Identification of Colon Cancer-Related RNAs Based on Heterogeneous Networks and Random Walk
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Expression Data
2.2. The Relationship of RNAs
2.3. Classification of RNA
2.4. Data Preprocessing
2.5. Differential Expression Analysis
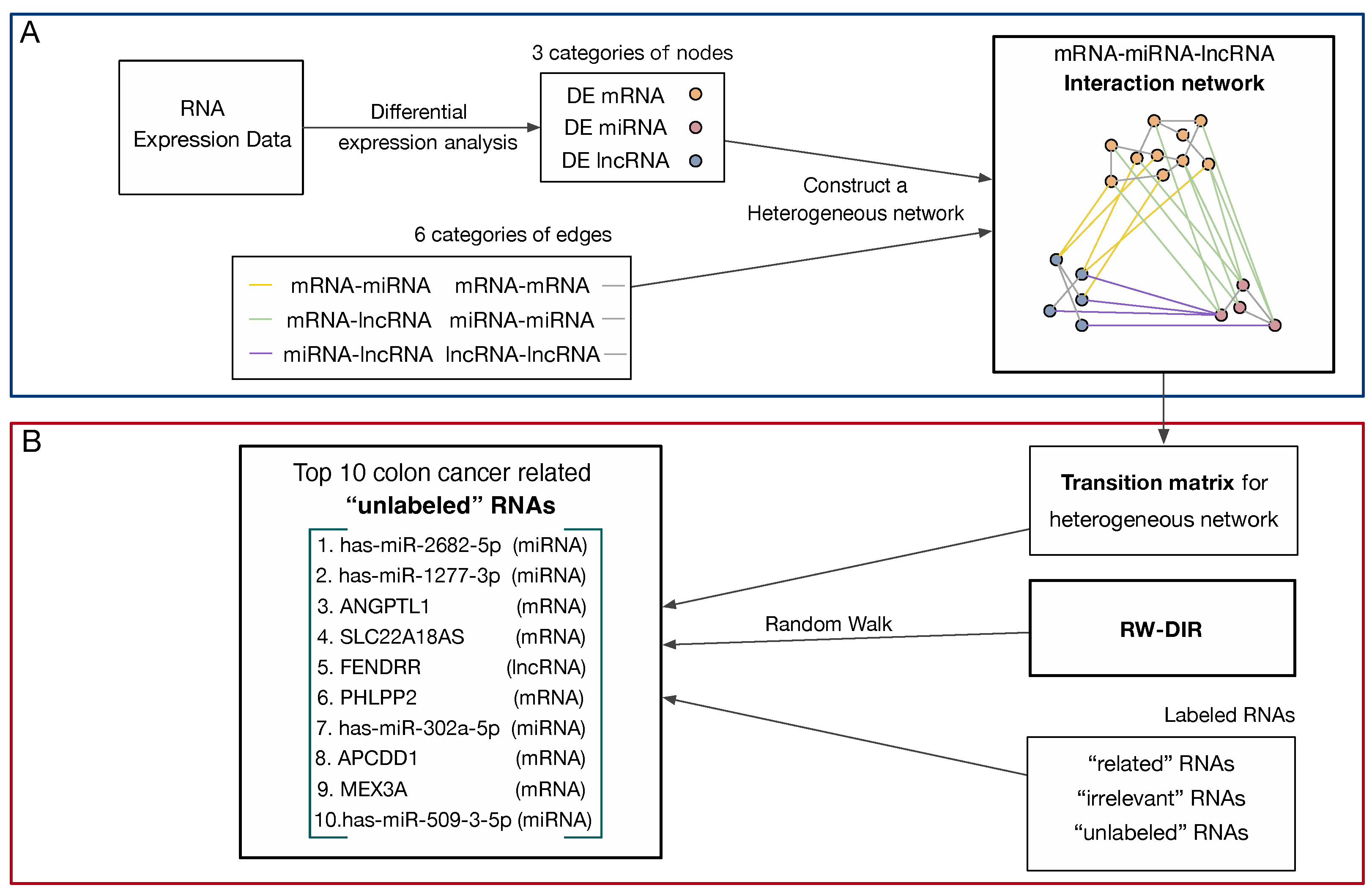
2.6. Construct Heterogeneous Network
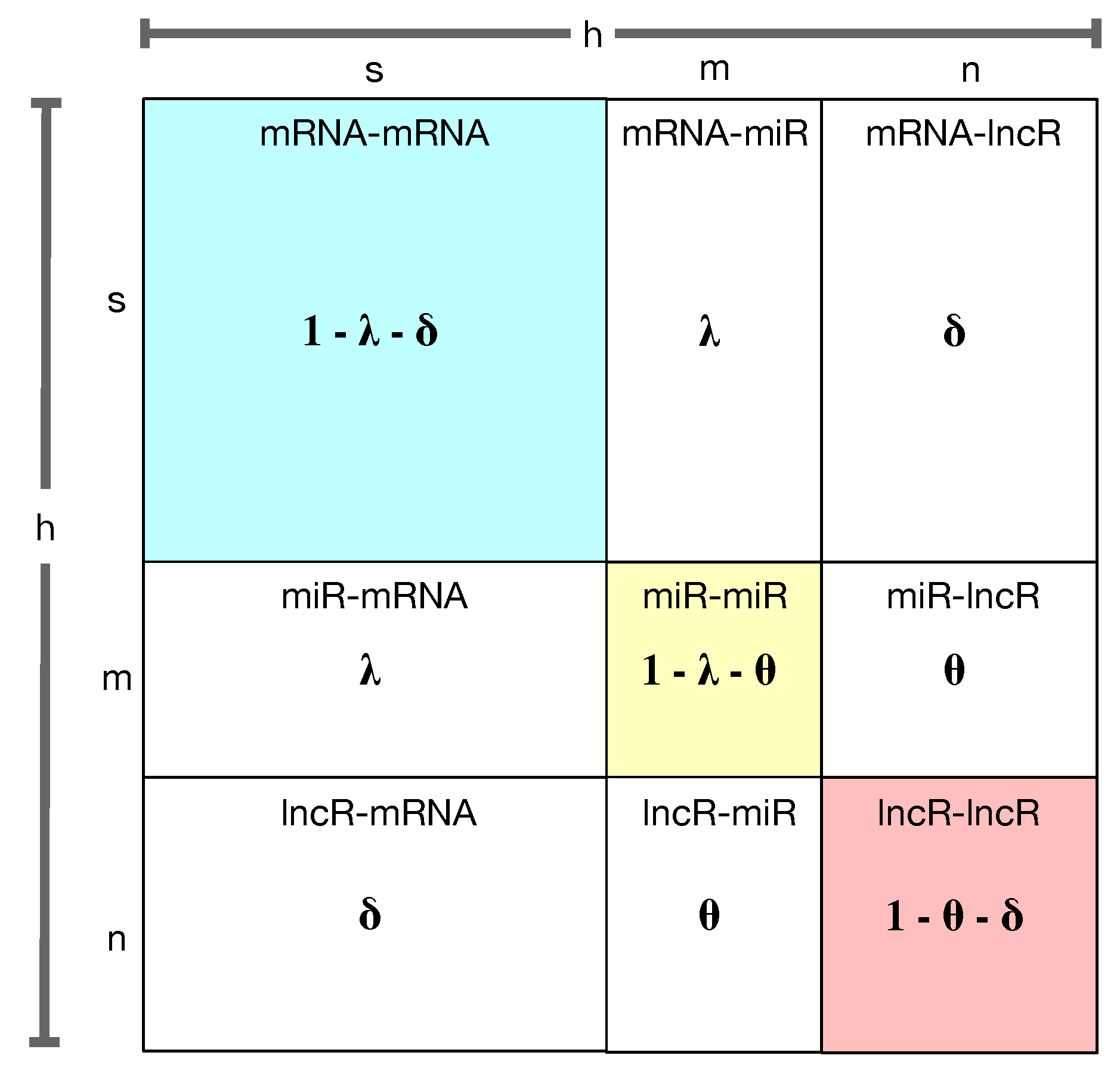
2.7. RW-DIR
| Algorithm 1 Random Walk with Dynamic Incentive Restart (RW-DIR) |
 |
3. Results
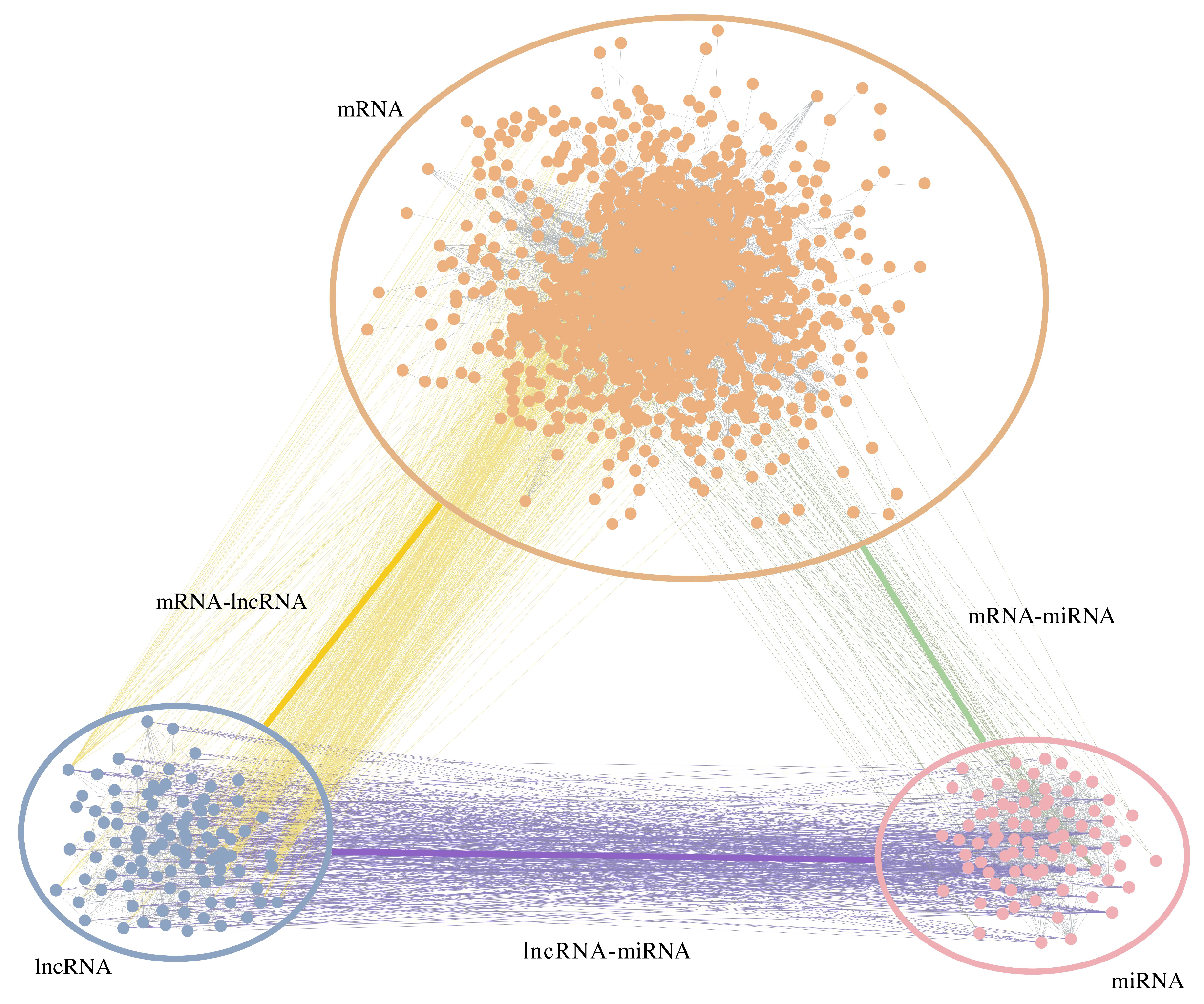
3.1. The Heterogeneous Network
3.2. The Result of RW-DIR
3.2.1. Colon Cancer Related mRNAs
3.2.2. Colon Cancer Related miRNAs
3.2.3. Colon Cancer Related lncRNAs
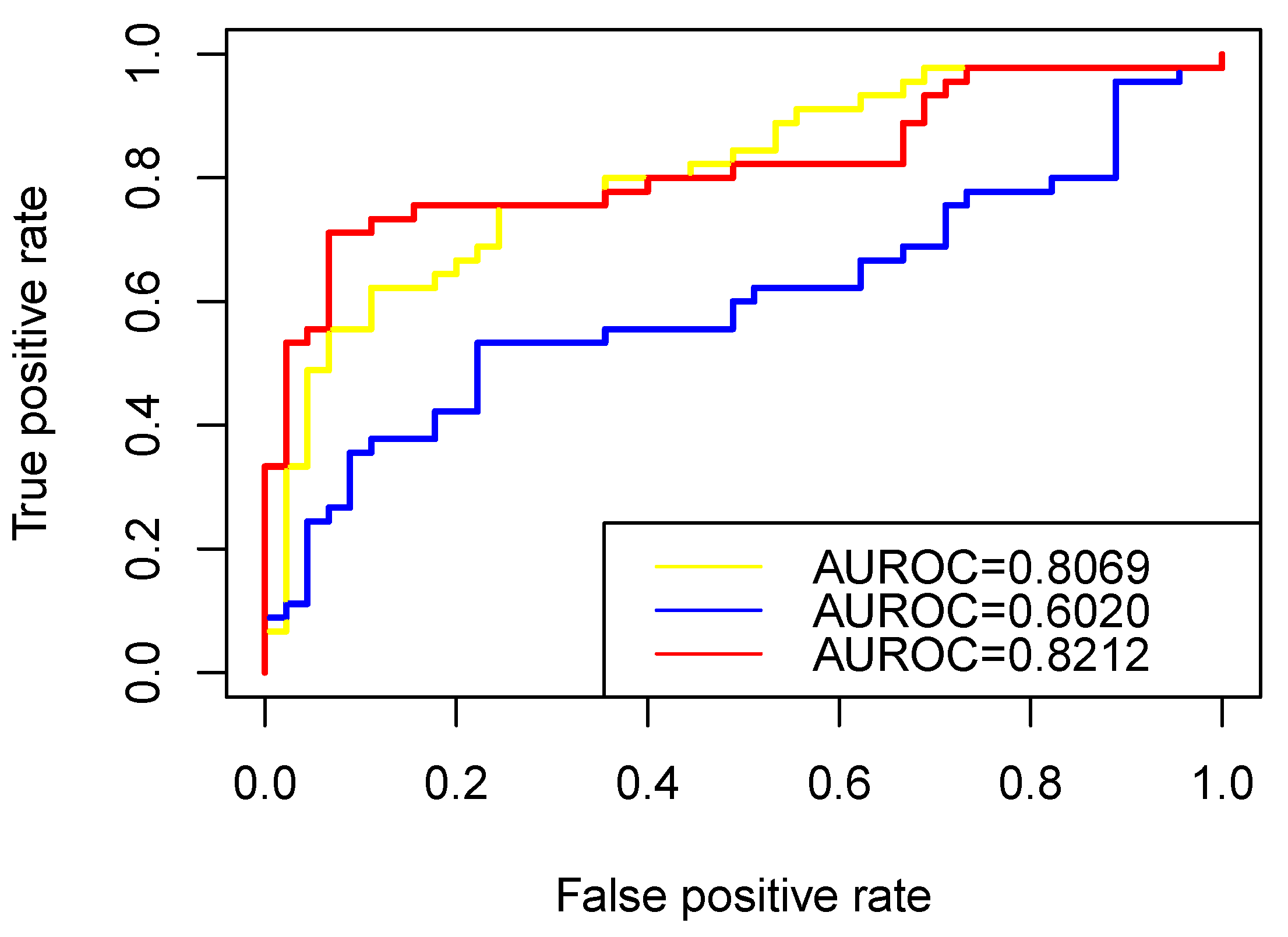
3.3. Performance of RW-DIR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2020, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ding, J.; Ma, Z.; Sun, R.; Seoane, J.A.; Scott Shaffer, J.; Suarez, C.J.; Berghoff, A.S.; Cremolini, C.; Falcone, A.; et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 2019, 51, 1113–1122. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xie, D.; Wang, L.; Zhao, Q.; You, Z.H.; Liu, H. BNPMDA: Bipartite network projection for MiRNA–disease association prediction. Bioinformatics 2018, 34, 3178–3186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Li, A.; Feng, H.; Wang, M. NTSMDA: Prediction of miRNA–disease associations by integrating network topological similarity. Mol. Biosyst. 2016, 12, 2224–2232. [Google Scholar] [CrossRef]
- Serviss, J.T.; Johnsson, P.; Grandér, D. An emerging role for long non-coding RNAs in cancer metastasis. Front. Genet. 2014, 5, 234. [Google Scholar] [CrossRef] [Green Version]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [Green Version]
- Lan, W.; Wu, X.; Chen, Q.; Peng, W.; Wang, J.; Chen, Y.P. GANLDA: Graph attention network for lncRNA-disease associations prediction. Neurocomputing 2022, 469, 384–393. [Google Scholar] [CrossRef]
- Xiao, X.; Zhu, W.; Liao, B.; Xu, J.; Gu, C.; Ji, B.; Yang, J. BPLLDA: Predicting lncRNA-disease associations based on simple paths with limited lengths in a heterogeneous network. Front. Genet. 2018, 9, 411. [Google Scholar] [CrossRef] [Green Version]
- Masuda, N.; Porter, M.A.; Lambiotte, R. Random walks and diffusion on networks. Phys. Rep. 2017, 716, 1–58. [Google Scholar] [CrossRef]
- Zhou, R.S.; Zhang, E.X.; Sun, Q.F.; Ye, Z.J.; Liu, J.W.; Zhou, D.H.; Tang, Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Yang, Y.; Hu, Q.; Shi, H. A label inference method based on maximal entropy random walk over graphs. In Proceedings of the Asia-Pacific Web Conference, Suzhou, China, 23–25 September 2016; pp. 506–518. [Google Scholar]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Flicek, P. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Theodorescu, D. The multiMiR R package and database: Integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2. 0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020, 48, D101–D110. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Mering, C.V. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; King, B.L.; McMorran, R.; Mattingly, C.J. The comparative toxicogenomics database: Update 2017. Nucleic Acids Res. 2017, 45, D972–D978. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [Green Version]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, C.W.; Alhamdoosh, M.; Su, S.; Dong, X.; Ritchie, M.E. Rna-seq analysis is easy as 1-2-3 with limma, glimma and edger. F1000Research 2016, 5, 1408. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovász, L. Random walks on graphs. In Combinatorics, Paul Erdos is Eighty; Yale University: New Haven, CT, USA, 1993. [Google Scholar]
- Page, L.; Brin, S.; Motwani, R.; Winograd, T. The PageRank Citation Ranking: Bringing Order to the Web; Technical Report; Stanford InfoLab: Stanford Digital Libraries Working Paper: Stanford, CA, USA, 1998. [Google Scholar]
- Chen, H.; Xiao, Q.; Hu, Y.; Chen, L.; Jiang, K.; Tang, Y.; Ding, K. ANGPTL1 attenuates colorectal cancer metastasis by up-regulating microRNA-138. J. Exp. Clin. Cancer Res. 2017, 36, 78. [Google Scholar] [CrossRef]
- Sun, R.; Yang, L.; Hu, Y.; Wang, Y.; Zhang, Q.; Zhang, Y.; Zhao, D. ANGPTL1 is a potential biomarker for differentiated thyroid cancer diagnosis and recurrence. Oncol. Lett. 2020, 20, 240. [Google Scholar] [CrossRef]
- Noguera-Uclés, J.F.; Boyero, L.; Salinas, A.; Cordero Varela, J.A.; Benedetti, J.C.; Bernabé-Caro, R.; Sánchez-Gastaldo, A.; Alonso, M.; Paz-Ares, L.; Molina-Pinelo, S. The Roles of Imprinted SLC22A18 and SLC22A18AS Gene Overexpression Caused by Promoter CpG Island Hypomethylation as Diagnostic and Prognostic Biomarkers for Non-Small Cell Lung Cancer Patients. Cancers 2020, 12, 2075. [Google Scholar] [CrossRef]
- Wang, H.; Gu, R.; Tian, F.; Liu, Y.; Fan, W.; Xue, G.; Cai, L.; Xing, Y. PHLPP2 as a novel metastatic and prognostic biomarker in non-small cell lung cancer patients. Thorac. Cancer 2019, 10, 2124–2132. [Google Scholar] [CrossRef]
- Cho, S.G. APC downregulated 1 inhibits breast cancer cell invasion by inhibiting the canonical WNT signaling pathway. Oncol. Lett. 2017, 14, 4845–4852. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhan, H.; Zhao, Y.; Wu, Y.; Li, L.; Wang, H. MEX3A contributes to development and progression of glioma through regulating cell proliferation and cell migration and targeting CCL2. Cell Death Dis. 2021, 12, 14. [Google Scholar] [CrossRef]
- Lu, N.; Yin, Y.; Yao, Y.; Zhang, P. SNHG3/miR-2682-5p/HOXB8 promotes cell proliferation and migration in oral squamous cell carcinoma. Oral Dis. 2021, 27, 1161–1170. [Google Scholar] [CrossRef]
- Chen, W.; Zhuang, X.; Qi, R.; Qiao, T. MiR-302a-5p suppresses cell proliferation and invasion in non-small cell lung carcinoma by targeting ITGA6. Am. J. Transl. Res. 2019, 11, 4348–4357. [Google Scholar] [PubMed]
- Ma, J.; Li, D.; Kong, F.F.; Yang, D.; Yang, H.; Ma, X.X. miR-302a-5p/367-3p-HMGA2 axis regulates malignant processes during endometrial cancer development. J. Exp. Clin. Cancer Res. 2018, 37, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.J.; Wang, J.Y.; Zhang, T.J.; An, G.S.; Ni, J.H.; Li, S.Y.; Jia, H.T. MiR-509-3-5p-NONHSAT112228. 2 Axis Regulates p21 and Suppresses Proliferation and Migration of Lung Cancer Cells. Curr. Top. Med. Chem. 2020, 20, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.P.; Huang, M.D.; Xia, R.; Liu, X.X.; Sun, M.; Yin, L.; Shu, Y.Q. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J. Hematol. Oncol. 2014, 7, 63. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, Q.; Yu, X.; He, Y.; Guo, W. FENDRR: A pivotal, cancer-related, long non-coding RNA. Biomed. Pharmacother. 2021, 137, 111390. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Mao, G.; Mu, Z.; Wu, D. Exosome-derived miR-2682-5p suppresses cell viability and migration by HDAC1-silence-mediated upregulation of ADH1A in non-small cell lung cancer. Hum. Exp. Toxicol. 2021, 40, S318–S330. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Zhang, L.; You, G.; Li, C.; Meng, B.; Zhang, M. LINC01006 promotes cell proliferation and metastasis in pancreatic cancer via miR-2682-5p/HOXB8 axis. Cancer Cell Int. 2019, 19, 320. [Google Scholar] [CrossRef] [Green Version]
| Types of RNA Associations | Database |
|---|---|
| mRNA-miRNA | multiMiR [16] |
| mRNA-lncRNA | starbase V3.0 [17] |
| miRNA-lncRNA | LncBase V2.0 [18] |
| mRNA-mRNA | STRING [19] |
| Types of RNA | Database |
|---|---|
| mRNA | Comparative Toxicogenomics Database [20] |
| miRNA | miR2disease [21] |
| lncRNA | LncRNADisease [22] |
| Type of RNA | Number of RNA | Number of Normal Samples | Number of Tumor Samples |
|---|---|---|---|
| mRNA | 116591 | 41 | 443 |
| miRNA | 302 | 8 | 444 |
| lncRNA | 1526 | 41 | 443 |
| Type of Edge | Number of Node | Number of Edge |
|---|---|---|
| mRNA-mRNA | 1300 (mRNA) | 7408 |
| miRNA- miRNA | 81 (miRNA) | 389 |
| lncRNA-lncRNA | 389 (lncRNA) | 604 |
| mRNA-miRNA | 56 (mRNA) - 326(miRNA) | 569 |
| mRNA-lncRNA | 33 (mRNA) - 70(lncRNA) | 94 |
| miRNA-lncRNA | 99 (miRNA) - 57(lncRNA) | 587 |
| Top 10 RNAs | Number of RNA | Related Diseases |
|---|---|---|
| hsa-miR-2682-5p | miRNA | Oral squamous cell carcinoma |
| hsa-miR-1277-3p | miRNA | / |
| ANGPTL1 | mRNA | Lung cancer, breast cancer, colorectal cancer |
| SLC22A18AS | mRNA | Lung adenocarcinoma |
| FENDRR | lncRNA | Gastric cancer, lung cancer, hepatocellular carcinoma (HCC), gastric cancer |
| PHLPP2 | mRNA | Diabetes, hepatic steatosis, and cancer |
| hsa-miR-302a-5p | miRNA | Endometrial carcinoma, glioma and breast cancer |
| APCDD1 | mRNA | Breast cancer |
| MEX3A | mRNA | Glioma |
| hsa-miR-509-3-5p | miRNA | Gastric cancer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Wang, T.; Zhang, J.; Zhang, S.; Shang, X. Identification of Colon Cancer-Related RNAs Based on Heterogeneous Networks and Random Walk. Biology 2022, 11, 1003. https://doi.org/10.3390/biology11071003
Chen B, Wang T, Zhang J, Zhang S, Shang X. Identification of Colon Cancer-Related RNAs Based on Heterogeneous Networks and Random Walk. Biology. 2022; 11(7):1003. https://doi.org/10.3390/biology11071003
Chicago/Turabian StyleChen, Bolin, Teng Wang, Jinlei Zhang, Shengli Zhang, and Xuequn Shang. 2022. "Identification of Colon Cancer-Related RNAs Based on Heterogeneous Networks and Random Walk" Biology 11, no. 7: 1003. https://doi.org/10.3390/biology11071003
APA StyleChen, B., Wang, T., Zhang, J., Zhang, S., & Shang, X. (2022). Identification of Colon Cancer-Related RNAs Based on Heterogeneous Networks and Random Walk. Biology, 11(7), 1003. https://doi.org/10.3390/biology11071003






