The Impact of Environmental Habitats and Diets on the Gut Microbiota Diversity of True Bugs (Hemiptera: Heteroptera)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
2.3. Bioinformatic Analyses
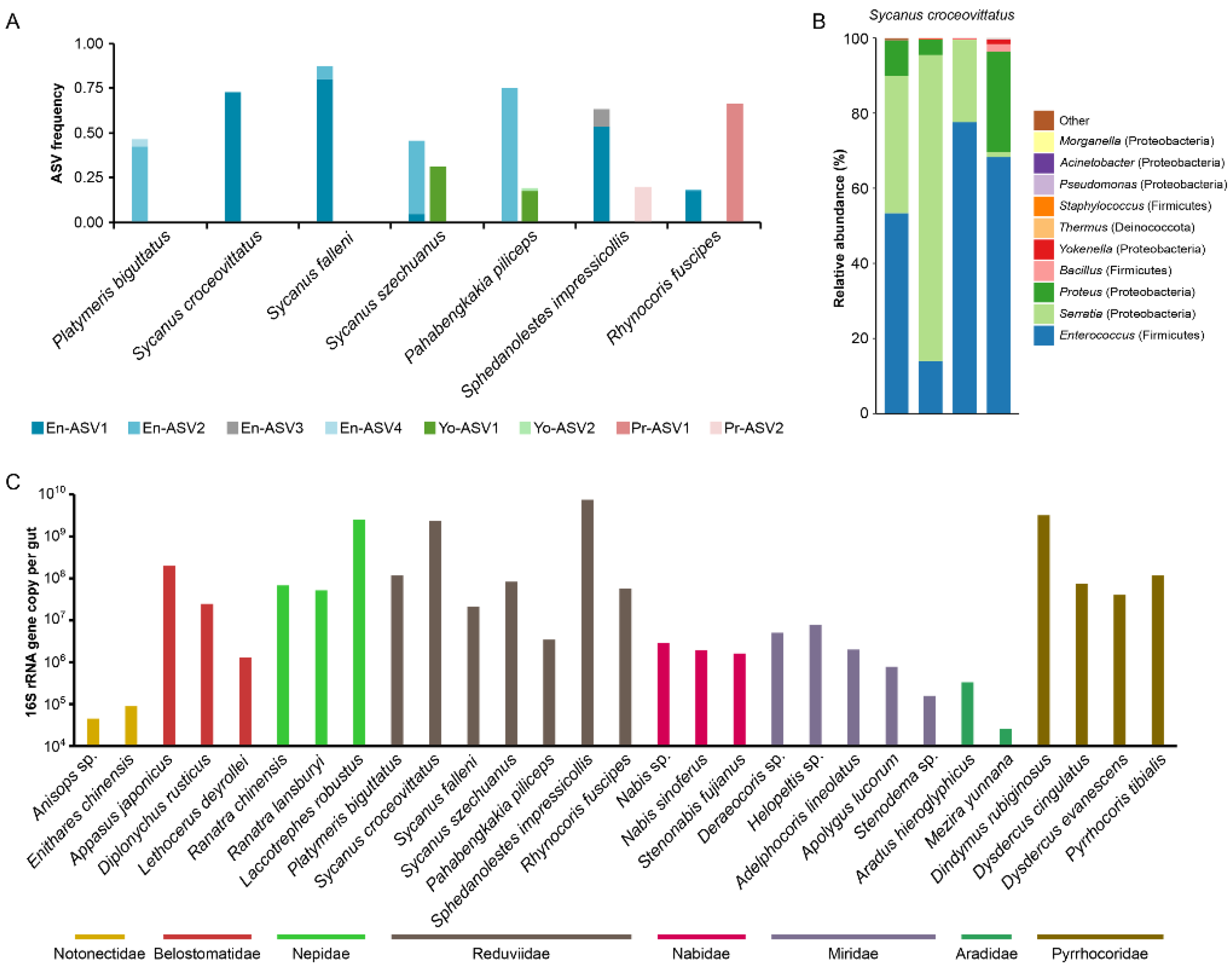
2.4. Determination of Gut Bacterial Number Using qPCR
3. Results
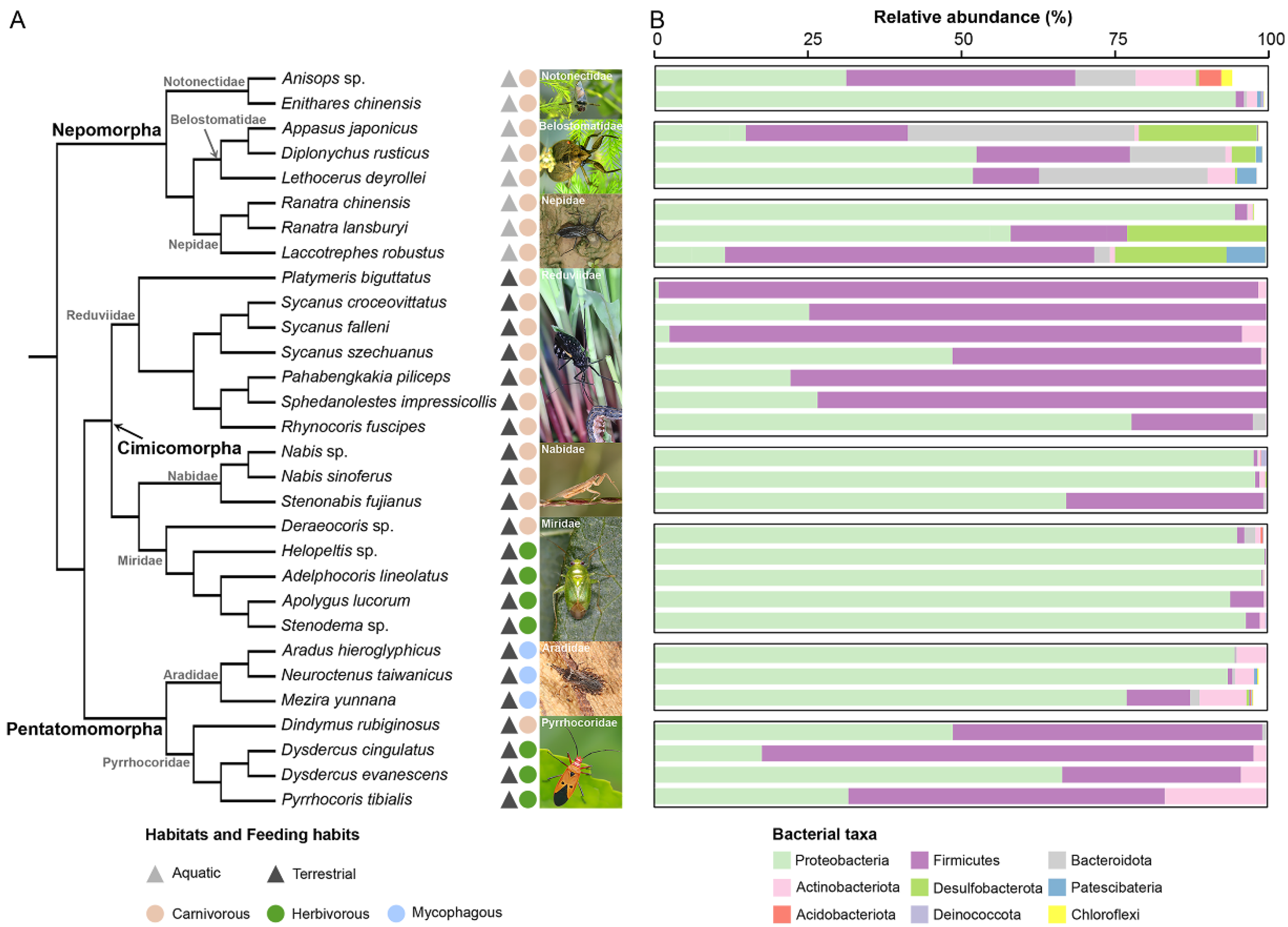
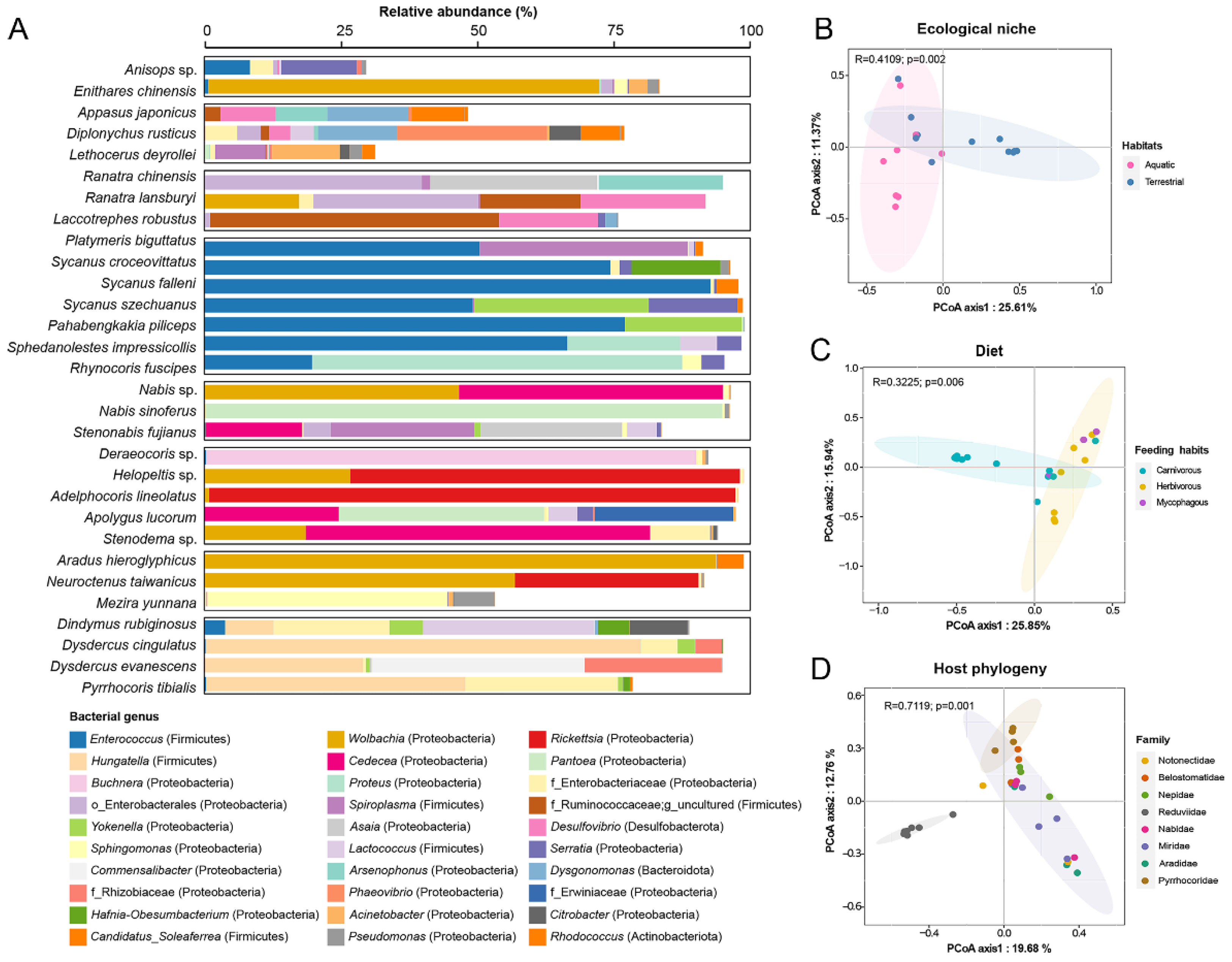
3.1. The Gut Bacterial Community Profile of Heteropteran Insects
3.2. Aquatic and Terrestrial Predatory True Bugs Had Distinct Gut Bacterial Community Profiles
3.3. The Association between Gut Microbiota and Feeding Habits
3.4. Assassin Bugs Had Characterized Gut Bacterial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reis, F.; Kirsch, R.; Pauchet, Y.; Bauer, E.; Bilz, L.C.; Fukumori, K.; Fukatsu, T.; Kölsch, G.; Kaltenpoth, M. Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles. Nat. Commun. 2020, 11, 2964. [Google Scholar] [CrossRef] [PubMed]
- Sudakaran, S.; Kost, C.; Kaltenpoth, M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017, 25, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.M.; McCutcheon, J.P.; MacDonald, B.R.; Romanovicz, D.; Moran, N.A. Differential genome evolution between companion symbionts in an Insect-Bacterial symbiosis. mBio 2014, 5, e01697-14. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Bittleston, L.S.; Hansen, A.K.; Sabree, Z.L.; Moran, N.A.; Almeida, R.P.P. Origin and examination of a leafhopper facultative endosymbiont. Curr. Microbiol. 2011, 62, 1565–1572. [Google Scholar] [CrossRef][Green Version]
- Grimaldi, D.A.; Engel, M.S. Evolution of the Insects; Cambridge University Press: New York: NY, USA, 2005. [Google Scholar]
- Terra, W.; Ferreira, C. Biochemistry of Digestion; Comprehensive Molecular Insect Science; Elsevier: Salt Lake City, UT, USA, 2005; Volume 4, pp. 171–224. [Google Scholar] [CrossRef]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef]
- Kuechler, S.M.; Gibbs, G.; Burckhardt, D.; Dettner, K.; Hartung, V. Diversity of bacterial endosymbionts and bacteria-host co-evolution in Gondwanan relict moss bugs (Hemiptera: Coleorrhyncha: Peloridiidae). Environ. Microbiol. 2013, 15, 2031–2042. [Google Scholar] [CrossRef]
- Matsuura, Y.; Moriyama, M.; Łukasik, P.; Vanderpool, D.; Tanahashi, M.; Meng, X.Y.; McCutcheon, J.P.; Fukatsu, T. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. USA 2018, 115, E5970–E5979. [Google Scholar] [CrossRef]
- Weirauch, C.; Schuh, R.T.; Cassis, G.; Wheeler, W.C. Revisiting habitat and lifestyle transitions in Heteroptera (Insecta: Hemiptera): Insights from a combined morphological and molecular phylogeny. Cladistics 2019, 35, 67–105. [Google Scholar] [CrossRef]
- Henry, T.J. Biodiversity of Heteroptera; Insect Biodiversity: Science and Society; Blackwell Publishing: Oxford, UK, 2009; pp. 223–263. [Google Scholar]
- Schuh, R.T.; Weirauch, C. True Bugs of the World (Hemiptera: Heteroptera): Classification and Natural History, 2nd ed.; Siri Scientific Press: Manchester, UK, 2020. [Google Scholar]
- Colman, D.R.; Toolson, E.C.; Takacs-Vesbach, C.D. Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 2012, 21, 5124–5137. [Google Scholar] [CrossRef]
- Figueroa, L.L.; Maccaro, J.J.; Krichilsky, E.; Yanega, D.; McFrederick, Q.S. Why did the bee eat the chicken? Symbiont gain, loss, and retention in the vulture bee microbiome. MBio 2021, 12, e02317-21. [Google Scholar] [CrossRef]
- Huang, K.; Wang, J.; Huang, J.; Zhang, S.; Vogler, A.P.; Liu, Q.; Li, Y.; Yang, M.; Li, Y.; Zhou, X. Host phylogeny and diet shape gut microbial communities within bamboo-feeding insects. Front. Microbiol. 2021, 12, 633075. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.H.; Roh, S.W.; Whon, T.W.; Jung, M.J.; Kim, M.S.; Park, D.S.; Yoon, C.; Nam, Y.; Kim, Y.; Choi, J.; et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 2014, 80, 5254–5264. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B 2017, 284, 20171223. [Google Scholar] [CrossRef] [PubMed]
- Kaiwa, N.; Hosokawa, T.; Nikoh, N.; Tanahashi, M.; Moriyama, M.; Meng, X.Y.; Maeda, T.; Yamaguchi, K.; Shigenobu, S.; Ito, M.; et al. Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr. Biol. 2014, 24, 2465–2470. [Google Scholar] [CrossRef]
- Kuechler, S.M.; Renz, P.; Dettner, K.; Kehl, S. Diversity of symbiotic organs and bacterial endosymbionts of: Lygaeoid bugs of the families Blissidae and Lygaeidae (Hemiptera: Heteroptera: Lygaeoidea). Appl. Environ. Microbiol. 2012, 78, 2648–2659. [Google Scholar] [CrossRef]
- Salem, H.; Bauer, E.; Strauss, A.S.; Vogel, H.; Marz, M.; Kaltenpoth, M. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. R. Soc. B 2014, 281, 20141838. [Google Scholar] [CrossRef]
- Santos-Garcia, D.; Silva, F.J.; Morin, S.; Dettner, K.; Kuechler, S.M. The all-rounder sodalis: A new bacteriome-associated endosymbiont of the lygaeoid bug Henestaris halophilus (heteroptera: Henestarinae) and a critical examination of its evolution. Genome Biol. Evol. 2017, 9, 2893–2910. [Google Scholar] [CrossRef]
- Hosokawa, T.; Hironaka, M.; Inadomi, K.; Mukai, H.; Nikoh, N.; Fukatsu, T. Diverse strategies for vertical symbiont transmission among subsocial stinkbugs. PLoS ONE 2013, 8, e65081. [Google Scholar] [CrossRef]
- Kaltenpoth, M.; Winter, S.A.; Kleinhammer, A. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 2009, 69, 373–383. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef]
- Kuechler, S.M.; Fukatsu, T.; Matsuura, Y. Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 2019, 21, 4378–4394. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y.; Kikuchi, Y.; Hosokawa, T.; Koga, R.; Meng, X.Y.; Kamagata, Y.; Nikoh, N.; Fukatsu, T. Evolution of symbiotic organs and endosymbionts in lygaeid stinkbugs. ISME J. 2012, 6, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Sudakaran, S.; Salem, H.; Kost, C.; Kaltenpoth, M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Mol. Ecol. 2012, 21, 6134–6151. [Google Scholar] [CrossRef] [PubMed]
- Eichler, S.; Schaub, G.A. Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 2002, 100, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef]
- Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 10257–10262. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open source, platform-independent, community- supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. 2018. Available online: http://www.mesquiteproject.org (accessed on 11 January 2021).
- Johnson, K.P.; Dietrich, C.H.; Friedrich, F.; Beutel, R.G.; Wipfler, B.; Peters, R.S.; Allen, J.M.; Petersen, M.; Donath, A.; Walden, K.K.O.; et al. Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. USA 2018, 115, 12775–12780. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Molecular phylogeny of the plant bugs (Heteroptera: Miridae) and the evolution of feeding habits. Cladistics 2012, 28, 50–79. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Ye, Z.; Damgaard, J.; Yang, H.; Hebsgaard, M.B.; Weir, T.; Bu, W. Phylogeny and diversification of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha). Cladistics 2020, 36, 72–87. [Google Scholar] [CrossRef]
- Naum, M.; Brown, E.W.; Mason-Gamer, R.J. Is 16S rDNA a reliable phylogenetic marker to characterize relationships below the family level in the enterobacteriaceae? J. Mol. Evol. 2008, 66, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.M.; Saltonstall, K.; Arias, C.F.; Chavarria, K.A.; Ramírez-Camejo, L.A.; Mejía, L.C.; De León, L.F. The microbiome of neotropical water striders and its potential role in codiversification. Insects 2020, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Dally, M.; Lalzar, M.; Belausov, E.; Gottlieb, Y.; Coll, M.; Zchori-Fein, E. Cellular localization of two Rickettsia symbionts in the digestive system and within the ovaries of the mirid bug, Macrolophous pygmaeus. Insects 2020, 11, 530. [Google Scholar] [CrossRef]
- Hirose, E.; Panizzi, A.R.; Souza, J.T.D.; Cattelan, A.J.; Aldrich, J.R. Bacteria in the gut of southern green stink bug (Heteroptera: Pentatomidae). Ann. Entomol. Soc. Am. 2006, 99, 91–95. [Google Scholar] [CrossRef]
- Matsuura, Y.; Kikuchi, Y.; Meng, X.Y.; Koga, R.; Fukatsu, T. Novel clade of Alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl. Environ. Microbiol. 2012, 78, 4149–4156. [Google Scholar] [CrossRef]
- Martiny, A.C.; Treseder, K.; Pusch, G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013, 7, 830–838. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef]
- Zheng, H.; Perreau, J.; Powell, J.E.; Han, B.; Zhang, Z.; Kwong, W.K.; Tringe, S.G.; Moran, N.A. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. USA 2019, 116, 25909–25916. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.A.; Yoon, J.H.; Ryu, J.H.; Lee, W.J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef]
- Lang, H.; Duan, H.; Wang, J.; Zhang, W.; Guo, J.; Zhang, X.; Hu, X.; Zheng, H. Specific strains of honeybee gut lactobacillus stimulate host immune system to protect against pathogenic Hafnia alvei. Microbiol. Spectr. 2022, 10, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Sudakaran, S.; Retz, F.; Kikuchi, Y.; Kost, C.; Kaltenpoth, M. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 2015, 9, 2587–2604. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Yeves, P.J.; Callieri, C.; Picazo, A.; Mehrshad, M.; Haro-Moreno, J.M.; Roda-Garcia, J.J.; Dzhembekova, N.; Slabakova, V.; Slabakova, N.; Moncheva, S.; et al. The microbiome of the Black Sea water column analyzed by shotgun and genome centric metagenomics. Environ. Microbiol. 2021, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Meyer, T.E.; Kyndt, J.A. Genome sequence of the unusual purple photosynthetic bacterium Phaeovibrio sulfidiphilus, only distantly related to Rhodospirillaceae, reveals unique genes for respiratory nitrate reduction and glycerol metabolism. Microbiol. Resour. Announc. 2020, 9, e01200-20. [Google Scholar] [CrossRef]
- Lakshmi, K.V.N.S.; Sasikala, C.; Ashok Kumar, G.V.; Chandrasekaran, R.; Ramana, C.V. Phaeovibrio sulfidiphilus gen. nov., sp. Nov., phototrophic alphaproteobacterial isolated from brackish water. Int. J. Syst. Evol. Microbiol. 2011, 61, 828–833. [Google Scholar] [CrossRef][Green Version]
- Salem, H.; Kreutzer, E.; Sudakaran, S.; Kaltenpoth, M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 2013, 15, 1956–1968. [Google Scholar] [CrossRef]
- Rodríguez-Ruano, S.M.; Škochová, V.; Rego, R.O.M.; Schmidt, J.O.; Roachell, W.; Hypša, V.; Nováková, E. Microbiomes of North American Triatominae: The grounds for Chagas disease epidemiology. Front. Microbiol. 2018, 9, 1167. [Google Scholar] [CrossRef]
- Chaston, J.M.; Murfin, K.E.; Heath-Heckman, E.A.; Goodrich-Blair, H. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cell. Microbiol. 2013, 15, 1545–1559. [Google Scholar] [CrossRef]
- Snyder, H.; Stock, S.P.; Kim, S.K.; Flores-Lara, Y.; Forst, S. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl. Environ. Microbiol. 2007, 73, 5338–5346. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Cheng, Y.; Guo, L.; Wang, A.; Lu, M.; Xu, L. Variation of gut microbiota caused by an imbalance diet is detrimental to bugs’ survival. Sci. Total Environ. 2021, 771, 144880. [Google Scholar] [CrossRef] [PubMed]
- Medina, V.; Sardoy, P.M.; Soria, M.; Vay, C.A.; Gutkind, G.O.; Zavala, J.A. Characterized non-transient microbiota from stinkbug (Nezara viridula) midgut deactivates soybean chemical defenses. PLoS ONE 2018, 13, e0200161. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.B.; Lehman, R.M.; Brözel, V.S.; Lundgren, J.G. An indigenous gut bacterium, Enterococcus faecalis (Lactobacillales: Enterococcaceae), increases seed consumption by Harpalus pensylvanicus (Coleoptera: Carabidae). Fla. Entomol. 2014, 97, 575–584. [Google Scholar] [CrossRef]
- Vilanova, C.; Baixeras, J.; Latorre, A.; Porcar, M. The generalist inside the specialist: Gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. Microbiol. 2016, 7, 1005. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, B.; Sun, C.; Ishida, K.; Hertweck, C.; Boland, W. Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Fla. Entomol. 2017, 24, 66–75. [Google Scholar] [CrossRef]
| Host Taxon | Host Species | Pielou’s Evenness | Chao1 | Shannon | Simpson’s Index of Diversity | |
|---|---|---|---|---|---|---|
| Infraorder | Family | |||||
| Nepomorpha | Notonectidae | Anisops sp. | 0.847 | 203.00 | 4.22 | 24.39 |
| Enithares chinensis | 0.344 | 90.50 | 1.55 | 1.93 | ||
| Belostomatidae | Appasus japonicus | 0.662 | 93.09 | 2.79 | 11.63 | |
| Diplonychus rusticus | 0.635 | 117.50 | 2.65 | 8.20 | ||
| Lethocerus deyrollei | 0.805 | 158.67 | 3.62 | 20.00 | ||
| Nepidae | Ranatra chinensis | 0.353 | 36.43 | 1.41 | 3.29 | |
| Ranatra lansburyi | 0.634 | 19.00 | 1.70 | 4.74 | ||
| Laccotrephes robustus | 0.527 | 55.75 | 1.75 | 3.09 | ||
| Cimicomorpha | Reduviidae | Platymeris biguttatus | 0.367 | 32.00 | 1.12 | 2.47 |
| Sycanus croceovittatus | 0.386 | 9.00 | 0.87 | 1.72 | ||
| Sycanus falleni | 0.261 | 10.00 | 0.36 | 1.16 | ||
| Sycanus szechuanus | 0.482 | 17.00 | 1.15 | 2.70 | ||
| Pahabengkakia piliceps | 0.210 | 26.60 | 0.63 | 1.56 | ||
| Sphedanolestes impressicollis | 0.606 | 5.00 | 0.99 | 2.03 | ||
| Rhynocoris fuscipes | 0.465 | 25.33 | 1.09 | 1.99 | ||
| Nabidae | Nabis sp. | 0.240 | 75.00 | 1.02 | 2.22 | |
| Nabis sinoferus | 0.374 | 56.38 | 0.38 | 1.12 | ||
| Stenonabis fujianus | 0.514 | 48.33 | 1.91 | 5.18 | ||
| Miridae | Deraeocoris sp. | 0.223 | 53.20 | 0.68 | 1.24 | |
| Helopeltis sp. | 0.232 | 37.25 | 0.72 | 1.72 | ||
| Adelphocoris lineolatus | 0.066 | 42.50 | 0.25 | 1.08 | ||
| Apolygus lucorum | 0.603 | 40.00 | 1.58 | 3.70 | ||
| Stenodema sp. | 0.555 | 27.00 | 1.24 | 2.25 | ||
| Pentatomomorpha | Aradidae | Aradus hieroglyphicus | 0.151 | 27.00 | 0.31 | 1.14 |
| Neuroctenus taiwanicus | 0.309 | 120.46 | 1.28 | 2.29 | ||
| Mezira yunnana | 0.598 | 134.91 | 2.68 | 4.67 | ||
| Pyrrhocoridae | Dindymus rubiginosus | 0.642 | 35.67 | 2.13 | 5.78 | |
| Dysdercus cingulatus | 0.392 | 28.60 | 0.89 | 1.56 | ||
| Dysdercus evanescens | 0.472 | 29.25 | 1.41 | 3.34 | ||
| Pyrrhocoris tibialis | 0.547 | 53.88 | 1.49 | 3.13 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Sun, J.; Meng, Y.; Yang, C.; Chen, Z.; Wu, Y.; Tian, L.; Song, F.; Cai, W.; Zhang, X.; et al. The Impact of Environmental Habitats and Diets on the Gut Microbiota Diversity of True Bugs (Hemiptera: Heteroptera). Biology 2022, 11, 1039. https://doi.org/10.3390/biology11071039
Li G, Sun J, Meng Y, Yang C, Chen Z, Wu Y, Tian L, Song F, Cai W, Zhang X, et al. The Impact of Environmental Habitats and Diets on the Gut Microbiota Diversity of True Bugs (Hemiptera: Heteroptera). Biology. 2022; 11(7):1039. https://doi.org/10.3390/biology11071039
Chicago/Turabian StyleLi, Guannan, Jingjing Sun, Yujie Meng, Chengfeng Yang, Zhuo Chen, Yunfei Wu, Li Tian, Fan Song, Wanzhi Cai, Xue Zhang, and et al. 2022. "The Impact of Environmental Habitats and Diets on the Gut Microbiota Diversity of True Bugs (Hemiptera: Heteroptera)" Biology 11, no. 7: 1039. https://doi.org/10.3390/biology11071039
APA StyleLi, G., Sun, J., Meng, Y., Yang, C., Chen, Z., Wu, Y., Tian, L., Song, F., Cai, W., Zhang, X., & Li, H. (2022). The Impact of Environmental Habitats and Diets on the Gut Microbiota Diversity of True Bugs (Hemiptera: Heteroptera). Biology, 11(7), 1039. https://doi.org/10.3390/biology11071039







