The Role of Serendipita indica (Piriformospora indica) in Improving Plant Resistance to Drought and Salinity Stresses
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Serendipita Indica (S. indica)
1.2. Drought Stress and S. indica
1.3. Salinity Stress and S. indica
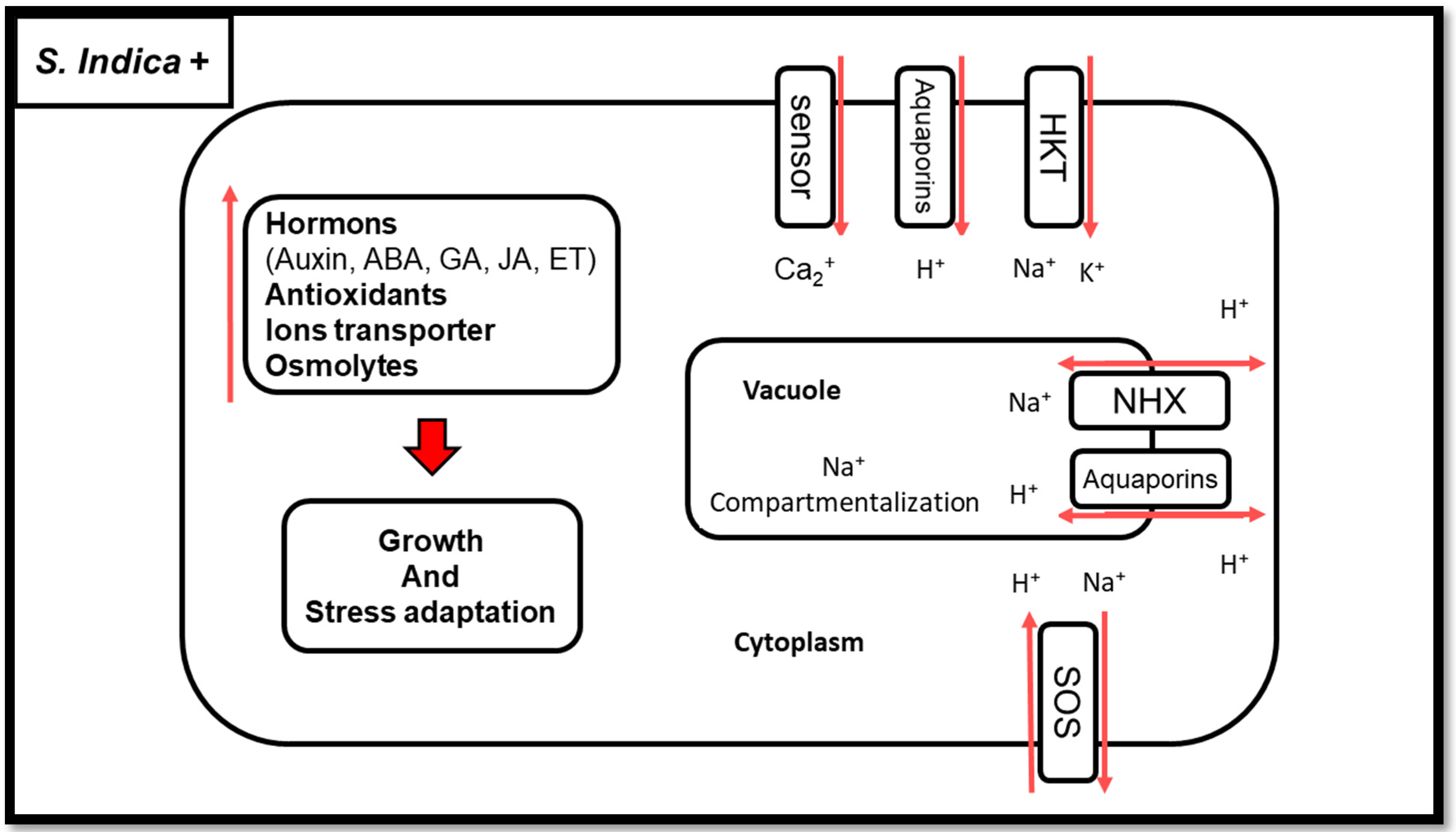
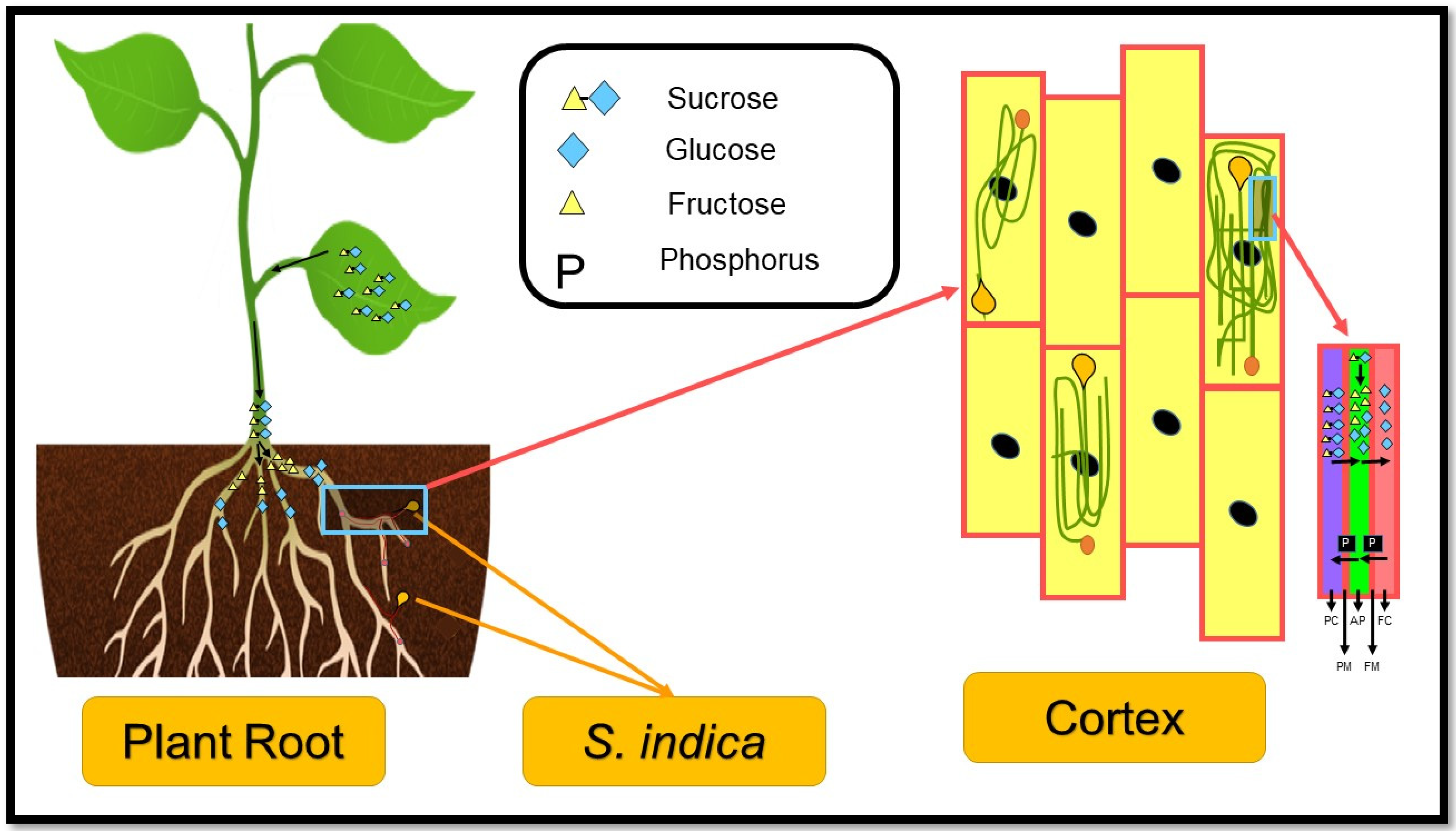
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, L.P.; Gill, S.S.; Tuteja, N. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal. Behav. 2011, 6, 175–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalid, M.; Rahman, S.U.; Huang, D. Molecular mechanism underlying Piriformospora indica-mediated plant improvement/protection for sustainable agriculture. Acta Biochim. Biophys. Sin. 2019, 51, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Singh, A.; Sahay, N.S.; Sharma, J.; Roy, A.; Kumari, M.; Rana, D.; Thakran, S.; Deka, D.; Bharti, K. Piriformospora indica: An axenically culturable mycorrhiza-like endosymbiotic fungus. In Fungal Associations; Springer: Berlin/Heidelberg, Germany, 2001; pp. 125–150. [Google Scholar]
- Ghorbani, A.; Razavi, S.; Omran, V.G.; Pirdashti, H. Piriformospora indica alleviates salinity by boosting redox poise and antioxidative potential of tomato. Russ. J. Plant Physiol. 2018, 65, 898–907. [Google Scholar] [CrossRef]
- Yadav, V.; Kumar, M.; Deep, D.K.; Kumar, H.; Sharma, R.; Tripathi, T.; Tuteja, N.; Saxena, A.K.; Johri, A.K. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J. Biol. Chem. 2010, 285, 26532–26544. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Varma, A.; Rexer, K.-H.; Hassel, A.; Kost, G.; Sarbhoy, A.; Bisen, P.; Bütehorn, B.; Franken, P. Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 1998, 90, 896–903. [Google Scholar] [CrossRef]
- Hassani, D.; Khalid, M.; Huang, D.; Zhang, Y.-D. Morphophysiological and molecular evidence supporting the augmentative role of Piriformospora indica in mitigation of salinity in Cucumis melo L. Acta Biochim. Biophys. Sin. 2019, 51, 301–312. [Google Scholar] [CrossRef]
- Opitz, M.W.; Daneshkhah, R.; Lorenz, C.; Ludwig, R.; Steinkellner, S.; Wieczorek, K. Serendipita indica changes host sugar and defense status in Arabidopsis thaliana: Cooperation or exploitation? Planta 2021, 253, 1–14. [Google Scholar] [CrossRef]
- Quoc, N.B.; Bao Chau, N.N. The role of cell wall degrading enzymes in pathogenesis of Magnaporthe oryzae. Curr. Protein Pept. Sci. 2017, 18, 1019–1034. [Google Scholar] [CrossRef]
- Giovannoni, M.; Gramegna, G.; Benedetti, M.; Mattei, B. Industrial use of cell wall degrading enzymes: The fine line between production strategy and economic feasibility. Front. Bioeng. Biotechnol. 2020, 8, 356. [Google Scholar] [CrossRef]
- Zuccaro, A.; Lahrmann, U.; Güldener, U.; Langen, G.; Pfiffi, S.; Biedenkopf, D.; Wong, P.; Samans, B.; Grimm, C.; Basiewicz, M. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 2011, 7, e1002290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, D.; Rovenich, H.; Jeena, G.; Nizam, S.; Tissier, A.; Balcke, G.U.; Mahdi, L.K.; Bonkowski, M.; Langen, G.; Zuccaro, A. The inconspicuous gatekeeper: Endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 2019, 224, 886–901. [Google Scholar] [CrossRef] [Green Version]
- Lanza, M.; Haro, R.; Conchillo, L.B.; Benito, B. The endophyte Serendipita indica reduces the sodium content of Arabidopsis plants exposed to salt stress: Fungal ENA ATPases are expressed and regulated at high pH and during plant co-cultivation in salinity. Environ. Microbiol. 2019, 21, 3364–3378. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zheng, X.; Lin, L.; An, Q.; Jiao, Y.; Li, Q.; Li, Z.; Hong, Y.; Zhang, K.; Xie, C. Remediation of soils co-contaminated with cadmium and dichlorodiphenyltrichloroethanes by king grass associated with Piriformospora indica: Insights into the regulation of root excretion and reshaping of rhizosphere microbial community structure. J. Hazard. Mater. 2022, 422, 126936. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, A.; Kumari, M.; Rai, M.K.; Varma, A. Biotechnological importance of Piriformospora indica Verma et al-a novel symbiotic Mycorrhiza-like fungus: An overview. NISCAIR-CSIR India 2003, 2, 65–75. [Google Scholar]
- Ghaffari, M.R.; Ghabooli, M.; Khatabi, B.; Hajirezaei, M.R.; Schweizer, P.; Salekdeh, G.H. Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant Mol. Biol. 2016, 90, 699–717. [Google Scholar] [CrossRef]
- Baltruschat, H.; Fodor, J.; Harrach, B.D.; Niemczyk, E.; Barna, B.; Gullner, G.; Janeczko, A.; Kogel, K.H.; Schäfer, P.; Schwarczinger, I. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 2008, 180, 501–510. [Google Scholar] [CrossRef]
- Hosseini, F.; Mosaddeghi, M.R.; Dexter, A.R.; Sepehri, M. Effect of endophytic fungus Piriformospora indica and PEG-induced water stress on maximum root growth pressure and elongation rate of maize. Plant Soil 2019, 435, 423–436. [Google Scholar] [CrossRef]
- Johnson, J.M.; Alex, T.; Oelmüller, R. Piriformospora indica: The versatile and multifunctional root endophytic fungus for enhanced yield and tolerance to biotic and abiotic stress in crop plants. J. Trop. Agric. 2014, 52, 103–122. [Google Scholar]
- Oelmüller, R.; Sherameti, I.; Tripathi, S.; Varma, A. Piriformospora indica, a cultivable root endophyte with multiple biotechnological applications. Symbiosis 2009, 49, 1–17. [Google Scholar] [CrossRef]
- Goh, C.-H.; Vallejos, D.F.V.; Nicotra, A.B.; Mathesius, U. The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J. Chem. Ecol. 2013, 39, 826–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padash, A.; Shahabivand, S.; Behtash, F.; Aghaee, A. A practicable method for zinc enrichment in lettuce leaves by the endophyte fungus Piriformospora indica under increasing zinc supply. Sci. Hortic. 2016, 213, 367–372. [Google Scholar] [CrossRef]
- Ahmadvand, G.; Hajinia, S. Effect of endophytic fungus Piriformospora indica on yield and some physiological traits of millet (Panicum miliaceum) under water stress. Crop Pasture Sci. 2018, 69, 594–605. [Google Scholar] [CrossRef]
- Meena, K.K.; Mesapogu, S.; Kumar, M.; Yandigeri, M.S.; Singh, G.; Saxena, A.K. Co-inoculation of the endophytic fungus Piriformospora indica with the phosphate-solubilising bacterium Pseudomonas striata affects population dynamics and plant growth in chickpea. Biol. Fertil. Soils 2010, 46, 169–174. [Google Scholar] [CrossRef]
- Tsai, H.-J.; Shao, K.-H.; Chan, M.-T.; Cheng, C.-P.; Yeh, K.-W.; Oelmüller, R.; Wang, S.-J. Piriformospora indica symbiosis improves water stress tolerance of rice through regulating stomata behavior and ROS scavenging systems. Plant Signal. Behav. 2020, 15, 1722447. [Google Scholar] [CrossRef]
- Sirrenberg, A.; Göbel, C.; Grond, S.; Czempinski, N.; Ratzinger, A.; Karlovsky, P.; Santos, P.; Feussner, I.; Pawlowski, K. Piriformospora indica affects plant growth by auxin production. Physiol. Plant. 2007, 131, 581–589. [Google Scholar] [CrossRef]
- Komis, G.; Luptovciak, I.; Doskocilova, A.; Samaj, J. Biotechnological aspects of cytoskeletal regulation in plants. Biotechnol. Adv. 2015, 33, 1043–1062. [Google Scholar] [CrossRef]
- Ganguly, A.; Dixit, R. Mechanisms for regulation of plant kinesins. Curr. Opin. Plant Biol. 2013, 16, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Del Carratore, R.; Podda, A.; Maserti, B. Fungal colonization improved growth and modulated the expression of myrosinase in black cabbage. Adv. Hortic. Sci. 2017, 31, 115–119. [Google Scholar]
- Serfling, A.; Wirsel, S.G.; Lind, V.; Deising, H.B. Performance of the biocontrol fungus Piriformospora indica on wheat under greenhouse and field conditions. Phytopathology 2007, 97, 523–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Pan, K.; Olatunji, O.A.; Graciano, C.; Li, Z.; Sun, F.; Sun, X.; Song, D.; Chen, W.; Zhang, A. Phosphorous application improves drought tolerance of Phoebe zhennan. Front. Plant Sci. 2017, 8, 1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Wei, Q.; Xu, L.; Li, H.; Oelmüller, R.; Zhang, W. Piriformospora indica enhances phosphorus absorption by stimulating acid phosphatase activities and organic acid accumulation in Brassica napus. Plant Soil 2018, 432, 333–344. [Google Scholar] [CrossRef]
- Azizi, M.; Fard, E.M.; Ghabooli, M. Piriformospora indica affect drought tolerance by regulation of genes expression and some morphophysiological parameters in tomato (Solanum lycopersicum L.). Sci. Hortic. 2021, 287, 110260. [Google Scholar] [CrossRef]
- Achatz, B.; Kogel, K.-H.; Franken, P.; Waller, F. Piriformospora indica mycorrhization increases grain yield by accelerating early development of barley plants. Plant Signal. Behav. 2010, 5, 1685–1687. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Kamal, S.; Shakil, N.A.; Sherameti, I.; Oelmüller, R.; Dua, M.; Tuteja, N.; Johri, A.K.; Varma, A. The root endophyte fungus Piriformospora indica leads to early flowering, higher biomass and altered secondary metabolites of the medicinal plant, Coleus forskohlii. Plant Signal. Behav. 2012, 7, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Bagde, U.S.; Prasad, R.; Varma, A. Impact of culture filtrate of Piriformospora indica on biomass and biosynthesis of active ingredient aristolochic acid in Aristolochia elegans Mart. Int. J. Biol. 2014, 6, 29. [Google Scholar] [CrossRef] [Green Version]
- Ghorbani, A.; Razavi, S.; Ghasemi Omran, V.; Pirdashti, H. Piriformospora indica inoculation alleviates the adverse effect of NaCl stress on growth, gas exchange and chlorophyll fluorescence in tomato (Solanum lycopersicum L.). Plant Biol. 2018, 20, 729–736. [Google Scholar] [CrossRef]
- Anith, K.; Aswini, S.; Varkey, S.; Radhakrishnan, N.; Nair, D.S. Root colonization by the endophytic fungus Piriformospora indica improves growth, yield and piperine content in black pepper (Piper nigurm L.). Biocatal. Agric. Biotechnol. 2018, 14, 215–220. [Google Scholar] [CrossRef]
- Tarraf, W.; Ruta, C.; De Cillis, F.; Tagarelli, A.; Tedone, L.; De Mastro, G. Effects of mycorrhiza on growth and essential oil production in selected aromatic plants. Ital. J. Agron. 2015, 10, 160–162. [Google Scholar] [CrossRef] [Green Version]
- Zolfaghari, M.; Nazeri, V.; Sefidkon, F.; Rejali, F. Effect of arbuscular mycorrhizal fungi on plant growth and essential oil content and composition of Ocimum basilicum L. Iran. J. Plant Physiol. 2013, 3, 643–650. [Google Scholar]
- Gill, S.S.; Gill, R.; Trivedi, D.K.; Anjum, N.A.; Sharma, K.K.; Ansari, M.W.; Ansari, A.A.; Johri, A.K.; Prasad, R.; Pereira, E. Piriformospora indica: Potential and significance in plant stress tolerance. Front. Microbiol. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawoof, A.; Ramchiary, N.; Abdin, M.Z. A high-throughput RNA-Seq approach to elucidate the transcriptional response of Piriformospora indica to high salt stress. Sci. Rep. 2021, 11, 1–15. [Google Scholar]
- Yaghoubian, Y.; Siadat, S.A.; Moradi Telavat, M.R.; Pirdashti, H.; Yaghoubian, I. Bio-removal of cadmium from aqueous solutions by filamentous fungi: Trichoderma spp. and Piriformospora indica. Environ. Sci. Pollut. Res. 2019, 26, 7863–7872. [Google Scholar] [CrossRef] [PubMed]
- Mohsenifard, E.; Ghabooli, M.; Mehri, N.; Bakhshi, B. Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. J. Plant Mol. Breed. 2017, 5, 10–18. [Google Scholar]
- Battie-Laclau, P.; Laclau, J.-P.; de Cassia Piccolo, M.; Arenque, B.C.; Beri, C.; Mietton, L.; Muniz, M.R.A.; Jordan-Meille, L.; Buckeridge, M.S.; Nouvellon, Y. Influence of potassium and sodium nutrition on leaf area components in Eucalyptus grandis trees. Plant Soil 2013, 371, 19–35. [Google Scholar] [CrossRef]
- Nanda, R.; Agrawal, V. Piriformospora indica, an excellent system for heavy metal sequestration and amelioration of oxidative stress and DNA damage in Cassia angustifolia Vahl under copper stress. Ecotoxicol. Environ. Saf. 2018, 156, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.; Hassani, D.; Bilal, M.; Liao, J.; Huang, D. Elevation of secondary metabolites synthesis in Brassica campestris ssp. chinensis L. via exogenous inoculation of Piriformospora indica with appropriate fertilizer. PLoS ONE 2017, 12, e0177185. [Google Scholar] [CrossRef] [PubMed]
- Saddique, M.A.B.; Ali, Z.; Khan, A.S.; Rana, I.A.; Shamsi, I.H. Inoculation with the endophyte Piriformospora indica significantly affects mechanisms involved in osmotic stress in rice. Rice 2018, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Aslam, M.M.; Karanja, J.; Bello, S.K. Piriformospora indica colonization reprograms plants to improved P-uptake, enhanced crop performance, and biotic/abiotic stress tolerance. Physiol. Mol. Plant Pathol. 2019, 106, 232–237. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S. Plant drought stress: Effects, mechanisms and management. Sustain. Agric. 2009, 29, 153–188. [Google Scholar]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Hussin, S.; Khalifa, W.; Geissler, N.; Koyro, H.W. Influence of the root endophyte Piriformospora indica on the plant water relations, gas exchange and growth of Chenopodium quinoa at limited water availability. J. Agron. Crop Sci. 2017, 203, 373–384. [Google Scholar] [CrossRef]
- Gohari, A.; Eslamian, S.; Abedi-Koupaei, J.; Bavani, A.M.; Wang, D.; Madani, K. Climate change impacts on crop production in Iran’s Zayandeh-Rud River Basin. Sci. Total Environ. 2013, 442, 405–419. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Bahrani, M.J.; Bahrami, H.; Haghighi, A.A.K. Effect of water stress on ten forage grasses native or introduced to Iran. Grassl. Sci. 2010, 56, 1–5. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E. Plant Physiology, 5th ed.; MA Sinauer Associates: Sunderland, UK, 2010. [Google Scholar]
- Lisar, S.; Motafakkerazad, R.; Hossain, M.M.; Rahman, I. Causes, Effects and Responses. In Water Stress; IntechOpen: London, UK, 2012; p. 25. [Google Scholar]
- Garg, B. Nutrient uptake and management under drought: Nutrient-moisture interaction. Curr. Agric. 2003, 27, 1–8. [Google Scholar]
- Reddy, A.R.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Takahashi, S.; Murata, N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008, 13, 178–182. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, D.P.; Belesky, D.P. Adaptations of endophyte-infected cool-season grasses to environmental stresses: Mechanisms of drought and mineral stress tolerance. Crop Sci. 2000, 40, 923–940. [Google Scholar] [CrossRef]
- Romanello, G.; Chuchra-Zbytniuk, K.; Vandermer, J.; Touchette, B. Morphological adjustments promote drought avoidance in the wetland plant Acorus americanus. Aquat. Bot. 2008, 89, 390–396. [Google Scholar] [CrossRef]
- Zlatev, Z.; Lidon, F.C. An overview on drought induced changes in plant growth, water relationsand photosynthesis. Emir. J. Food Agric. 2012, 24, 57–72. [Google Scholar]
- Hosseini, F.; Mosaddeghi, M.R.; Dexter, A.R.; Sepehri, M. Maize water status and physiological traits as affected by root endophytic fungus Piriformospora indica under combined drought and mechanical stresses. Planta 2018, 247, 1229–1245. [Google Scholar] [CrossRef] [PubMed]
- Fahramand, M.; Mahmoody, M.; Keykha, A.; Noori, M.; Rigi, K. Influence of abiotic stress on proline, photosynthetic enzymes and growth. Int. Res. J. Appl. Basic Sci. 2014, 8, 257–265. [Google Scholar]
- Hosseini, F.; Mosaddeghi, M.R.; Dexter, A.R. Effect of the fungus Piriformospora indica on physiological characteristics and root morphology of wheat under combined drought and mechanical stresses. Plant Physiol. Biochem. 2017, 118, 107–120. [Google Scholar] [CrossRef]
- Salehi-lisar, S.; Motafakkerazad, R.; Hossain, M.; Rahman, I. Water Stress in Plants: Causes, Effects and Responses; InTech: Rijeka, Croatia, 2012. [Google Scholar]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, J.; Xu, L.; Wang, A.; Huang, L.; Du, H.; Qiu, L.; Oelmüller, R. Drought stress responses in maize are diminished by Piriformospora indica. Plant Signal. Behav. 2018, 13, e1414121. [Google Scholar] [CrossRef] [Green Version]
- Bengough, A.G.; Bransby, M.F.; Hans, J.; McKenna, S.J.; Roberts, T.J.; Valentine, T.A. Root responses to soil physical conditions; growth dynamics from field to cell. J. Exp. Bot. 2006, 57, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferdous, J.; Hussain, S.S.; Shi, B.J. Role of micro RNA s in plant drought tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.-J.; McGrath, S.P.; Meharg, A.A. Arsenic as a food chain contaminant: Mechanisms of plant uptake and metabolism and mitigation strategies. Annu. Rev. Plant Biol. 2010, 61, 535–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Running, S.W. Drought-induced reduction in global terrestrial net primary production from 2000 through 2009. Science 2010, 329, 940–943. [Google Scholar] [CrossRef] [Green Version]
- Mutum, R.D.; Kumar, S.; Balyan, S.; Kansal, S.; Mathur, S.; Raghuvanshi, S. Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci. Rep. 2016, 6, 30786. [Google Scholar] [CrossRef] [Green Version]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Valentín, R.; Campos, F.; Battaglia, M.; Solórzano, R.M.; Rosales, M.A.; Covarrubias, A.A. Group 6 Late embryogenesis abundant (LEA) proteins in monocotyledonous plants: Genomic organization and transcript accumulation patterns in response to stress in Oryza sativa. Plant Mol. Biol. Report. 2014, 32, 198–208. [Google Scholar] [CrossRef] [Green Version]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Hu, Y.; Zhou, Z.; Cai, X.; Wang, X.; Hou, Y.; Wang, K. Cotton late embryogenesis abundant (LEA2) genes promote root growth and confer drought stress tolerance in transgenic Arabidopsis thaliana. G3 Genes Genomes Genet. 2018, 8, 2781–2803. [Google Scholar] [CrossRef] [Green Version]
- Brundrett, M.C. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002, 154, 275–304. [Google Scholar] [CrossRef]
- Ghaffari, M.R.; Mirzaei, M.; Ghabooli, M.; Khatabi, B.; Wu, Y.; Zabet-Moghaddam, M.; Mohammadi-Nejad, G.; Haynes, P.A.; Hajirezaei, M.R.; Sepehri, M. Root endophytic fungus Piriformospora indica improves drought stress adaptation in barley by metabolic and proteomic reprogramming. Environ. Exp. Bot. 2019, 157, 197–210. [Google Scholar] [CrossRef]
- Tyagi, J.; Varma, A.; Pudake, R.N. Evaluation of comparative effects of arbuscular mycorrhiza (Rhizophagus intraradices) and endophyte (Piriformospora indica) association with finger millet (Eleusine coracana) under drought stress. Eur. J. Soil Biol. 2017, 81, 1–10. [Google Scholar] [CrossRef]
- Xu, L.; Wang, A.; Wang, J.; Wei, Q.; Zhang, W. Piriformospora indica confers drought tolerance on Zea mays L. through enhancement of antioxidant activity and expression of drought-related genes. Crop J. 2017, 5, 251–258. [Google Scholar] [CrossRef] [Green Version]
- Swetha, S.; Padmavathi, T. Mitigation of Drought Stress by Piriformospora indica in Solanum melongena L. cultivars. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2020, 90, 585–593. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Johnson, J.M.; Cai, D.; Sherameti, I.; Oelmüller, R.; Lou, B. Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. J. Plant Physiol. 2010, 167, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Blanco, N.E.; Ceccoli, R.D.; Segretin, M.E.; Poli, H.O.; Voss, I.; Melzer, M.; Bravo-Almonacid, F.F.; Scheibe, R.; Hajirezaei, M.R.; Carrillo, N. Cyanobacterial flavodoxin complements ferredoxin deficiency in knocked-down transgenic tobacco plants. Plant J. 2011, 65, 922–935. [Google Scholar] [CrossRef]
- Abo-Doma, A.; Edrees, S.; Abdel-Aziz, S. The effect of mycorrhiza growth and expression of some genes in barley. Egypt J. Genet Cytol. 2016, 40, 301–313. [Google Scholar] [CrossRef] [Green Version]
- Peskan-Berghöfer, T.; Vilches-Barro, A.; Müller, T.M.; Glawischnig, E.; Reichelt, M.; Gershenzon, J.; Rausch, T. Sustained exposure to abscisic acid enhances the colonization potential of the mutualist fungus Piriformospora indica on Arabidopsis thaliana roots. New Phytol. 2015, 208, 873–886. [Google Scholar] [CrossRef]
- Dexter, A. Mechanics of root growth. Plant Soil 1987, 98, 303–312. [Google Scholar] [CrossRef]
- Azam, G.; Grant, C.D.; Misra, R.K.; Murray, R.S.; Nuberg, I.K. Growth of tree roots in hostile soil: A comparison of root growth pressures of tree seedlings with peas. Plant Soil 2013, 368, 569–580. [Google Scholar] [CrossRef]
- Greacen, E.L.; Oh, J.S. Physics of root growth. Nat. New Biol. 1972, 235, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Verma, S.; Sudha Sahay, N.; Bütehorn, B.; Franken, P. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 1999, 65, 2741–2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidarianpour, M.B.; Aliasgharzad, N.; Olsson, P.A. Positive effects of co-inoculation with Rhizophagus irregularis and Serendipita indica on tomato growth under saline conditions, and their individual colonization estimated by signature lipids. Mycorrhiza 2020, 30, 455–466. [Google Scholar] [CrossRef]
- Grümberg, B.C.; Urcelay, C.; Shroeder, M.A.; Vargas-Gil, S.; Luna, C.M. The role of inoculum identity in drought stress mitigation by arbuscular mycorrhizal fungi in soybean. Biol. Fertil. Soils 2015, 51, 1–10. [Google Scholar] [CrossRef]
- Zhu, B.; Su, J.; Chang, M.; Verma, D.P.S.; Fan, Y.-L.; Wu, R. Overexpression of a Δ1-pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water-and salt-stress in transgenic rice. Plant Sci. 1998, 139, 41–48. [Google Scholar] [CrossRef]
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Atia, M.A.; Mahmoud, A.W.M.; Saad, M.M.; Hirt, H. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Sci. Hortic. 2019, 256, 108532. [Google Scholar] [CrossRef]
- Porcel, R.; Aroca, R.; Ruiz-Lozano, J.M. Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 2012, 32, 181–200. [Google Scholar] [CrossRef] [Green Version]
- Shelden, M.C.; Dias, D.A.; Jayasinghe, N.S.; Bacic, A.; Roessner, U. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 2016, 67, 3731–3745. [Google Scholar] [CrossRef] [Green Version]
- Mesbah, N.M.; Wiegel, J. Life under multiple extreme conditions: Diversity and physiology of the halophilic alkalithermophiles. Appl. Environ. Microbiol. 2012, 78, 4074–4082. [Google Scholar] [CrossRef] [Green Version]
- Latef, A.A.H.A.; Chaoxing, H. Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salinity stress. Sci. Hortic. 2011, 127, 228–233. [Google Scholar] [CrossRef]
- Sharma, P.; Kharkwal, A.C.; Abdin, M.; Varma, A. Piriformospora indica-mediated salinity tolerance in Aloe vera plantlets. Symbiosis 2017, 72, 103–115. [Google Scholar] [CrossRef]
- Munns, R.; Cramer, G.R.; Ball, M.C. Interactions between rising CO2, soil salinity, and plant growth. In Carbon Dioxide and Environmental Stress; Elsevier: Amsterdam, The Netherlands, 1999; pp. 139–167. [Google Scholar]
- Porcel, R.; Aroca, R.; Azcon, R.; Ruiz-Lozano, J.M. Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na+ root-to-shoot distribution. Mycorrhiza 2016, 26, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, F.; Aroca, R.; Porcel, R.; Ruiz-Lozano, J.M. Influence of salinity on the in vitro development of Glomus intraradices and on the in vivo physiological and molecular responses of mycorrhizal lettuce plants. Microb. Ecol. 2008, 55, 45. [Google Scholar] [CrossRef] [PubMed]
- Yun, P.; Xu, L.; Wang, S.-S.; Shabala, L.; Shabala, S.; Zhang, W.-Y. Piriformospora indica improves salinity stress tolerance in Zea mays L. plants by regulating Na+ and K+ loading in root and allocating K+ in shoot. Plant Growth Regul. 2018, 86, 323–331. [Google Scholar] [CrossRef]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.; Hassani, D.; Liao, J.; Xiong, X.; Bilal, M.; Huang, D. An endosymbiont Piriformospora indica reduces adverse effects of salinity by regulating cation transporter genes, phytohormones, and antioxidants in Brassica campestris ssp. Chinensis. Environ. Exp. Bot. 2018, 153, 89–99. [Google Scholar] [CrossRef]
- Ahmad, P. Growth and antioxidant responses in mustard (Brassica juncea L.) plants subjected to combined effect of gibberellic acid and salinity. Arch. Agron. Soil Sci. 2010, 56, 575–588. [Google Scholar] [CrossRef]
- Loha, A.; Kashyap, A.K.; Sharma, P. A putative cyclin, SiPHO80 from root endophytic fungus Serendipita indica regulates phosphate homeostasis, salinity and heavy metal toxicity tolerance. Biochem. Biophys. Res. Commun. 2018, 507, 414–419. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizwan, M.; Ali, S.; Ibrahim, M.; Farid, M.; Adrees, M.; Bharwana, S.A.; Zia-ur-Rehman, M.; Qayyum, M.F.; Abbas, F. Mechanisms of silicon-mediated alleviation of drought and salt stress in plants: A review. Environ. Sci. Pollut. Res. 2015, 22, 15416–15431. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar] [CrossRef] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Ghorbani, A.; Omran, V.O.G.; Razavi, S.M.; Pirdashti, H.; Ranjbar, M. Piriformospora indica confers salinity tolerance on tomato (Lycopersicon esculentum Mill.) through amelioration of nutrient accumulation, K+/Na+ homeostasis and water status. Plant Cell Rep. 2019, 38, 1151–1163. [Google Scholar] [CrossRef]
- Barragán, V.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernández, J.A.; Cubero, B.; Pardo, J.M. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [Green Version]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef]
- Gálvez, F.J.; Baghour, M.; Hao, G.; Cagnac, O.; Rodríguez-Rosales, M.P.; Venema, K. Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiol. Biochem. 2012, 51, 109–115. [Google Scholar] [CrossRef]
- Wu, S.-J.; Ding, L.; Zhu, J.-K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansour, M.M.F. Protection of plasma membrane of onion epidermal cells by glycinebetaine and proline against NaCl stress. Plant Physiol. Biochem. 1998, 36, 767–772. [Google Scholar] [CrossRef]
- Hashem, A.; Abd_Allah, E.; Ahmad, P. Effect of AM fungi on growth, physio-biochemical attributes, lipid peroxidation, antioxidant enzymes and plant growth regulators in Lycopersicon esculantum Mill. subjected to different concentration of NaCl. Pak. J. Bot. 2015, 47, 327–340. [Google Scholar]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and non-enzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Ahmad, P.; Hashem, A.; Abd-Allah, E.F.; Alqarawi, A.; John, R.; Egamberdieva, D.; Gucel, S. Role of Trichoderma harzianum in mitigating NaCl stress in Indian mustard (Brassica juncea L) through antioxidative defense system. Front. Plant Sci. 2015, 6, 868. [Google Scholar] [CrossRef] [Green Version]
- Rasool, S.; Hameed, A.; Azooz, M.; Siddiqi, T.; Ahmad, P. Salt stress: Causes, types and responses of plants. In Ecophysiology and Responses of Plants under Salt Stress; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–24. [Google Scholar]
- Sarwat, M.; Hashem, A.; Ahanger, M.A.; Abd_Allah, E.F.; Alqarawi, A.; Alyemeni, M.N.; Ahmad, P.; Gucel, S. Mitigation of NaCl stress by arbuscular mycorrhizal fungi through the modulation of osmolytes, antioxidants and secondary metabolites in mustard (Brassica juncea L.) plants. Front. Plant Sci. 2016, 7, 869. [Google Scholar] [CrossRef] [Green Version]
- Maurel, C.; Verdoucq, L.; Luu, D.-T.; Santoni, V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [Green Version]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [Green Version]
- Sepehri, M.; Ghaffari, M.R.; Khayam Nekoui, M.; Sarhadi, E.; Moghadam, A.; Khatabi, B.; Hosseini Salekdeh, G. Root endophytic fungus Serendipita indica modulates barley leaf blade proteome by increasing the abundance of photosynthetic proteins in response to salinity. J. Appl. Microbiol. 2021, 131, 1870–1889. [Google Scholar] [CrossRef]
- Kord, H.; Fakheri, B.; Ghabooli, M.; Solouki, M.; Emamjomeh, A.; Khatabi, B.; Sepehri, M.; Salekdeh, G.H.; Ghaffari, M.R. Salinity-associated microRNAs and their potential roles in mediating salt tolerance in rice colonized by the endophytic root fungus Piriformospora indica. Funct. Integr. Genom. 2019, 19, 659–672. [Google Scholar] [CrossRef] [PubMed]
- de Zélicourt, A.; Synek, L.; Saad, M.M.; Alzubaidy, H.; Jalal, R.; Xie, Y.; Andres-Barrao, C.; Rolli, E.; Guerard, F.; Mariappan, K.G. ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genet. 2018, 14, e1007273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jogawat, A.; Vadassery, J.; Verma, N.; Oelmüller, R.; Dua, M.; Nevo, E.; Johri, A.K. PiHOG1, a stress regulator MAP kinase from the root endophyte fungus Piriformospora indica, confers salinity stress tolerance in rice plants. Sci. Rep. 2016, 6, 36765. [Google Scholar] [CrossRef] [PubMed]
- Zarea, M.; Hajinia, S.; Karimi, N.; Goltapeh, E.M.; Rejali, F.; Varma, A. Effect of Piriformospora indica and Azospirillum strains from saline or non-saline soil on mitigation of the effects of NaCl. Soil Biol. Biochem. 2012, 45, 139–146. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Zhang, X.; Tang, M. Arbuscular mycorrhizal symbiosis alleviates salt stress in black locust through improved photosynthesis, water status, and K+/Na+ homeostasis. Front. Plant Sci. 2017, 8, 1739. [Google Scholar] [CrossRef] [PubMed]
- Gazara, R.K.; Khan, S.; Iqrar, S.; Ashrafi, K.; Abdin, M.Z. Comparative transcriptome profiling of rice colonized with beneficial endophyte, Piriformospora indica, under high salinity environment. Mol. Biol. Rep. 2020, 47, 7655–7673. [Google Scholar]
- Varma, A.; Bakshi, M.; Lou, B.; Hartmann, A.; Oelmueller, R. Piriformospora indica: A novel plant growth-promoting mycorrhizal fungus. Agric. Res. 2012, 1, 117–131. [Google Scholar] [CrossRef]
- Abdelaziz, M.E.; Kim, D.; Ali, S.; Fedoroff, N.V.; Al-Babili, S. The endophytic fungus Piriformospora indica enhances Arabidopsis thaliana growth and modulates Na+/K+ homeostasis under salt stress conditions. Plant Sci. 2017, 263, 107–115. [Google Scholar] [CrossRef] [Green Version]
- Nomura, H.; Komori, T.; Kobori, M.; Nakahira, Y.; Shiina, T. Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 2008, 53, 988–998. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boorboori, M.R.; Zhang, H.-Y. The Role of Serendipita indica (Piriformospora indica) in Improving Plant Resistance to Drought and Salinity Stresses. Biology 2022, 11, 952. https://doi.org/10.3390/biology11070952
Boorboori MR, Zhang H-Y. The Role of Serendipita indica (Piriformospora indica) in Improving Plant Resistance to Drought and Salinity Stresses. Biology. 2022; 11(7):952. https://doi.org/10.3390/biology11070952
Chicago/Turabian StyleBoorboori, Mohammad Reza, and Hai-Yang Zhang. 2022. "The Role of Serendipita indica (Piriformospora indica) in Improving Plant Resistance to Drought and Salinity Stresses" Biology 11, no. 7: 952. https://doi.org/10.3390/biology11070952
APA StyleBoorboori, M. R., & Zhang, H.-Y. (2022). The Role of Serendipita indica (Piriformospora indica) in Improving Plant Resistance to Drought and Salinity Stresses. Biology, 11(7), 952. https://doi.org/10.3390/biology11070952





