Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Culture Conditions, Plant Material and Treatments
2.2. Estimation of MDA Content and O2.- Production Rate
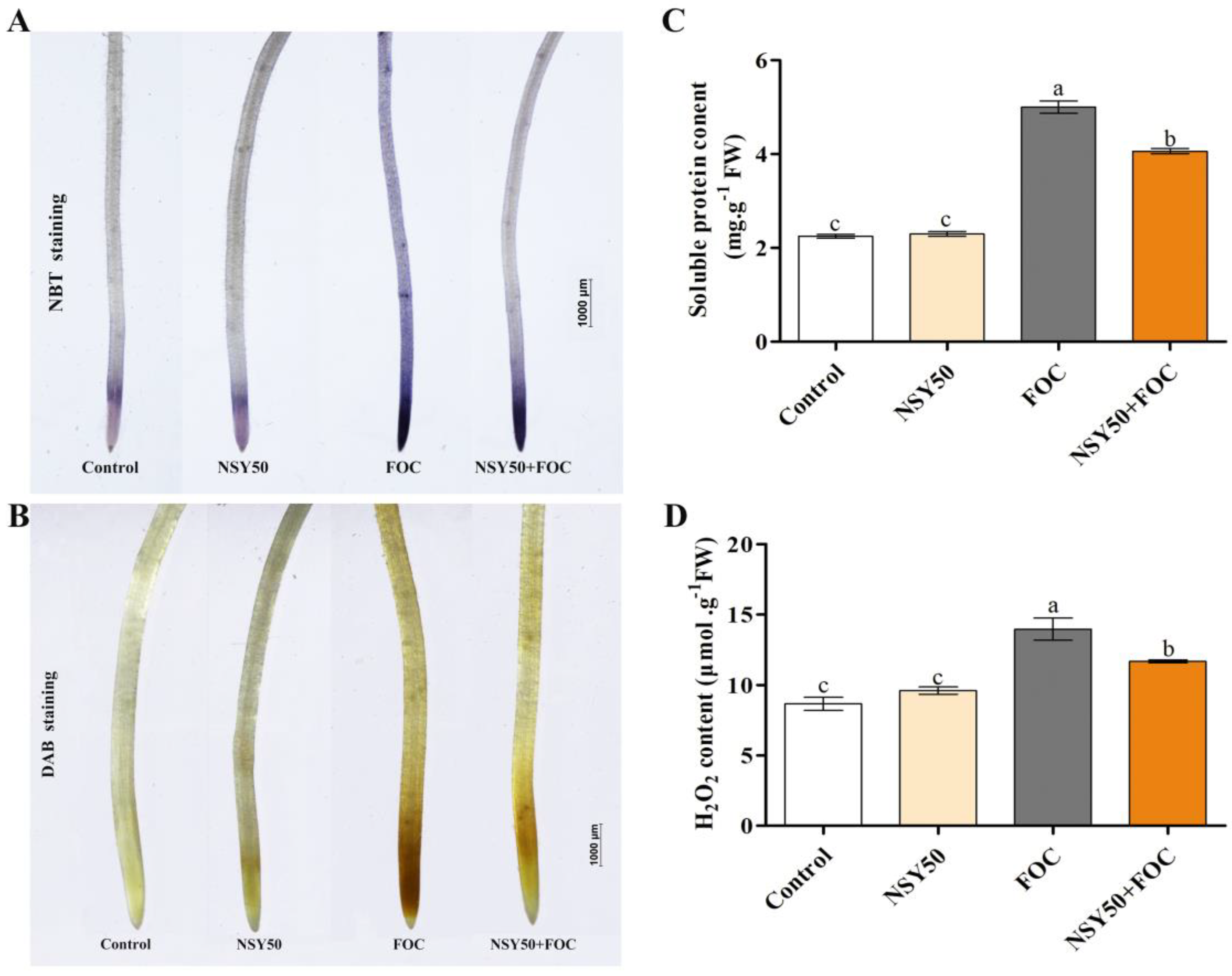
2.3. Determination of H2O2 and Soluble Protein Contents and Histochemical Detection of H2O2 and O2.- in Roots
2.4. Metabolite Analyses
2.5. Measurements of GSH Content and Enzyme Activities
2.6. RNA Extraction, and Gene Expression Analysis by qRT-PCR
2.7. Statistical Analysis
3. Results
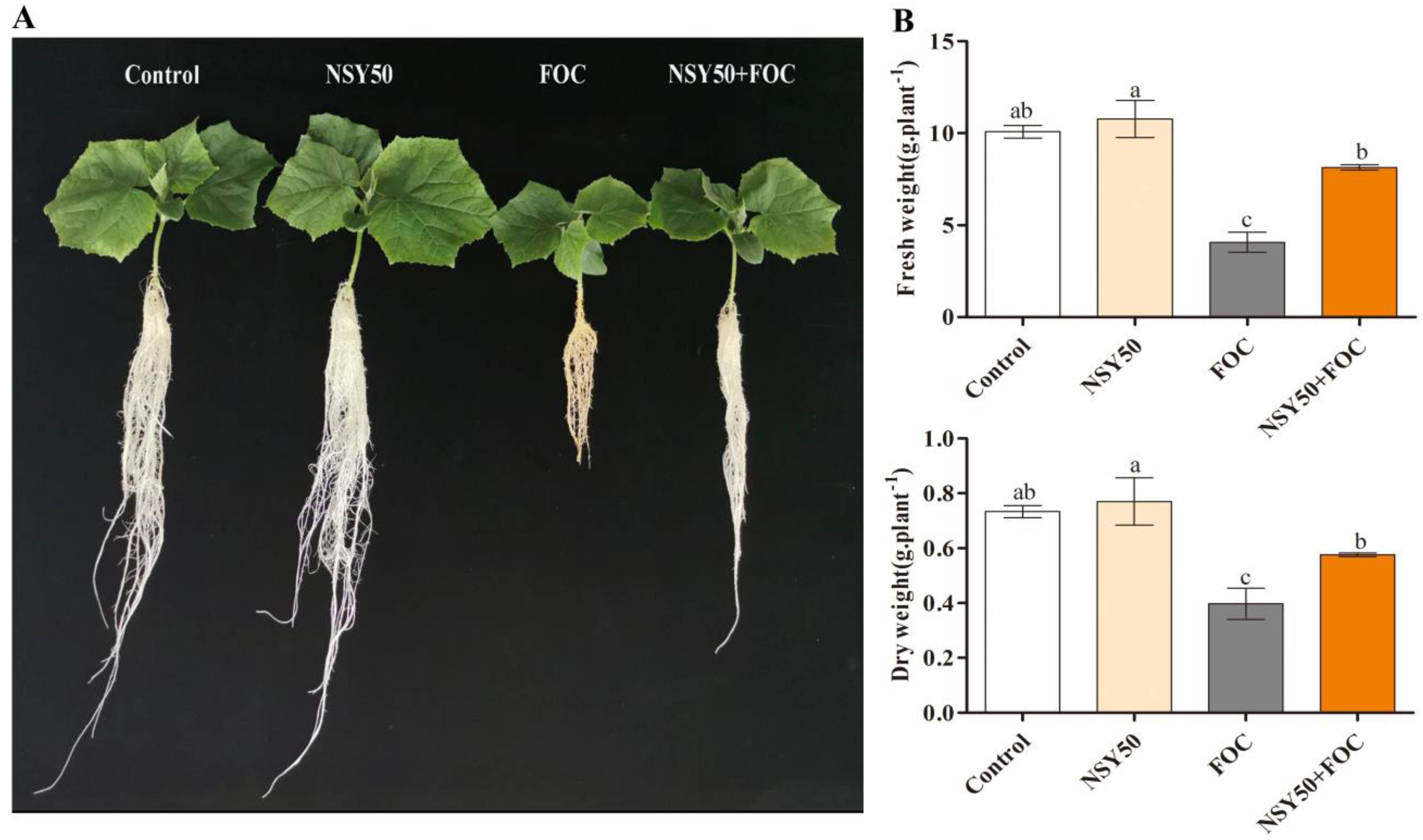
3.1. Characteristics of Plant Growth under Different Treatments
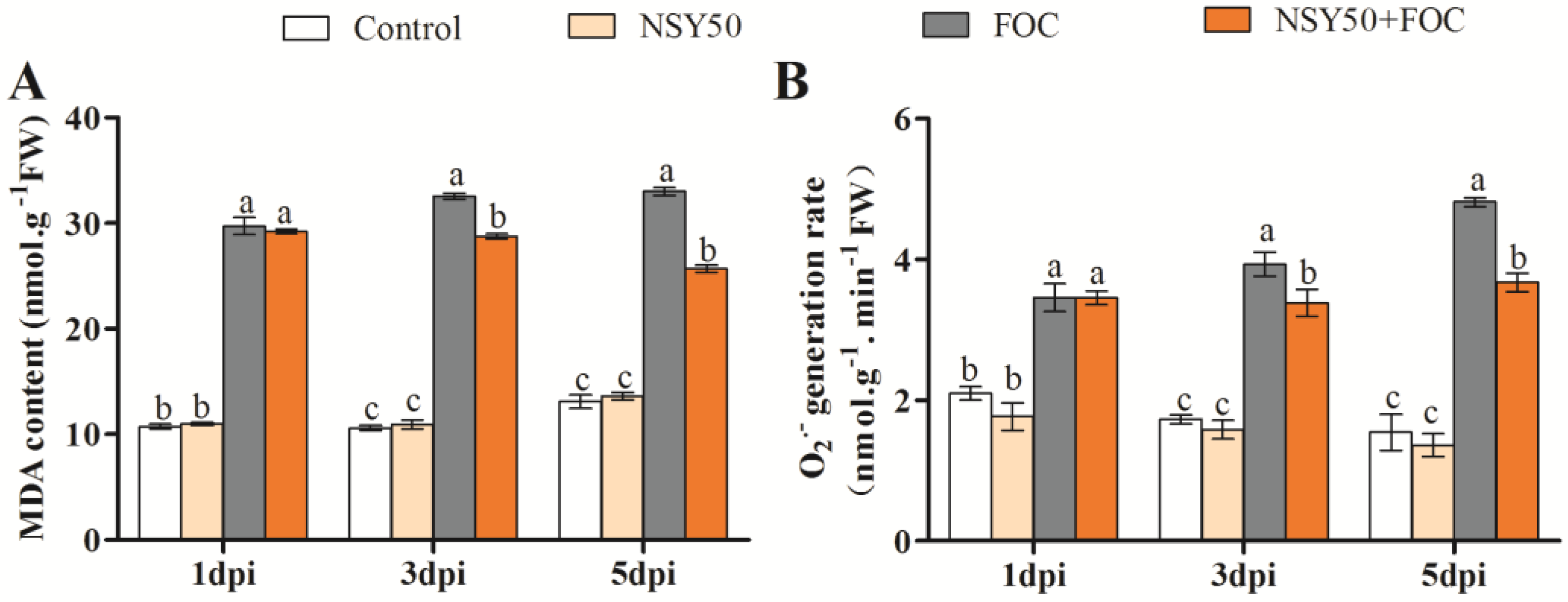
3.2. Lipid Peroxidation
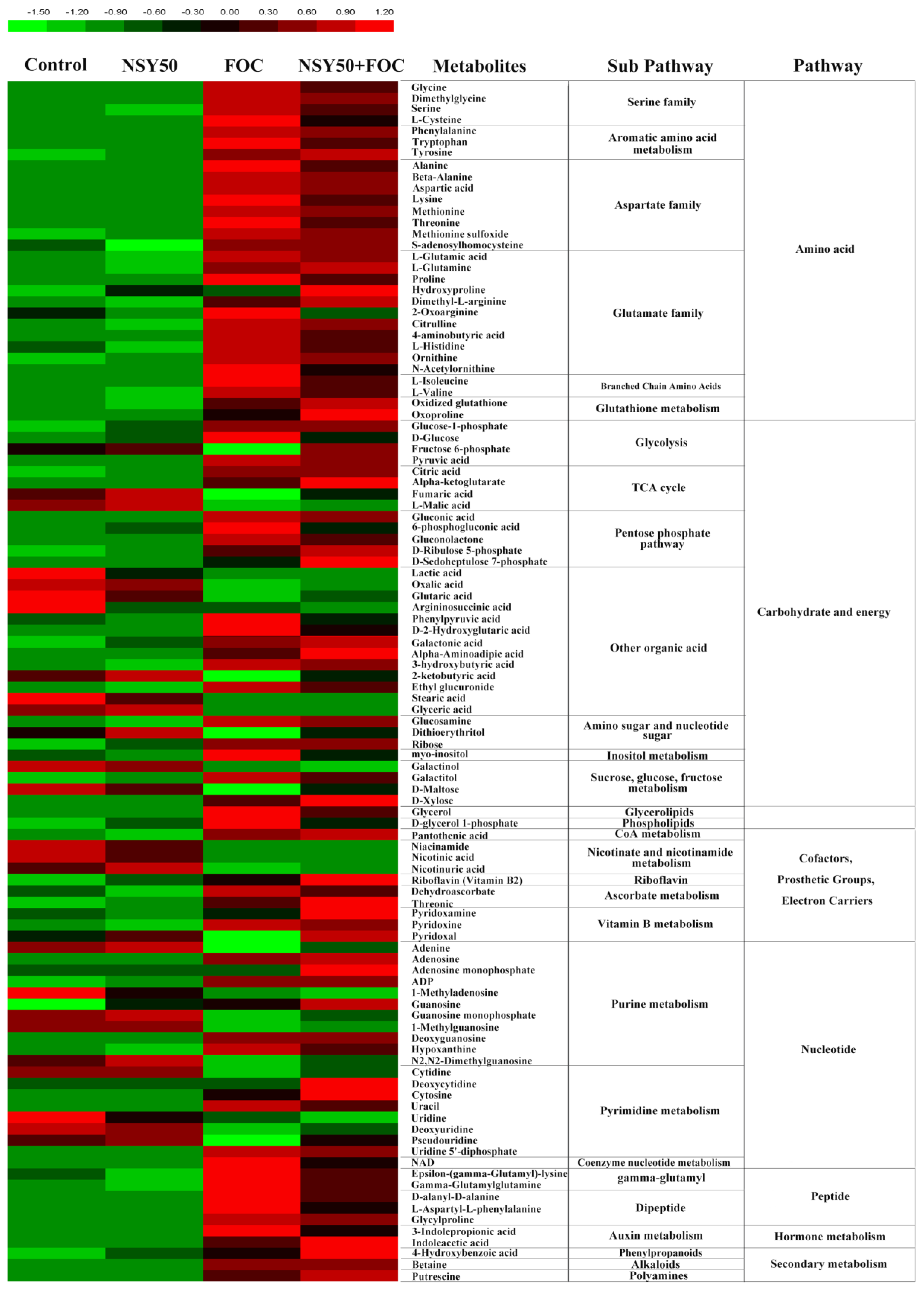
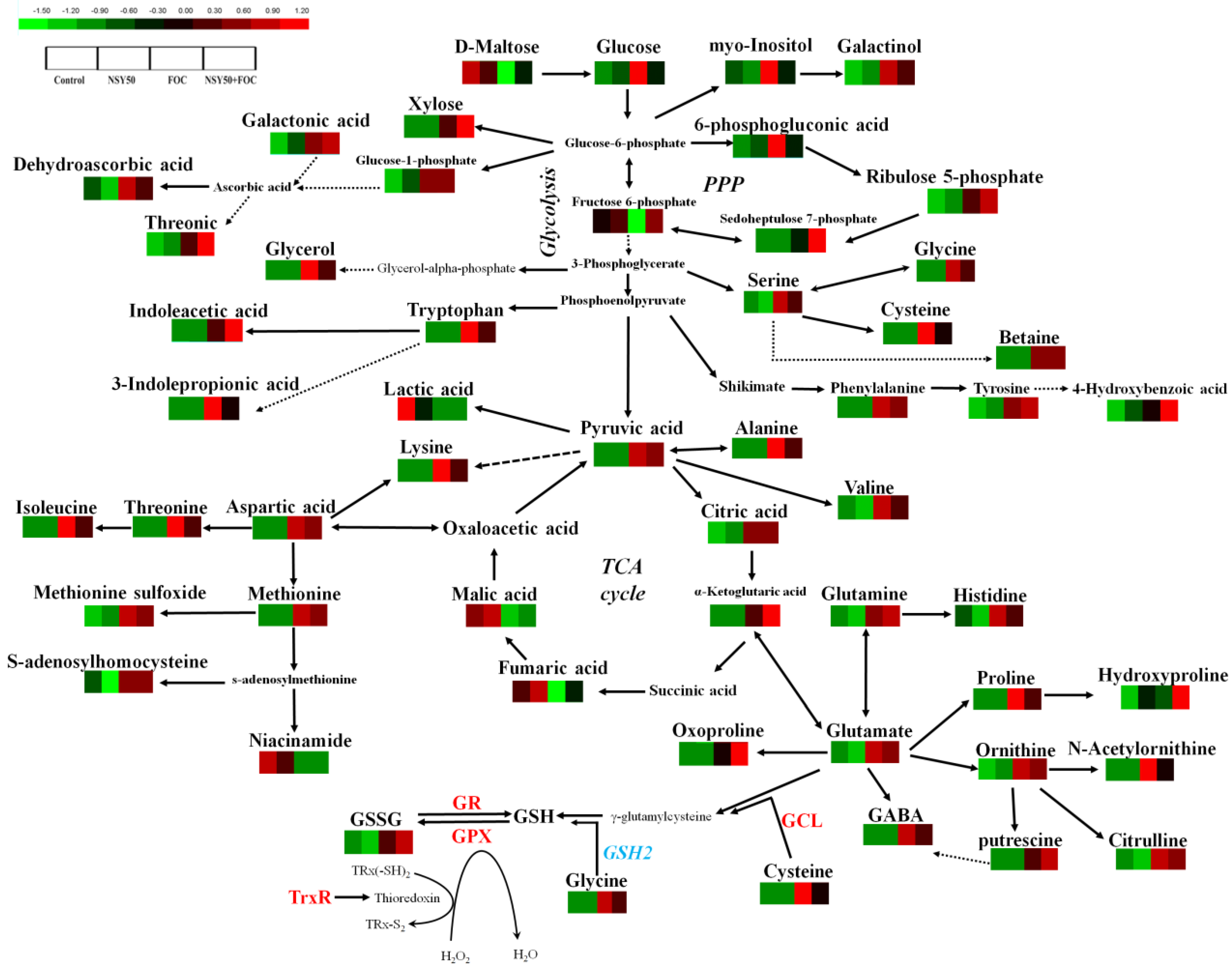
3.3. Metabolomic Analysis
3.4. GSH Content and Activities of GSH-Related Enzymes in Response to NSY50 and FOC Inoculation
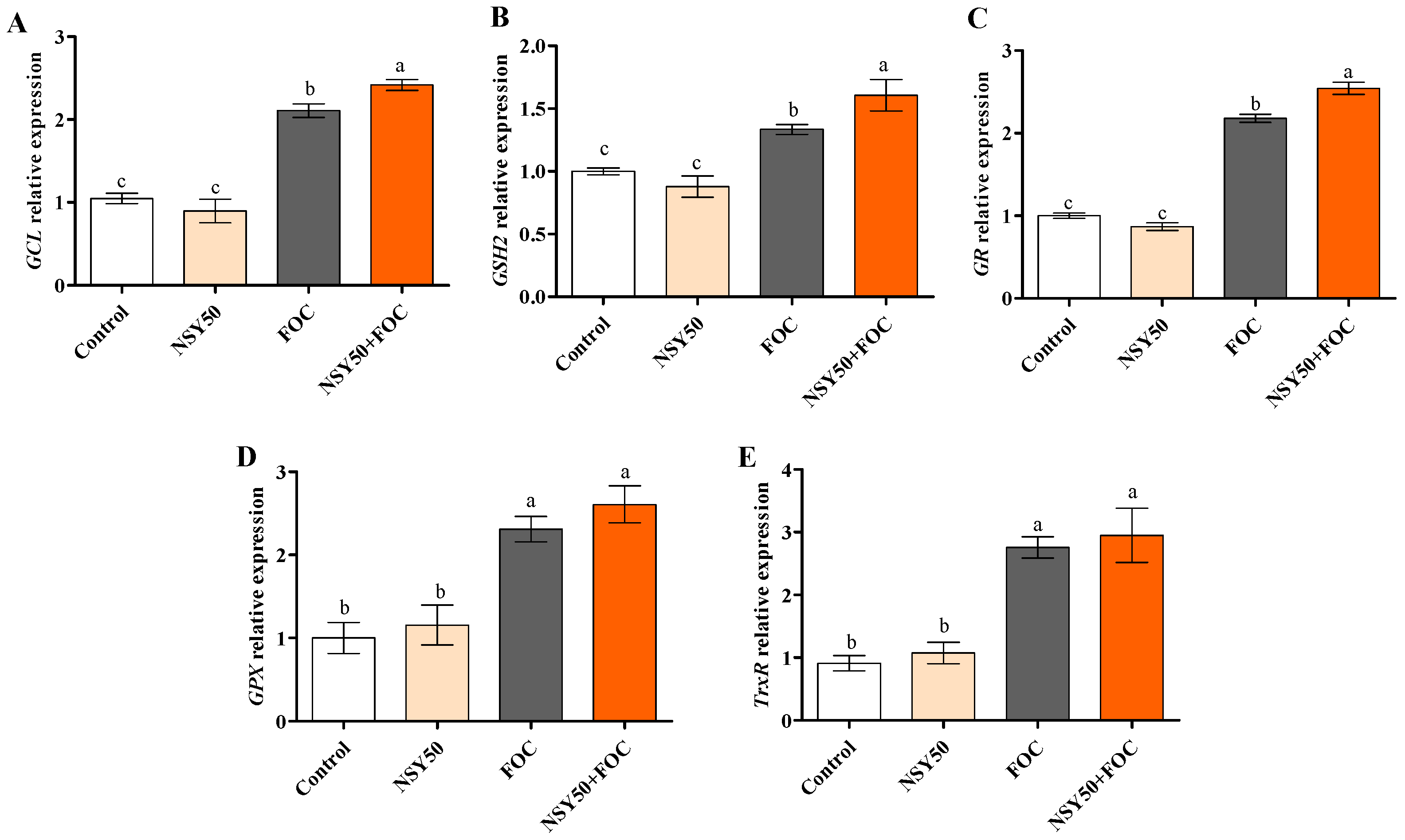
3.5. Expression Levels of Genes Related to GSH Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, N.; Shi, L.; Du, L.; Yuan, Y.; Li, B.; Sang, T.; Guo, S. Effect of vinegar residue compost amendments on cucumber growth and Fusarium wilt. Environ. Sci. Pollut. Res. 2015, 22, 19133–19141. [Google Scholar] [CrossRef]
- Huang, X.; Shi, D.; Sun, F.; Lu, H.; Liu, J.; Wu, W. Efficacy of sludge and manure compost amendments against Fusarium wilt of cucumber. Environ. Sci. Pollut. Res. 2012, 19, 3895–3905. [Google Scholar] [CrossRef]
- Shi, L.; Du, N.; Shu, S.; Sun, J.; Li, S.; Guo, S. Paenibacillus polymyxa NSY50 suppresses Fusarium wilt in cucumbers by regulating the rhizospheric microbial community. Sci. Rep. 2017, 7, 41234. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Wang, Y.; Xian, Q.; Chen, X.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant. Biol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Chen, D.; Liu, X.; Li, C.; Tian, W.; Shen, Q.; Shen, B. Isolation of Bacillus amyloliquefaciens S20 and its application in control of eggplant bacterial wilt. J. Environ. Manag. 2014, 137, 120–127. [Google Scholar] [CrossRef]
- Zhang, H.; Mallik, A.; Zeng, R. Control of Panama disease of banana by rotating and intercropping with Chinese chive (Allium tuberosum Rottler): Role of plant volatiles. J. Chem. Ecol. 2013, 39, 243–252. [Google Scholar] [CrossRef]
- Nel, B.; Steinberg, C.; Labuschagne, N.; Viljoen, A. Evaluation of fungicides and sterilants for potential application in the management of Fusarium wilt of banana. Crop. Prot. 2007, 26, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xu, Z.; Ning, L.; Yuan, Y.; Yang, X.; Chen, L.; Shen, B.; Shen, Q. Isolation and identification of lipopeptides produced by B. subtilis SQR 9 for suppressing Fusarium wilt of cucumber. Sci. Hortic. 2012, 35, 32–39. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Abdel-Motaal, F.; El-Sayed, M.; Jogaiah, S.; Shigyo, M.; Ito, S.I.; Tran, L.S. Dissection of Trichoderma longibrachiatum-induced defense in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Sci. 2016, 246, 128–138. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhang, R.; Wang, D.; Qiu, M.; Feng, H.; Zhang, N.; Shen, Q. Enhanced control of cucumber wilt disease by Bacillus amyloliquefaciens SQR9 by altering the regulation of Its DegU phosphorylation. Appl. Environ. Microbiol. 2014, 80, 2941–2950. [Google Scholar] [CrossRef] [Green Version]
- Sudisha, J.; Mostafa, A.; Phan, L.S.; Ito, S.I. Characterization of rhizosphere fungi that mediate resistance in tomato against bacterial wilt disease. J. Exp. Bot. 2013, 64, 3829–3842. [Google Scholar]
- Babu, N.A.; Jogaiah, S.; Ito, S.; Nagaraj, A.K.; Tran, L.P. Improvement of growth, fruit weight and early blight disease protection of tomato plants by rhizosphere bacteria is correlated with their beneficial traits and induced biosynthesis of antioxidant peroxidase and polyphenol oxidase. Plant Sci. 2015, 231, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Saberi Riseh, R.; Skorik, Y.A.; Thakur, V.K.; Moradi Pour, M.; Tamanadar, E.; Noghabi, S.S. Encapsulation of Plant Biocontrol Bacteria with Alginate as a Main Polymer Material. Int. J. Mol. Sci. 2021, 22, 11165. [Google Scholar] [CrossRef]
- Timmusk, S.; Grantcharova, N.; Wagner, E.G.H. Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 2005, 71, 7292–7300. [Google Scholar] [CrossRef] [Green Version]
- Raza, W.; Yuan, J.; Wu, Y.C.; Rajer, F.U.; Huang, Q.; Qirong, S. Biocontrol traits of two Paenibacillus polymyxa strains SQR-21 and WR-2 in response to fusaric acid, a phytotoxin produced by Fusarium species. Plant Pathol. 2015, 64, 1041–1052. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kotnala, B.; Kim, Y.H.; Jeon, Y. Biological characteristics of Paenibacillus polymyxa GBR-1 involved root rot of stored Korean ginseng. J. Ginseng. Res. 2016, 40, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Guo, J.S.; Zhu, L.; Xiao, X.; Xie, Y.; Zhu, J.; Ma, Z.Y.; Wang, J. Paenibacillus polymyxa BFKC01 enhances plant iron absorption via improved root systems and activated iron acquisition mechanisms. Plant Physiol. Biochem. 2016, 105, 162–173. [Google Scholar] [CrossRef]
- Beatty, P.H.; Jensen, S.E. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can. J. Microbiol. 2002, 48, 159–169. [Google Scholar] [CrossRef]
- He, Z.; Kisla, D.; Zhang, L.; Yuan, C.; Green-Church, K.B.; Yousef, A.E. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 2007, 73, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Ariza, A.; Eklof, J.M.; Spadiut, O.; Offen, W.A.; Roberts, S.M.; Besenmatter, W.; Friis, E.P.; Skjot, M.; Wilson, K.S.; Brumer, H.; et al. Structure and activity of Paenibacillus polymyxa xyloglucanase from glycoside hydrolase family 44. J. Biol. Chem. 2011, 286, 33890–33900. [Google Scholar] [CrossRef] [Green Version]
- Du, N.; Shi, L.; Yuan, Y.; Sun, J.; Shu, S.; Guo, S. Isolation of a potential biocontrol agent Paenibacillus polymyxa NSY50 from vinegar waste compost and its induction of host defense responses against Fusarium wilt of cucumber. Microbiol. Res. 2017, 202, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.E.; Kwon, S.Y.; Park, J.M. Biocontrol activity of Paenibacillus polymyxa AC-1 against Pseudomonas syringae and its interaction with Arabidopsis thaliana. Microbiol. Res. 2016, 185, 13–21. [Google Scholar] [CrossRef]
- Khan, Z.; Kim, S.G.; Jeon, Y.H.; Khan, H.U.; Son, S.H.; Kim, Y.H. A plant growth promoting rhizobacterium, Paenibacillus polymyxa strain GBR-1, suppresses root-knot nematode. Bioresour. Technol. 2008, 99, 3016–3023. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cho, Y.E.; Park, S.H.; Balaraju, K.; Park, J.W.; Lee, S.W.; Park, K. An antibiotic fusaricidin: A cyclic depsipeptide from Paenibacillus polymyxa E681 induces systemic resistance against Phytophthora blight of red-pepper. Phytoparasitica 2013, 41, 49–58. [Google Scholar] [CrossRef]
- Mahmood, A.; Kataoka, R. Metabolite profiling reveals a complex response of plants to application of plant growth-promoting endophytic bacteria. Microbiol. Res. 2020, 234, 126421. [Google Scholar] [CrossRef]
- Du, N.; Shi, L.; Yuan, Y.; Li, B.; Shu, S.; Sun, J.; Guo, S. Proteomic Analysis Reveals the Positive Roles of the Plant-Growth-Promoting Rhizobacterium NSY50 in the Response of Cucumber Roots to Fusarium oxysporum f. sp. cucumerinum Inoculation. Front. Plant Sci. 2016, 7, 1859. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.S.; Lee, D.Y.; Rakwal, R.; Baek, S.B.; Lee, J.H.; Kwak, Y.S.; Seo, J.S.; Chung, W.S.; Bae, D.W.; Kim, S.G. Proteomic analyses of the interaction between the plant—Growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 2016, 16, 122–135. [Google Scholar] [CrossRef]
- Xie, J.; Shi, H.; Du, Z.; Wang, T.; Liu, X.; Chen, S. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016, 6, 21329. [Google Scholar] [CrossRef]
- Liu, S.L.; Yang, R.J.; Ma, M.D.; Dan, F.; Zhao, Y.; Jiang, P.; Wang, M.H. Effects of exougenous NO on the growth, mineral nutrient content, antioxidant system, and ATPase activities of Trifolium repens L. plants under cadmium stress. Acta Physiol. Plant. 2015, 37, 1721. [Google Scholar] [CrossRef]
- Elstner, E.F.; Heipel, A. Inhibition of nitrite formation from hydroxyl ammonium chloride: A simple assay for superoxide dismutase. Anal. Biochem. 1976, 70, 616–620. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, M.; Du, N.; Shu, S.; Sun, J.; Guo, S. Putrescine enhances salt tolerance of cucumber seedlings by regulating ion homeostasis. Environ. Exp. Bot. 2019, 165, 70–82. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–252. [Google Scholar] [CrossRef]
- Frahry, G.; Schopfer, P. NADH-stimulated, cyanide-resistant superoxide production in maize coleoptiles analyzed with a tetrazolium-based assay. Planta 2001, 212, 175–183. [Google Scholar] [CrossRef]
- Guo, Z.; Lv, J.; Zhang, H.; Hu, C.; Qin, Y.; Dong, H.; Zhang, T.; Dong, X.; Du, N.; Piao, F. Red and blue light function antagonistically to regulate cadmium tolerance by modulating the photosynthesis, antioxidant defense system and Cd uptake in cucumber (Cucumis sativus L.). J. Hazard. Mater. 2022, 429, 128412. [Google Scholar] [CrossRef]
- Liu, B.; Peng, X.; Han, L.; Hou, L.; Li, B. Effects of Exogenous Spermidine on Root Metabolism of Cucumber Seedlings under Salt Stress by GC-MS. Agronomy 2020, 10, 459. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cao, W.; Xi, P.; Li, L.; Qiao, S.; Luo, H.; Zhang, J.; Liu, X.; Du, N. S-glycosylation of Fluensulfone in Tomatoes: An Important Way of Fluensulfone Metabolism. J. Agric. Food Chem. 2021, 69, 12974–12984. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Wang, Y.; Hou, K.; Shu, S.; Sun, J.; Guo, S.R. TGase positively regulates photosynthesis via activation of Calvin cycle enzymes in tomato. Hortic. Res. 2019, 6, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B.; Foyer, C.H. Ascorbic acid, metal ions and the superoxide radical. Biochem. J. 1976, 155, 697–700. [Google Scholar] [CrossRef]
- Quesada, M.P.; Macheix, J.J. Caractérisation d’une peroxidase impliquée spécifiquement dans la lignification, en relation avec l’incompatibilité au greffage chez l’abricotier. Physiol. Vég. 1984, 22, 533–540. [Google Scholar]
- Dhinsa, R.S. Inhibition of protein synthesis by products of lipid peroxidation. Phytochemistry 1982, 21, 309–313. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lin, X.; Wang, J.; Shen, W.; Wu, S.; Peng, S.; Mao, T. Arbuscular Mycorrhizal Fungal Inoculation Enhances Suppression of Cucumber Fusarium Wilt in Greenhouse Soils. Pedosphere 2010, 20, 586–593. [Google Scholar] [CrossRef]
- Zhai, Y.; Zhu, J.X.; Tan, T.M.; Xu, J.P.; Wei, L. Isolation and characterization of antagonistic Paenibacillus polymyxa HX-140 and its biocontrol potential against Fusarium wilt of cucumber seedlings. BMC Microbiol. 2021, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Kejela, T.; Thakkar, V.R.; Patel, R.R. A novel strain of Pseudomonas inhibits Colletotrichum gloeosporioides and Fusarium oxysporum infections and promotes germination of coffee. Rhizosphere 2017, 4, 9–15. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Maji, D.; Chanotiya, C.S.; Kalra, A. 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol. Bioch. 2012, 58, 227–235. [Google Scholar] [CrossRef]
- Patel, R.R.; Patel, D.D.; Bhatt, J.; Thakor, P.; Triplett, L.R.; Thakkar, V.R. Induction of pre-chorismate, jasmonate and salicylate pathways by Burkholderia sp. RR18 in peanut seedlings. J. Appl. Microbiol. 2021, 131, 1417–1430. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, F.G. Redox Homeostasis and Antioxidant Signaling: A Metabolic Interface between Stress Perception and Physiological Responses. Plant Cell. 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Hamilton, C.E.; Gundel, P.E.; Helander, M.; Saikkonen, K. Endophytic mediation of reactive oxygen species and antioxidant activity in plants: A review. Fungal Divers. 2012, 54, 1–10. [Google Scholar] [CrossRef]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 2018, 154, 134–142. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, H.; Wang, Z.; Zheng, C.; Zhao, P.; Guan, Z.; Qin, H.; Liu, A.; Lin, X.; Ahammed, G.J. Trichoderma harzianum-induced resistance against Fusarium oxysporum involves regulation of nuclear DNA content, cell viability and cell cycle-related genes expression in cucumber roots. Eur. J. Plant Pathol. 2017, 147, 43–53. [Google Scholar] [CrossRef]
- Li, M.; Ma, G.; Lian, H.; Su, X.; Tian, Y.; Huang, W.; Mei, J.; Jiang, X. The effects of Trichoderma on preventing cucumber fusarium wilt and regulating cucumber physiology. J. Integr. Agric. 2019, 18, 607–617. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Ren, J.; Zhao, H.; Wang, X.; Wang, T.; Jin, S.; Wang, Z.; Li, C.; Liu, A.; Lin, X. Trichoderma harzianum Improves Defense Against Fusarium oxysporum by Regulating ROS and RNS Metabolism, Redox Balance, and Energy flow in Cucumber Roots. Phytopathology 2019, 109, 972–982. [Google Scholar] [CrossRef]
- Yuan, P.; Pan, H.; Boak, E.N.; Pierson, L.S.; Pierson, E.A. Phenazine-Producing Rhizobacteria Promote Plant Growth and Reduce Redox and Osmotic Stress in Wheat Seedlings Under Saline Conditions. Front. Plant Sci. 2020, 11, 1442. [Google Scholar] [CrossRef]
- Gao, C.; Ma, G.; Lian, H.; Liu, M.; Zhang, C.; Qu, H. Effect of Trichoderma on the Growth of Cucumber Seedlings, Membrane Lipid Indexes and Control Effect against Fusarium Wilt. Chin. J. Biol. Control. 2018, 34, 762–770. (In Chinese) [Google Scholar]
- Mundim, F.M.; Pringle, E.G. Whole-Plant Metabolic Allocation Under Water Stress. Front. Plant Sci. 2018, 9, 852. [Google Scholar]
- Carrera, F.P.; Noceda, C.; Maridueña-Zavala, M.G.; Cevallos-Cevallos, J.M. Metabolomics, a Powerful Tool for Understanding Plant Abiotic Stress. Agronomy 2021, 11, 824. [Google Scholar] [CrossRef]
- Bittleston, L.S.; Brockmann, F.; Wcislo, W.; Van Bael, S.A. Endophytic fungi reduce leaf-cutting ant damage to seedlings. Biol Lett. 2011, 7, 30–32. [Google Scholar] [CrossRef]
- Zuluaga, M.; Milani, K.; Miras-Moreno, M.B. The adaptive metabolomic profile and functional activity of tomato rhizosphere are revealed upon PGPB inoculation under saline stress. Environ. Exp. Bot. 2021, 189, 104552. [Google Scholar] [CrossRef]
- He, T.; Xu, Z.J.; Wang, J.F.; Wang, F.P.; Zhou, X.F.; Wang, L.L.; Li, Q.S. Improving cadmium accumulation by Solanum nigrum L. via regulating rhizobacterial community and metabolic function with phosphate-solubilizing bacteria colonization. Chemosphere 2022, 287, 132209. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhao, H.; Liang, C.; Li, M.; Liu, R. Effects and Mechanisms of Symbiotic Microbial Combination Agents to Control Tomato Fusarium Crown and Root Rot Disease. Front. Microbiol. 2021, 12, 1555. [Google Scholar] [CrossRef]
- Williams, T.C.R.; Poolman, M.G.; Howden, A.J.M.; Schwarzlander, M.; Fell, D.A.; Ratcliffe, R.G. A genomescale metabolic model accurately predicts fluxes in central carbon metabolism under stress conditions. Plant Physiol. 2010, 154, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savchenko, T.; Tikhonov, K. Oxidative Stress-Iduced Alteration of Plant Central Metabolism. Life 2021, 11, 304. [Google Scholar] [CrossRef]
- Qu, X.; Wang, H.; Chen, M.; Liao, J.; Yuan, J.; Niu, G. Drought stress induced physiological and metabolic changes in leaves of two oil tea cultivars. J. Am. Soc. Hortic. Sci. 2019, 144, 439–447. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Qu, Y.; Han, H.; Chen, F.; Li, L.; Tang, H.; Che, D.; Zhang, X. Unraveling Physiological and Metabolomic Responses Involved in Phlox subulata L. Tolerance to Drought stress. Plant. Mol. Biol. Rep. 2021, 39, 98–111. [Google Scholar] [CrossRef]
- Huan, L.; Xie, X.; Zheng, Z.; Sun, F.; Wu, S.; Li, M.; Wang, G. Positive correlation between PSI response and oxidative pentose phosphate pathway activity during salt stress in an intertidal macroalga. Plant Cell Physiol. 2021, 55, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Orwat, J.; Sarkar, D.; Osorno, J.; Shetty, K. Improved Salinity Resilience in Black Bean by Seed Elicitation Using Organic Compounds. Agron. J. 2017, 109, 1991–2003. [Google Scholar] [CrossRef]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Gomez-Rodriguez, M.V.; Chaki, M.; Pedrajas, J.R.; Barroso, J.B. The dehydrogenase mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant Cell Environ. 2006, 29, 1449–1459. [Google Scholar] [CrossRef]
- Koç, I.; Yuksel, I.; Caetano-Anollés, G. Metabolite-Centric Reporter Pathway and Tripartite Network Analysis of Arabidopsis Under Cold Stress. Front. Bioeng. Biotechnol. 2018, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Hu, L.; Du, Z.; Sun, X.; Amombo, E.; Fan, J.; Fu, J. Effects of cadmium exposure on growth and metabolic profile of bermudagrass [Cynodon dactylon (L.) Pers.]. PLoS ONE 2014, 9, e115279. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, J.; Peng, Y.; Huang, B. Metabolic pathways regulated by γ-aminobutyric acid (GABA) contributing to heat tolerance in creeping bentgrass (Agrostis stolonifera). Sci. Rep. 2016, 6, 30338. [Google Scholar] [CrossRef] [Green Version]
- Nautiyal, C.S.; Bhadauria, S.; Kumar, P.; Lal, H.; Mondal, R.; Verma, D. Stress induced phosphate solubilization in bacteria isolated from alkaline soils. FEMS Microbiol. Lett. 2000, 182, 291–296. [Google Scholar] [CrossRef]
- Srivastava, S.; Chaudhry, V.; Mishra, A.; Chauhan, P.S.; Rehman, A.; Yadav, A.; Tuteja, N.; Nautiyal, C.S. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012, 7, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Limami, A.M.; Gl´evarec, G.; Ricoult, C.; Cliquet, J.B.; Planchet, E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 2008, 59, 2325–2335. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, Y.; Du, Y.; Chen, S.; Tang, H. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 2011, 10, 1904–1914. [Google Scholar] [CrossRef]
- López-Gómez, M.; Hidalgo-Castellanos, J.; Iribarne, C.; Lluch, C. Proline accumulation has prevalence over polyamines in nodules of Medicago sativa in symbiosis with Sinorhizobium meliloti during the initial response to salinity. Plant Soil. 2014, 374, 149–159. [Google Scholar] [CrossRef]
- Trovato, M.; Mattioli, R.; Costantino, P. Multiple roles of proline in plant stress tolerance and development. Rend. Lincei Sci. Fis. 2008, 19, 325–346. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [Green Version]
- Restrepo Rubio, J.S.; López Carrascal, C.E.; Melgarejo, L.M. Physiological behavior of cassava plants (Manihot esculenta Crantz) in response to infection by Xanthomonas axonopodis pv. manihotis under greenhouse conditions. Physiol. Mol. Plant Pathol. 2017, 100, 136–141. [Google Scholar] [CrossRef]
- Deepak, S.; Shailasree, S.; Kini, R.K.; Muck, A.; Mithöfer, A.; Shetty, S.H. Hydroxyproline-rich Glycoproteins and Plant Defense. J. Phytopathol. 2010, 158, 585–593. [Google Scholar]
- Chen, S.; White, C.E.; George, C.; Zhang, Y.; Stogios, P.J.; Savchenko, A.; Finan, T.M. L-Hydroxyproline and D-proline catabolism in Sinorhizobium meliloti. J. Bacteriol. 2016, 198, 1171–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Cao, X.; Tan, C.; Deng, Y.; Bai, J. Analysis of the effect of cadmium stress on root exudates of Sedum plumbizincicola based on metabolomics. Ecotoxicol. Environ. Saf. 2020, 205, 111152. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xing, Y.; Fu, X.; Ji, L.; Li, T.; Wang, J.; Chen, G.; Qi, Z.; Zhang, Q. Biochemical mechanisms of rhizospheric Bacillus subtilis-facilitated phytoextraction by alfalfa under cadmium stress-Microbial diversity and metabolomics analyses. Ecotoxicol. Environ. Saf. 2021, 212, 112016. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Pandey, P.; Singh, J.; Achary, V.M.; Reddy, M.K. Redox homeostasis via gene families of ascorbate-glutathione pathway. Front. Environ. Sci. 2015, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Kuźniak, E.; Kopczewski, T.; Chojak-Koźniewska, J. Ascorbate-Glutathione Cycle and Biotic Stress Tolerance in Plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Springer: Berlin/Heidelberg, Germany, 2017; pp. 201–231. [Google Scholar]
- Dubreuil-Maurizi, C.; Poinssot, B. Role of glutathione in plant signaling under biotic stress. Plant Signal. Behav. 2012, 7, 210–212. [Google Scholar] [CrossRef] [Green Version]
- Bolter, C.; Brammall, R.A.; Cohen, R.; Lazarovits, G. Glutathione alterations in melon and tomato roots following treatment with chemicals which induce disease resistance to Fusarium wilt. Physiol. Mol. Plant Pathol. 1993, 42, 321–336. [Google Scholar] [CrossRef]
- May, M.J.; Hammond-Kosack, K.E.; Jones, J.D.G. Involvement of Reactive Oxygen Species, Glutathione Metabolism and Lipid Peroxidation in the Cf-Gene-Dependent Defence Response of Tomato Cotyledons Induced by Race-Specific Elicitors of Cladosporium fulvum. Plant Physiol. 1996, 110, 1367–1379. [Google Scholar] [CrossRef] [Green Version]
- Schnaubelt, D.; Queval, G.; Dong, Y.; Diaz-Vivancos, P.; Makgopa, M.E.; Howell, G.; De Simone, A.; Bai, J.; Hannah, M.A.; Foyer, C.H. Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil-Maurizi, C.; Vitecek, J.; Marty, L.; Branciard, L.; Frettinger, P.; Wendehenne, D.; Meyer, A.J.; Mauch, F.; Poinssot, B. Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 2011, 157, 2000–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, X.; Guo, S.; Tian, W.; Chen, Y.; Han, H.; Chen, E.; Li, B.; Li, Y.; Chen, Z. Effects of Plant Growth-Promoting Bacteria (PGPB) Inoculation on the Growth, Antioxidant Activity, Cu Uptake, and Bacterial Community Structure of Rape (Brassica napus L.) Grown in Cu-Contaminated Agricultural Soil. Front. Microbiol. 2019, 10, 1455. [Google Scholar] [CrossRef]
- Berglund, T.; Ohlsson, A.B. The Glutathione Biosynthesis Inhibitor Buthioninesulfoximine (BSO) Induces Cardenolide Accumulation in Digitalis lanata Tissue Culture. J. Plant Physiol. 1993, 142, 248–250. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, N.; Yang, Q.; Guo, H.; Xue, L.; Fu, R.; Dong, X.; Dong, H.; Guo, Z.; Zhang, T.; Piao, F.; et al. Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling. Biology 2022, 11, 1028. https://doi.org/10.3390/biology11071028
Du N, Yang Q, Guo H, Xue L, Fu R, Dong X, Dong H, Guo Z, Zhang T, Piao F, et al. Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling. Biology. 2022; 11(7):1028. https://doi.org/10.3390/biology11071028
Chicago/Turabian StyleDu, Nanshan, Qian Yang, Hui Guo, Lu Xue, Ruike Fu, Xiaoxing Dong, Han Dong, Zhixin Guo, Tao Zhang, Fengzhi Piao, and et al. 2022. "Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling" Biology 11, no. 7: 1028. https://doi.org/10.3390/biology11071028
APA StyleDu, N., Yang, Q., Guo, H., Xue, L., Fu, R., Dong, X., Dong, H., Guo, Z., Zhang, T., Piao, F., & Shen, S. (2022). Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling. Biology, 11(7), 1028. https://doi.org/10.3390/biology11071028






