M. Myo1c is designed for the adaptation response in the inner ear. EMBO J. 2004, 23, 1433–1440. [Google Scholar] [CrossRef]
- Holt, J.R.; Gillespie, S.K.; Provance, D.; Shah, K.; Shokat, K.M.; Corey, D.P.; Mercer, J.; Gillespie, P.G. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell 2002, 108, 371–381. [Google Scholar] [CrossRef]
- Donaudy, F.; Ferrara, A.; Esposito, L.; Hertzano, R.; Ben-David, O.; Bell, R.E.; Melchionda, S.; Zelante, L.; Avraham, K.B.; Gasparini, P. Multiple mutations ofMYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am. J. Hum. Genet. 2003, 72, 1571–1577. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-D.; Yan, B.; Chalovich, J.M.; Brenner, B. Theoretical kinetic studies of models for binding myosin subfragment-1 to regulated actin: Hill model versus Geeves model. Biophys. J. 2001, 80, 2338–2349. [Google Scholar] [CrossRef][Green Version]
- Baek, J.-I.; Oh, S.-K.; Kim, D.-B.; Choi, S.-Y.; Kim, U.-K.; Lee, K.-Y.; Lee, S.-H. Targeted massive parallel sequencing: The effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J. Rare Dis. 2012, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Ripatti, S.; Kettunen, J.; Lyytikäinen, L.-P.; Oksala, N.; Laurila, P.-P.; Kangas, A.J.; Soininen, P.; Savolainen, M.J.; Viikari, J.; et al. Novel loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012, 8, e1002907. [Google Scholar] [CrossRef]
- Tassopoulou-Fishell, M.; Deeley, K.; Harvey, E.M.; Sciote, J.; Vieira, A.R. Genetic variation in Myosin 1H contributes to mandibular prognathism. Am. J. Orthod. Dentofac. Orthop. 2012, 141, 51–59. [Google Scholar] [CrossRef]
- Toyoda, T.; An, D.; Witczak, C.A.; Koh, H.-J.; Hirshman, M.F.; Fujii, N.; Goodyear, L.J. Myo1c regulates glucose uptake in mouse skeletal muscle. J. Biol. Chem. 2011, 286, 4133–4140. [Google Scholar] [CrossRef]
- Sun, R.; Wang, Y.; Jin, M.; Chen, L.; Cao, Y.; Chen, F. Identification and functional studies of MYO1H for mandibular prognathism. J. Dent. Res. 2018, 97, 1501–1509. [Google Scholar] [CrossRef]
- Tyska, M.J.; Mackey, A.T.; Huang, J.-D.; Copeland, N.G.; Jenkins, N.A.; Mooseker, M.S. Myosin-1a is critical for normal brush border structure and composition. Mol. Biol. Cell 2005, 16, 2443–2457. [Google Scholar] [CrossRef]
- Arif, E.; Solanki, A.K.; Srivastava, P.; Rahman, B.; Tash, B.R.; Holzman, L.B.; Janech, M.G.; Martin, R.; Knölker, H.-J.; Fitzgibbon, W.R.; et al. The motor protein Myo1c regulates transforming growth factor-β–signaling and fibrosis in podocytes. Kidney Int. 2019, 96, 139–158. [Google Scholar] [CrossRef]
- Chase, S.E.; Encina, C.V.; Stolzenburg, L.R.; Tatum, A.H.; Holzman, L.B.; Krendel, M. Podocyte-specific knockout of myosin 1e disrupts glomerular filtration. Am. J. Physiol. Ren. Physiol. 2012, 303, F1099–F1106. [Google Scholar] [CrossRef][Green Version]
- Kim, S.V.; Mehal, W.Z.; Dong, X.; Heinrich, V.; Pypaert, M.; Mellman, I.; Dembo, M.; Mooseker, M.S.; Wu, D.; Flavell, R.A. Modulation of cell adhesion and motility in the immune system by Myo1f. Science 2006, 314, 136–139. [Google Scholar] [CrossRef]
- Spielmann, M.; Hernandez-Miranda, L.R.; Ceccherini, I.; Weese-Mayer, D.E.; Kragesteen, B.K.; Harabula, I.; Krawitz, P.; Birchmeier, C.; Leonard, N.; Mundlos, S. Mutations in MYO1H cause a recessive form of central hypoventilation with autonomic dysfunction. J. Med. Genet. 2017, 54, 754–761. [Google Scholar] [CrossRef]
- Ruppert, C.; Kroschewski, R.; Bähler, M. Identification, characterization and cloning of myr 1, a mammalian myosin-I. J. Cell Biol. 1993, 120, 1393–1403. [Google Scholar] [CrossRef]
- Bähler, M.; Kroschewski, R.; E Stöffler, H.; Behrmann, T. Rat myr 4 defines a novel subclass of myosin I: Identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity. J. Cell Biol. 1994, 126, 375–389. [Google Scholar] [CrossRef]
- Köhler, D.; Struchholz, S.; Bähler, M. The two IQ-motifs and Ca2+/calmodulin regulate the rat myosin 1d ATPase activity: Regulation of Myo1d ATPase activity. FEBS J. 2005, 272, 2189–2197. [Google Scholar] [CrossRef]
- Patino-Lopez, G.; Aravind, L.; Dong, X.; Kruhlak, M.J.; Ostap, E.M.; Shaw, S. Myosin 1G is an abundant class I Myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent Pleckstrin Homology (PH) domain (Myo1PH). J. Biol. Chem. 2010, 285, 8675–8686. [Google Scholar] [CrossRef]
- Gupta, P.; Gauthier, N.C.; Cheng-Han, Y.; Zuanning, Y.; Pontes, B.; Ohmstede, M.; Martin, R.; Knölker, H.-J.; Döbereiner, H.-G.; Krendel, M.; et al. Myosin 1E localizes to actin polymerization sites in lamellipodia, affecting actin dynamics and adhesion formation. Biol. Open 2013, 2, 1288–1299. [Google Scholar] [CrossRef]
- Münnich, S.; Taft, M.H.; Manstein, D.J. Crystal structure of human myosin 1c—The motor in GLUT4 exocytosis: Implications for Ca2+ regulation and 14-3-3 binding. J. Mol. Biol. 2014, 426, 2070–2081. [Google Scholar] [CrossRef]
- Dart, A.E.; Tollis, S.; Bright, M.D.; Frankel, G.; Endres, R.G. The motor protein myosin 1G functions in FcγR-mediated phagocytosis. J. Cell Sci. 2012, 125, 6020–6029. [Google Scholar] [CrossRef]
- Swanson, J.; Johnson, M.; Beningo, K.; Post, P.; Mooseker, M.; Araki, N. A contractile activity that closes phagosomes in macrophages. J. Cell Sci. 1999, 112, 307–316. [Google Scholar] [CrossRef]
- Nambiar, R.; McConnell, R.E.; Tyska, M.J. Control of cell membrane tension by myosin-I. Proc. Natl. Acad. Sci. USA 2009, 106, 11972–11977. [Google Scholar] [CrossRef] [PubMed]
- Gérard, A.; Patino-Lopez, G.; Beemiller, P.; Nambiar, R.; Ben-Aissa, K.; Liu, Y.; Totah, F.J.; Tyska, M.J.; Shaw, S.; Krummel, M.F. Detection of rare antigen-presenting cells through T cell-intrinsic meandering motility, mediated by Myo1g. Cell 2014, 158, 492–505. [Google Scholar] [CrossRef] [PubMed]
- López-Ortega, O.; Ovalle-García, E.; Ortega-Blake, I.; Antillón, A.; Chávez-Munguía, B.; Patiño-López, G.; Fragoso-Soriano, R.; Santos-Argumedo, L. Myo1g is an active player in maintaining cell stiffness in B-lymphocytes: Myo1g mediates cell stiffness in B-lymphocytes. Cytoskeleton 2016, 73, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Diz-Munoz, A.; Krieg, M.; Bergert, M.; Ibarlucea-Benitez, I.; Muller, D.J.; Paluch, E.; Heisenberg, C.-P. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol. 2010, 8, e1000544. [Google Scholar] [CrossRef]
- Cordonnier, M.-N.; Dauzonne, D.; Louvard, D.; Coudrier, E. Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol. Biol. Cell 2001, 12, 4013–4029. [Google Scholar] [CrossRef]
- Raposo, G.; Cordonnier, M.-N.; Tenza, D.; Menichi, B.; Dürrbach, A.; Louvard, D.; Coudrier, E. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol. Biol. Cell 1999, 10, 1477–1494. [Google Scholar] [CrossRef]
- Salas-Cortes, L.; Ye, F.; Tenza, D.; Wilhelm, C.; Theos, A.; Louvard, D.; Raposo, G.; Coudrier, E. Myosin Ib modulates the morphology and the protein transport within multi-vesicular sorting endosomes. J. Cell Sci. 2005, 118, 4823–4832. [Google Scholar] [CrossRef]
- McIntosh, B.; Pyrpassopoulos, S.; Holzbaur, E.L.; Ostap, E.M. Opposing kinesin and myosin-I motors drive membrane deformation and tubulation along engineered cytoskeletal networks. Curr. Biol. 2018, 28, 236–248.e5. [Google Scholar] [CrossRef]
- Chugh, P.; Paluch, E.K. The actin cortex at a glance. J. Cell Sci. 2018, 131, jcs186254. [Google Scholar] [CrossRef]
- Wang, F.-S.; Liu, C.-W.; Diefenbach, T.J.; Jay, D.G. Modeling the role of myosin 1c in neuronal growth cone turning. Biophys. J. 2003, 85, 3319–3328. [Google Scholar] [CrossRef]
- Maravillas-Montero, J.L.; Gillespie, P.G.; Patiño-López, G.; Shaw, S.; Santos-Argumedo, L. Myosin 1c participates in B cell cytoskeleton rearrangements, is recruited to the immunologic synapse, and contributes to antigen presentation. J. Immunol. 2011, 187, 3053–3063. [Google Scholar] [CrossRef]
- Schauer, K.; Duong, T.; Bleakley, K.; Bardin, S.; Bornens, M.; Goud, B. Probabilistic density maps to study global endomembrane organization. Nat. Methods 2010, 7, 560–566. [Google Scholar] [CrossRef]
- Grossier, J.-P.; Xouri, G.; Goud, B.; Schauer, K. Cell adhesion defines the topology of endocytosis and signaling. EMBO J. 2014, 33, 35–45. [Google Scholar] [CrossRef]
- McIntosh, B.B.; Holzbaur, E.L.; Ostap, E.M. Control of the initiation and termination of kinesin-1-driven transport by myosin-Ic and nonmuscle tropomyosin. Curr. Biol. 2015, 25, 523–529. [Google Scholar] [CrossRef]
- Tang, N.; Ostap, E. Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 2001, 11, 1131–1135. [Google Scholar] [CrossRef]
- Collins, K.; Sellers, J.R.; Matsudaira, P. Calmodulin dissociation regulates brush border myosin I (110-kD-calmodulin) mechanochemical activity in vitro. J. Cell Biol. 1990, 110, 1137–1147. [Google Scholar] [CrossRef]
- Kee, A.J.; Yang, L.; Lucas, C.A.; Greenberg, M.J.; Martel, N.; Leong, G.M.; Hughes, W.E.; Cooney, G.J.; James, D.E.; Ostap, E.M.; et al. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic 2015, 16, 691–711. [Google Scholar] [CrossRef]
- Joensuu, M.; Belevich, I.; Rämö, O.; Nevzorov, I.; Vihinen, H.; Puhka, M.; Witkos, T.M.; Lowe, M.; Vartiainen, M.K.; Jokitalo, E. ER sheet persistence is coupled to myosin 1c–regulated dynamic actin filament arrays. Mol. Biol. Cell 2014, 25, 1111–1126. [Google Scholar] [CrossRef]
- Soldati, T. Unconventional myosins, actin dynamics and endocytosis: A ménage à trois?: Myosins, actin dynamics and endocytosis. Traffic 2003, 4, 358–366. [Google Scholar] [CrossRef]
- Jung, G.; Remmert, K.; Wu, X.; Volosky, J.M.; Hammer, J.A. The dictyostelium carmil protein links capping protein and the Arp2/3 complex to type I myosins through their Sh3 domains. J. Cell Biol. 2001, 153, 1479–1498. [Google Scholar] [CrossRef]
- Geli, M.I.; Lombardi, R.; Schmelzl, B.; Riezman, H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000, 19, 4281–4291. [Google Scholar] [CrossRef] [PubMed]
- Manenschijn, H.E.; Picco, A.; Mund, M.; Rivier-Cordey, A.-S.; Ries, J.; Kaksonen, M. Type-I myosins promote actin polymerization to drive membrane bending in endocytosis. eLife 2019, 8, e44215. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, R.D.; Taft, M.H.; Giese, S.; Thiel, C.; Balázs, Z.; Giese, H.; Manstein, D.J.; Sondergaard, T.E. Phenamacril is a reversible and noncompetitive inhibitor of Fusarium class I myosin. J. Biol. Chem. 2019, 294, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
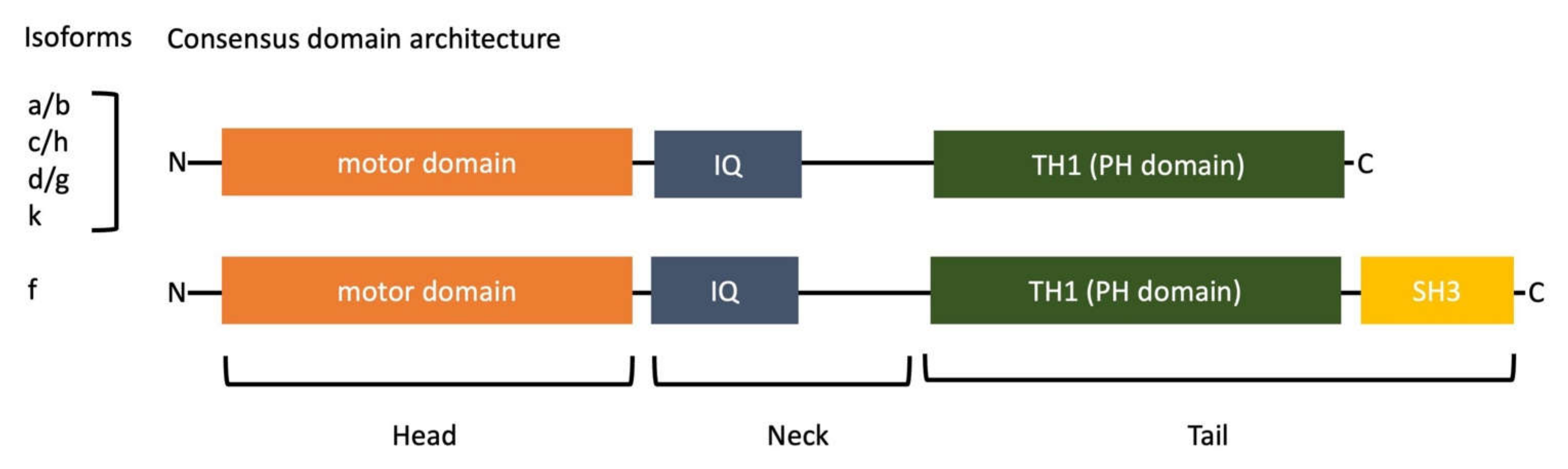
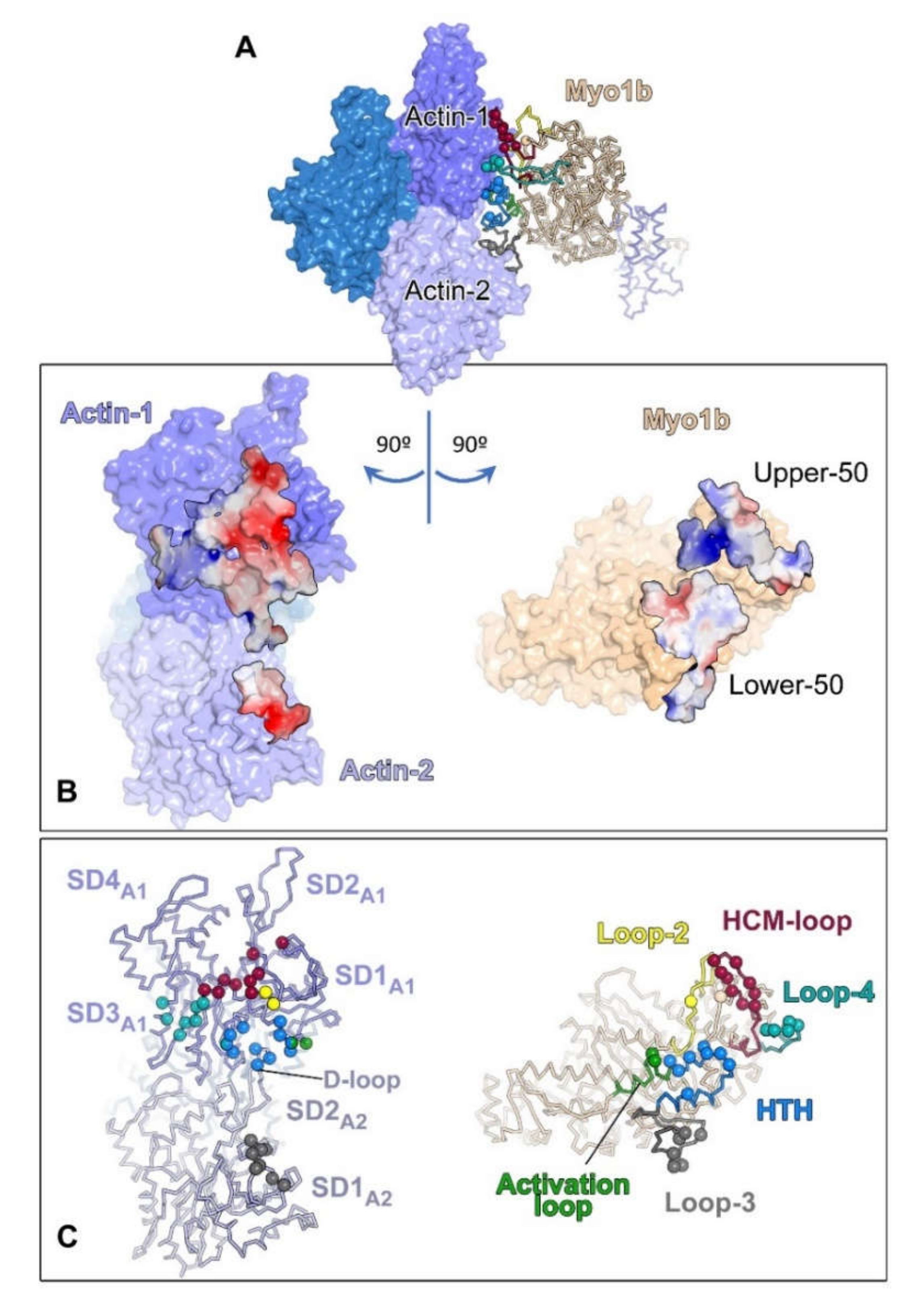
| Myosin Name | Localizations | Functions | References |
|---|---|---|---|
| Myo1A | Intestine | Linker between actin bundles and the microvillus membrane in mice | Benesh, A.E. [16] |
| Myo1B | Lung, liver, heart, and brain | Interacts with EPHB2 and controls cell repulsion and morphology (Hek293T or HCT116 cells) | Prospéri, M.-T. [17] Prospéri, M.-T. [18] |
| Regulates the formation of filopodia in growth cones in neurons | Iuliano, O. [19] | ||
| Promotes the formation of tubules at the Trans-Golgi Network (HeLa cells) | Almeida, C.G. [20] | ||
| Facilitates tube extraction (in vitro, rat Myo1B) | Yamada, A. [21] | ||
| Acts as an actin depolymerase (in vitro, rat Myo1B) | Pernier, J. [22] | ||
| Affects branched F-actin (in vitro, rat Myo1B) | Almeida, C.G. [20] Pernier, J. [23] | ||
| Myo1C | Ubiquitously expressed | Mechanical transduction in the ear (mouse Myo1c) | Lin, T. [24] |
| Regulation of cellular membrane tension (Dictyostelium) | Dai, J. [25] | ||
| Cholesterol transport (HeLa cells) | Brandstaetter, H. [26] | ||
| Tethering of vesicles (mouse Myo1c, hTERT-RPE1 cells) | Boguslavsky, S. [27] Capmany, A. [28] | ||
| Autophagosome-lysosome fusion (HeLa cells) | Brandstaetter, H. [29] | ||
| Chirality establishment in Drosophila | Lebreton, G. [30] Pyrpassopoulos, S. [31] | ||
| Docking of GLUT4-containing vesicles to the plasma membrane in muscle and adipocytes | Boguslavsky, S. [27] Bose, A. [32] Bose, A. [33] Yip, M.F. [34] Chen, X.-W. [35] Huang, S. [36] | ||
| Controls exocytosis in secretory cells | Tiwari, A. [37] Kittelberger, N. [38] | ||
| Chromatin modifications to gene transcription and cell cycle progression in nucleus (HeLa cells) | Sarshad, A. [39] | ||
| Myo1D | Widely expressed, high expression in brain | Influences the establishment of rotational planar cell polarity in epithelial cells of the trachea | Hegan, P.S. [40] |
| Myo1E | Hematopoietic cells | Involved in clathrin-mediated endocytosis | Lechler, T. [41] Cheng, J. [42] |
| Myo1F | Hematopoietic cells | Regulates adhesion | Kim, S.V. [15] |
| Myo1G | Hematopoietic cells | Involved in phagocytosis and exocytosis processes | Maravillas-Montero, J.L. [43] |
| Myosin Name | Amino Acid Mutation | Effect and Associated Disease | Reference |
|---|---|---|---|
| Myo1A | Insertion between 349 and 350 | Deafness | Donaudy, F. [51] |
| V306M | Deafness | ||
| E385D | Affect ATPase activity Deafness | ||
| G662E | Deafness | ||
| G647D | Deafness | ||
| S797F | Deafness | ||
| S910P | Deafness | ||
| Myo1C | 690STOP | No recruitment of Neph1, glomerular disease | Arif, E. [44] |
| Y61G | Increased sensitivity to inhibition, hair cells defect | Holt, J.R. [50] | |
| R156W | Decreased myosin duty ratio and force sensitivity, hearing loss | Lin, T. [24] | |
| K111A (in Mouse) | Glucose uptake defects | Toyoda, T. [56] | |
| Myo1E | A159P | Abnormal localization and function, glomerulosclerosis | Mele, C. [46] |
| Y695STOP | Loss of calmodulin binding at the tail domain of Myo1E, glomerulosclerosis | ||
| A159P | No efficient assembly of actin cables along cell-cell junctions, glomerular disease | Bi, J. [48] | |
| Myo1F | I502V | Destabilized actin binding site and ATP binding site, hearing loss | Baek, J.-I. [53] |
| Myo1H | P1001L | Mandibular prognathism | Sun, R. [57] |
| Myosin Name | Genetic Modification | Phenotypes | References |
|---|---|---|---|
| Myo1A | Complete knockout | No overt phenotypes at the whole animal, defects in microvillar membrane morphology and in brush-border organization | Tyska, M.J. [58] |
| Myo1B | - | - | |
| Myo1C | Complete knockout | Retinal phenotypes only | Arif, E. [59] |
| Myo1C | Podocytes-specific knockout | Downregulation of canonical and non-canonical TGF-β pathways | Arif, E. [59] |
| Myo1D | Complete knockout | Perturbed planar cell polarity of epithelial cells of the trachea, change of velocity and linearity of cilia-driven movement, loss of asymmetric clustering of cilia of ependymal cells and left-right positioning of the clusters, no obvious motor defects of rats, no obvious differences in kidney and liver morphology | Hegan, P.S. [40] |
| Myo1E | Complete knockout | Nephrotic syndrome and focal segmental glomerulosclerosis | Krendel, M. [45] |
| Myo1E | Podocytes-specific knockout | Proteinuria, podocyte foot process effacement, glomerular basement membrane disorganization, anormal glomerular filtration | Chase, S.E. [60] |
| Myo1F | Complete knockout | Increased susceptibility to infection by Listeria monocytogenes and an impaired neutrophil response | Kim, S.V. [61] |
| Myo1G | Complete knockout | Abnormalities in the adhesion ability and chemokine-induced directed migration in B lymphocytes | Maravillas-Montero, J.L. [43] |
| Myo1H | Complete knockout | Severe cyanosis and death within the first four postnatal hours | Spielmann, M. [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pernier, J.; Schauer, K. Does the Actin Network Architecture Leverage Myosin-I Functions? Biology 2022, 11, 989. https://doi.org/10.3390/biology11070989
Pernier J, Schauer K. Does the Actin Network Architecture Leverage Myosin-I Functions? Biology. 2022; 11(7):989. https://doi.org/10.3390/biology11070989
Chicago/Turabian StylePernier, Julien, and Kristine Schauer. 2022. "Does the Actin Network Architecture Leverage Myosin-I Functions?" Biology 11, no. 7: 989. https://doi.org/10.3390/biology11070989
APA StylePernier, J., & Schauer, K. (2022). Does the Actin Network Architecture Leverage Myosin-I Functions? Biology, 11(7), 989. https://doi.org/10.3390/biology11070989






