Taxonomic and Phylogenetic Insights into Novel Ascomycota from Forest Woody Litter
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolates and Specimens
2.2. Morphological Observations
2.3. DNA Extraction, PCR Amplifications and Sequencing
2.4. Molecular Phylogenetic Analyses
2.4.1. Sequencing and Sequence Alignment
2.4.2. Phylogenetic Analyses
3. Results
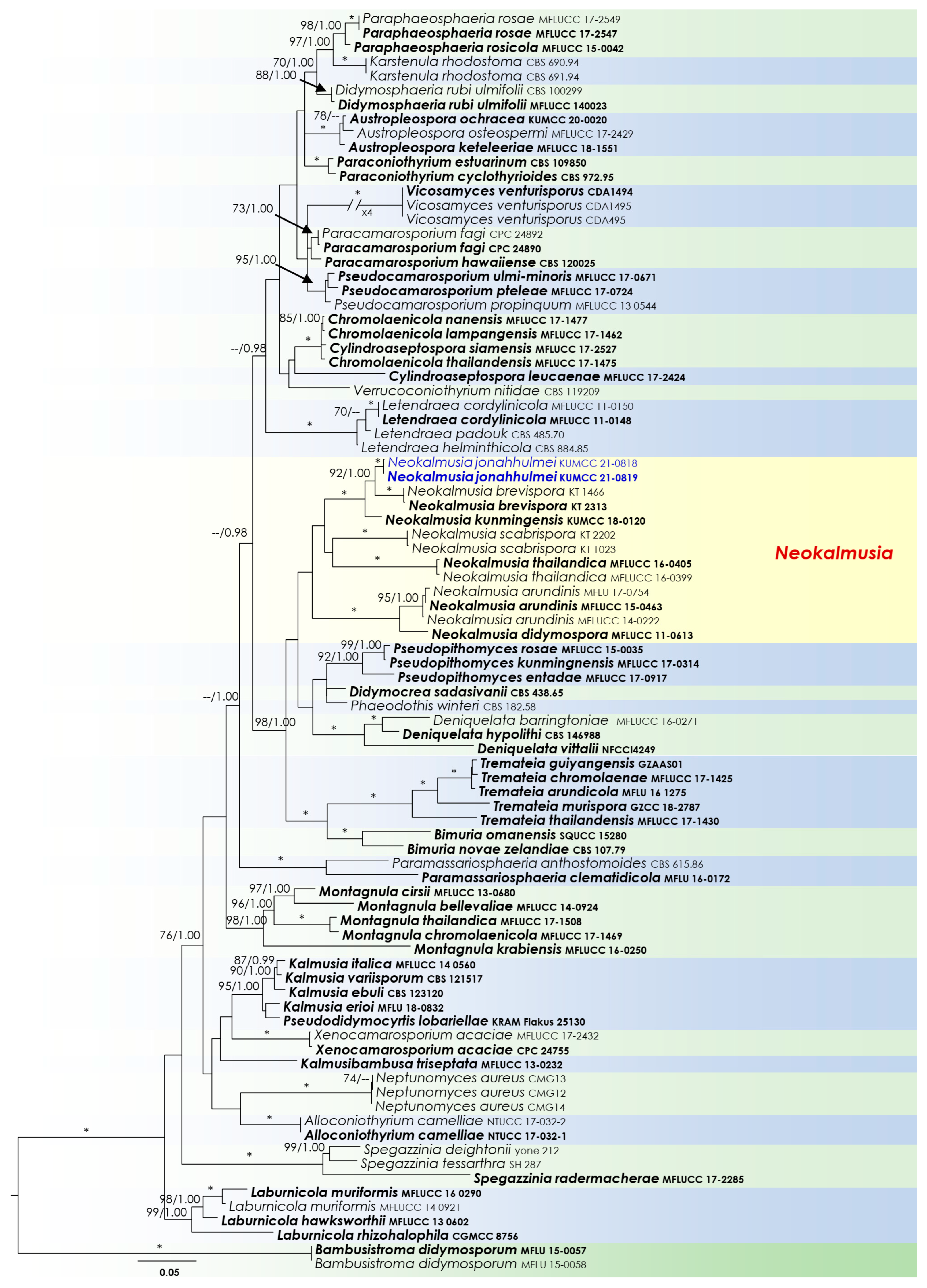
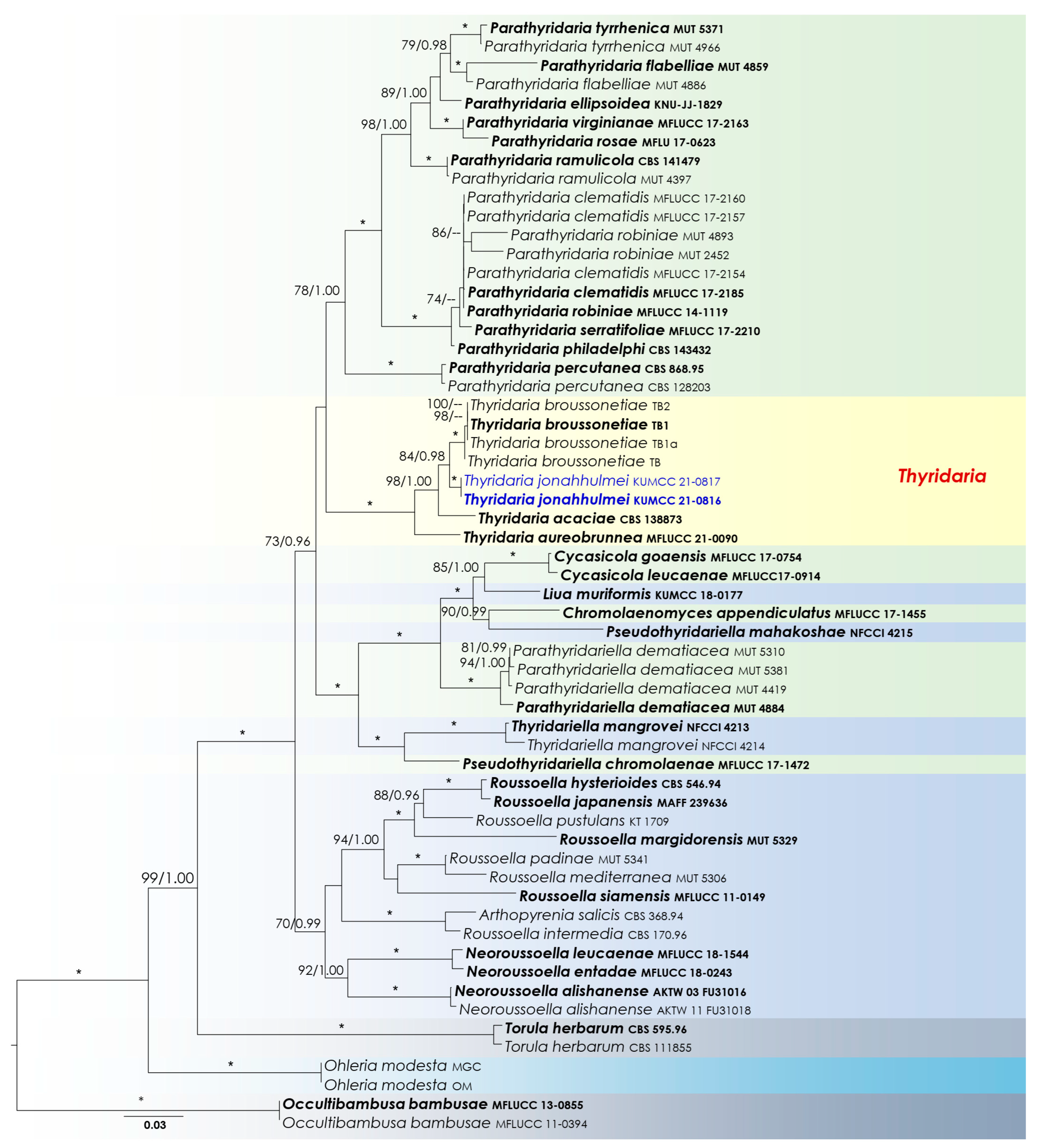
3.1. Phylogenetic Analyses
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, J.R.; Gorham, E. Litter production in forests of the world. Adv. Ecol. Res. 1964, 2, 101–157. [Google Scholar] [CrossRef]
- Lowman, M.D. Litter fall and leaf decay in three Australian rainforest formations. J. Ecol. 1988, 76, 451–465. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.M.; Tsukamoto, J.; Tokumoto, Y.; Shuvo, M.A.R. The role of quantitative traits of leaf litter on decomposition and nutrient cycling of the forest ecosystems. J. For. Sci. 2013, 29, 38–48. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Zhou, G.; Xu, Z. Forest litter fall and its composition: A new data set of observational data from China. Ecology 2016, 97, 1365. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Lang, X.; Du, B.; Zhang, H.; Liu, H.; Zhang, Y.; Shang, L. Litter fall and nutrient return in moist evergreen broad-leaved primary forest and mixed subtropical secondary deciduous broad-leaved forest in China. Eur. J. For. Res. 2016, 135, 77–86. [Google Scholar] [CrossRef]
- Giweta, M. Role of litter production and its decomposition, and factors affecting the processes in a tropical forest ecosystem: A review. J. Ecol. Environ. 2020, 44, 11. [Google Scholar] [CrossRef]
- Bao, D.F.; Luo, Z.L.; Liu, J.K.; Bhat, D.J.; Sarunya, N.; Li, W.L.; Su, H.Y.; Hyde, K.D. Lignicolous freshwater fungi in China III: Three new species and a new record of Kirschsteiniothelia from northwestern Yunnan province. Mycosphere 2018, 9, 755–768. [Google Scholar] [CrossRef]
- Huang, S.K.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bhat, D.J.; Chomnunti, P.; Hyde, K.D.; Lumyong, S. Morphological and molecular taxonomy of Jahnula dianchia sp. nov. (Jahnulales) from submerged wood in Dianchi Lake, Yunnan China. Mycol. Prog. 2018, 17, 547–555. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Bhat, D.J.; Bao, D.F.; Li, W.L.; Su, H.W.Q. Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 2018, 9, 444–461. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Wijayawardene, N.N.; Xu, J.; Cheewangkoon, R.; Mortimer, P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 2020, 15, e0235855. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Mortimer, P.E.; Xu, J. Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China). J. Fungi 2021, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, P.E.; Jeewon, R.; Xu, J.C.; Lumyong, S.; Wanasinghe, D.N. Morpho-phylo taxonomy of novel dothideomycetous fungi associated with dead woody twigs in Yunnan Province, China. Front. Microbiol. 2021, 12, 654683. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Hua, Z.; Lv, R.; Yu, Z. Neodactylariales, Neodactylariaceae (Dothideomycetes, Ascomycota): New order and family, with a new species from China. MycoKeys 2020, 73, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Rathnayaka, A.R.; Dayarathne, M.C.; Maharachchikumbura, S.S.N.; Liu, J.K.; Tennakoon, D.S.; Hyde, K.D. Introducing Seriascoma yunnanense sp. nov. (Occultibambusaceae, Pleosporales) based on evidence from morphology and phylogeny. AJOM 2019, 2, 245–253. [Google Scholar] [CrossRef]
- Thiyagaraja, V.; Wanasinghe, D.N.; Worthy, F.; Karunarathna, S.C. Addition to Melanommataceae: A new geographical record of Alpinaria rhododendri from Shangri La, China. AJOM 2020, 3, 335–344. [Google Scholar] [CrossRef]
- Yasanthika, E.; Dissanayake, L.S.; Wanasinghe, D.N.; Karunarathna, S.C.; Mortimer, P.E.; Samarakoon, B.C.; Monkai, J.; Hyde, K.D. Lonicericola fuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa 2020, 446, 103–113. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols Appl. 1990, 18, 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. News from the Sea: A New Genus and Seven New Species in the Pleosporalean Families Roussoellaceae and Thyridariaceae. Diversity 2020, 12, 144. [Google Scholar] [CrossRef] [Green Version]
- Samarakoon, B.C.; Wanasinghe, D.N.; Samarakoon, M.C.; Phookamsak, R.; McKenzie, E.H.C.; Chomnunti, P.; Hyde, K.D.; Lumyong, S.; Karunarathna, S.C. Multi-gene phylogenetic evidence suggests Dictyoarthrinium belongs in Didymosphaeriaceae (Pleosporales, Dothideomycetes) and Dictyoarthrinium musae sp. nov. on Musa from Thailand. MycoKeys 2020, 71, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nylander, J.A.A.; Wilgenbusch, J.C.; Warren, D.L.; Swofford, D.L. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 2008, 24, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. FigTree Version 1.4.0. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 23 January 2022).
- Ariyawansa, H.A.; Tanaka, K.; Thambugala, K.M.; Phookamsak, R.; Tian, Q.; Camporesi, E.; Hongsanan, S.; Monkai, J.; Wanasinghe, D.N.; Mapook, A.; et al. A molecular phylogenetic reappraisal of the Didymosphaeriaceae (=Montagnulaceae). Fungal Divers. 2014, 68, 69–104. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef] [Green Version]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Ariyawansa, H.A.; Goonasekara, I.D.; Phookamsak, R.; Dissanayake, A.; et al. Mycosphere notes 1-50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Dai, D.Q.; Bahkali, A.H.; Ariyawansa, H.A.; Li, W.J.; Bhat, D.J.; Hyde, K.D. Neokalmusia didymospora (Didymosphaeriaceae), a new species from bamboo. Sydowia 2016, 68, 17–25. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Summerell, B.A.; Giraldo, A.; Gené, J.; Guarro, J.; Wanasinghe, D.N.; Hyde, K.D.; Camporesi, E.; et al. Fungal Planet Description Sheets: 281–319. Persoonia 2014, 33, 212–289. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, G.E.B.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016, 85, 35–64. [Google Scholar] [CrossRef] [Green Version]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Rehm, H. Ascomycetes philippinenses—VIII. Leafl. Philipp. Bot. 1916, 8, 2945. [Google Scholar]
- Petrak, F. Ein Beitrag zur Pilzflora von Hawai. Sydowia 1952, 6, 363–371. [Google Scholar]
- Saccardo, P.A. Sylloge fungorum XXIV, sect. II. Abellini. Sylloge Fungorum. 1928, 24, 705–1438. [Google Scholar]
- Wehmeyer, L.E. The genus Thyridaria (Pyrenomycetes). Lloydia 1941, 4, 241–262. [Google Scholar]
- Barr, M.E. Melanommatales (Loculoascomycetes). N. Am. Flora Ser. II 1990, 13, 1–129. [Google Scholar]
- Munk, A. The system of the Pyrenomycetes. Dansk botanisk Arkiv. 1953, 15, 1–163. [Google Scholar]
- Gonçalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Neptunomyces aureus gen. et sp. nov. (Didymosphaeriaceae, Pleosporales) isolated from algae in Ria de Aveiro, Portugal. MycoKeys 2019, 60, 31–44. [Google Scholar] [CrossRef]
- Flakus, A.; Etayo, J.; Miadlikowska, J.; Lutzoni, F.; Kukwa, M.; Matura, N.; Rodriguez-Flakus, P. Biodiversity assessment of ascomycetes inhabiting Lobariella lichens in Andean cloud forests led to one new family, three new genera and 13 new species of lichenicolous fungi. Plant Fungal Syst. 2019, 64, 283–344. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Gareth, J.E.B.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Species Fungorum. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 23 January 2022).
- Wehmeyer, L.E. A revision of Melanconis, Pseudovalsa, Prosthecium and Titania. Univ. Mich. Stud. Sci. Ser. 1941, 14, 1–161. [Google Scholar]
- Wehmeyer, L.E. The Pyrenomycetous Fungi. Mycologia Memoir No. 6. The New York Botanical Garden; J. Cramer Publishing: Lehre, Germany, 1975; pp. 1–250. [Google Scholar]
- Munk, A. Danish pyrenomycetes: A preliminary flora. Dansk Botanisk Arkiv 1957, 17, 491. [Google Scholar]
- Luttrell, E.S. Loculoascomycetes. In The Fungi. An Advanced Treatise, a Taxonomic Review with Keys: Ascomycetes and Fungi Imperfecti; Ainsworth, G.C., Sparrow, F.K., Sussman, A.S., Eds.; Academic Press: New York, NY, USA, 1973; pp. 135–219. [Google Scholar]
- Müller, E.; von Arx, J.A. Pyrenomycetes: Meliolales, Coronophorales, Sphaeriales. New Exot. Fungi. Grevillea 1973, 17, 42–43. [Google Scholar]
- Dennis, R.W.G. British Ascomycetes, 3rd ed.; Cramer, J.: Vaduz, Liechtenstein, 1978; pp. 1–585. [Google Scholar]
- Barr, M.E. A classification of Loculoascomycetes. Mycologia 1979, 71, 935–995. [Google Scholar] [CrossRef]
- Barr, M.E. On the Massariaceae in North America. Mycotaxon 1979, 9, 17–37. [Google Scholar]
- Barr, M.E. The affinities of Thyridaria. Mycotaxon 2003, 88, 271–278. [Google Scholar]
- Dai, Y.C.; Cui, B.K.; Yuan, H.S.; He, S.H.; Wei, Y.L.; Qin, W.M.; Zhou, L.W.; Li, H.J. Wood-inhabiting fungi in southern China 4. Polypores from Hainan province. Ann. Bot. Fenn. 2011, 48, 219–231. [Google Scholar] [CrossRef]
- Dai, Y.; Härkönen, M.; Niemelä, T. Wood-inhabiting fungi in southern China 1. Polypores from Hunan province. Ann. Bot. Fenn. 2003, 40, 381–393. [Google Scholar]
- Dai, Y.; Wei, Y.; Wang, Z. Wood-inhabiting fungi in southern China 2. Polypores from Sichuan province. Ann. Bot. Fenn. 2004, 41, 319–329. [Google Scholar]
- Cui, B.K.; Dai, Y.; Bao, H.Y. Wood-inhabiting fungi in southern China 3. A new species of Phellinus (Hymenochaetales) from tropical China. Mycotaxon 2009, 110, 125–130. [Google Scholar] [CrossRef]
- Wang, B.; Cui, B.K.; Li, H.J.; Du, P.; Jia, B.S. Wood-rotting fungi in eastern China 5. Polypore diversity in Jiangxi Province. Ann. Bot. Fenn. 2011, 48, 237–246. [Google Scholar] [CrossRef]
- Yuan, H.; Dai, Y. Wood-inhabiting fungi in southern China. 6. Polypores from Guangxi autonomous region. Ann. Bot. Fenn. 2012, 49, 341–351. [Google Scholar] [CrossRef]
- Promputtha, I.; Lumyong, S.; Dhanasekaran, V.; McKenzie, E.H.C.; Hyde, K.D.; Jeewon, R. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb. Ecol. 2007, 53, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Photita, W.; Lumyong, S.; Lumyong, P.; McKenzie, E.H.C.; Hyde, K.D. Are some endophytes from Musa acuminate latent pathogens? Fungal Divers. 2004, 16, 131–140. [Google Scholar]
- Luplertlop, N. Pseudallescheria/Scedosporium complex species: From saprobic to pathogenic fungus. J. Mycol. Med. 2018, 2, 249–256. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Nilsson, R.H.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]




| Species | Strain | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1 | rpb2 | ||
| Alloconiothyrium camelliae | NTUCC 17-032-1 | MT112294 | MT071221 | MT071270 | MT232967 | - |
| Alloconiothyrium camelliae | NTUCC 17-032-2 | MT112295 | MT071222 | MT071271 | MT232965 | - |
| Austropleospora keteleeriae | MFLUCC 18-1551 | NR_163349 | MK347910 | NG_070075 | MK360045 | MK434909 |
| Austropleospora ochracea | KUMCC 20-0020 | MT799859 | MT808321 | MT799860 | MT872714 | - |
| Austropleospora osteospermi | MFLUCC 17-2429 | MK347757 | MK347863 | MK347974 | MK360044 | MK434884 |
| Bambusistroma didymosporum | MFLU 15-0057 | KP761733 | KP761737 | KP761730 | KP761727 | KP761720 |
| Bambusistroma didymosporum | MFLU 15-0058 | KP761734 | KP761738 | KP761731 | KP761728 | KP761721 |
| Bimuria novae-zelandiae | CBS 107.79 | MH861181 | AY016338 | AY016356 | DQ471087 | DQ470917 |
| Bimuria omanensis | SQUCC 15280 | NR_173301 | - | NG_071257 | MT279046 | - |
| Chromolaenicola lampangensis | MFLUCC 17-1462 | MN325016 | MN325010 | MN325004 | MN335649 | MN335654 |
| Chromolaenicola nanensis | MFLUCC 17-1477 | MN325014 | MN325008 | MN325002 | MN335647 | MN335653 |
| Chromolaenicola thailandensis | MFLUCC 17-1475 | MN325019 | MN325013 | MN325007 | MN335652 | MN335656 |
| Cylindroaseptospora leucaenae | MFLUCC 17-2424 | NR_163333 | MK347856 | NG_066310 | MK360047 | - |
| Cylindroaseptospora siamensis | MFLUCC 17-2527 | NR_163337 | MK347866 | NG_066311 | MK360048 | - |
| Deniquelata barringtoniae | MFLUCC 16-0271 | MH275059 | - | MH260291 | MH412766 | MH412753 |
| Deniquelata hypolithi | CBS 146988 | MZ064429 | - | NG_076735 | MZ078250 | MZ078201 |
| Deniquelata vittalii | NFCCI4249 | MF406218 | MF622059 | MF182395 | MF182398 | MF168942 |
| Didymocrea sadasivanii | CBS 438.65 | MH858658 | DQ384066 | DQ384103 | - | - |
| Didymosphaeria rubi ulmifolii | CBS 100299 | MH862698 | AY642523 | JX496124 | - | - |
| Didymosphaeria rubi ulmifolii | MFLUCC 140023 | KJ436586 | NG_063557 | KJ436586 | - | - |
| Kalmusia ebuli | CBS 123120 | KF796674 | JN851818 | JN644073 | - | - |
| Kalmusia erioi | MFLU 18-0832 | MN473058 | MN473046 | MN473052 | MN481599 | - |
| Kalmusia italica | MFLUCC 14 0560 | KP325440 | KP325442 | KP325441 | - | - |
| Kalmusia variisporum | CBS 121517 | NR_145165 | - | JX496143 | - | - |
| Kalmusibambusa triseptata | MFLUCC 13-0232 | KY682697 | KY682696 | KY682695 | - | - |
| Karstenula rhodostoma | CBS 690.94 | - | GU296154 | GU301821 | GU349067 | GU371788 |
| Karstenula rhodostoma | CBS 691.94 | LC014559 | AB797241 | AB807531 | AB808506 | - |
| Laburnicola hawksworthii | MFLUCC 13 0602 | KU743194 | KU743196 | KU743195 | - | - |
| Laburnicola muriformis | MFLUCC 16 0290 | KU743197 | KU743199 | KU743198 | KU743213 | - |
| Laburnicola muriformis | MFLUCC 14 0921 | KU743200 | KU743202 | KU743201 | - | - |
| Laburnicola rhizohalophila | CGMCC 8756 | KJ125522 | - | KJ125523 | KJ125525 | KJ125524 |
| Letendraea cordylinicola | MFLUCC 11 0150 | KM213996 | KM214002 | KM213999 | - | - |
| Letendraea cordylinicola | MFLUCC 11 0148 | NR_154118 | KM214001 | NG_059530 | - | - |
| Letendraea helminthicola | CBS 884.85 | MK404145 | AY016345 | AY016362 | MK404174 | MK404164 |
| Letendraea padouk | CBS 485.70 | - | GU296162 | AY849951 | - | - |
| Montagnula bellevaliae | MFLUCC 14 0924 | NR_155377 | KT443904 | KT443902 | KX949743 | - |
| Montagnula chromolaenicola | MFLUCC 17-1469 | NR_168866 | NG_070157 | NG_070948 | MT235773 | MT235809 |
| Montagnula cirsii | MFLUCC 13 0680 | KX274242 | KX274255 | KX274249 | KX284707 | - |
| Montagnula krabiensis | MFLUCC 16-0250 | MH275070 | MH260343 | MH260303 | MH412776 | - |
| Montagnula thailandica | MFLUCC 17-1508 | MT214352 | NG_070158 | NG_070949 | MT235774 | MT235810 |
| Neokalmusia arundinis | MFLU 17-0754 | MT649882 | MT649880 | MT649878 | MT663766 | - |
| Neokalmusia arundinis | MFLUCC 15-0463 | NR_165852 | NG_068372 | NG_068237 | KY244024 | - |
| Neokalmusia arundinis | MFLUCC 14-0222 | KX965731 | KX986344 | KX954400 | KY271091 | - |
| Neokalmusia brevispora | KT 2313 | LC014574 | AB524460 | AB524601 | AB539113 | - |
| Neokalmusia brevispora | KT 1466 | LC014573 | AB524459 | AB524600 | AB539112 | - |
| Neokalmusia didymospora | MFLUCC 11-0613 | - | KP091435 | KP091434 | - | - |
| Neokalmusia jonahhulmei | KUMCC 21-0818 | ON007043 | ON007039 | ON007048 | ON009133 | ON009137 |
| Neokalmusia jonahhulmei | KUMCC 21-0819 | ON007044 | ON007040 | ON007049 | ON009134 | ON009138 |
| Neokalmusia kunmingensis | KUMCC 18-0120 | MK079886 | MK079887 | MK079889 | MK070172 | - |
| Neokalmusia scabrispora | KT 1023 | LC014575 | AB524452 | AB524593 | AB539106 | - |
| Neokalmusia scabrispora | KT 2202 | LC014576 | AB524453 | AB524594 | AB539107 | - |
| Neokalmusia thailandica | MFLUCC 16-0405 | NR_154255 | KY706137 | NG_059792 | KY706145 | KY706148 |
| Neokalmusia thailandica | MFLUCC 16-0399 | KY706141 | KY706136 | KY706131 | - | - |
| Neptunomyces aureus | CMG12 | MK912121 | - | - | MK948000 | - |
| Neptunomyces aureus | CMG13 | MK912122 | - | - | MK948001 | - |
| Neptunomyces aureus | CMG14 | MK912123 | - | - | MK948002 | - |
| Paracamarosporium fagi | CPC 24890 | NR_154318 | - | NG_070630 | - | - |
| Paracamarosporium fagi | CPC 24892 | KR611887 | - | KR611905 | - | - |
| Paracamarosporium hawaiiense | CBS 120025 | JX496027 | EU295655 | JX496140 | - | - |
| Paraconiothyrium cyclothyrioides | CBS 972.95 | JX496119 | AY642524 | JX496232 | - | - |
| Paraconiothyrium estuarinum | CBS 109850 | JX496016 | AY642522 | JX496129 | - | - |
| Paramassariosphaeria anthostomoides | CBS 615.86 | MH862005 | GU205246 | GU205223 | - | - |
| Paramassariosphaeria clematidicola | MFLU 16-0172 | KU743206 | KU743208 | KU743207 | - | - |
| Paraphaeosphaeria rosae | MFLUCC 17-2547 | MG828935 | MG829150 | MG829044 | MG829222 | - |
| Paraphaeosphaeria rosae | MFLUCC 17-2549 | MG828937 | MG829152 | MG829046 | MG829223 | - |
| Paraphaeosphaeria rosicola | MFLUCC 15-0042 | NR_157528 | MG829153 | MG829047 | - | - |
| Phaeodothis winteri | CBS 182.58 | - | GU296183 | GU301857 | - | - |
| Pseudocamarosporium propinquum | MFLUCC 13 0544 | KJ747049 | KJ819949 | KJ813280 | - | - |
| Pseudocamarosporium pteleae | MFLUCC 17-0724 | NR_157536 | MG829166 | MG829061 | MG829233 | - |
| Pseudocamarosporium ulmi-minoris | MFLUCC 17-0671 | NR_157537 | MG829167 | MG829062 | - | - |
| Pseudodidymocyrtis lobariellae | KRAM Flakus 25130 | NR_169714 | NG_070349 | NG_068933 | - | - |
| Pseudopithomyces entadae | MFLUCC 17-0917 | MK347835 | NG_066305 | MK360083 | MK434899 | |
| Pseudopithomyces kunmingnensis | MFLUCC 17-0314 | MF173607 | MF173606 | MF173605 | - | - |
| Pseudopithomyces rosae | MFLUCC 15-0035 | MG828953 | MG829168 | MG829064 | - | - |
| Spegazzinia deightonii | yone 212 | AB797292 | AB807582 | AB808558 | - | |
| Spegazzinia radermacherae | MFLUCC 17-2285 | MK347740 | MK347848 | MK347957 | MK360088 | MK434893 |
| Spegazzinia tessarthra | SH 287 | JQ673429 | AB797294 | AB807584 | AB808560 | - |
| Tremateia arundicola | MFLU 16 1275 | KX274241 | KX274254 | KX274248 | KX284706 | - |
| Tremateia chromolaenae | MFLUCC 17-1425 | NR_168868 | NG_070160 | NG_068710 | MT235778 | MT235816 |
| Tremateia guiyangensis | GZAAS01 | KX274240 | KX274253 | KX274247 | KX284705 | - |
| Tremateia murispora | GZCC 18-2787 | NR_165916 | MK972750 | MK972751 | MK986482 | - |
| Tremateia thailandensis | MFLUCC 17-1430 | NR_168869 | NG_070161 | NG_068711 | MT235781 | MT235819 |
| Verrucoconiothyrium nitidae | CBS 119209 | EU552112 | - | EU552112 | - | - |
| Vicosamyces venturisporus | CDA1494 | MF802825 | - | MF802828 | - | - |
| Vicosamyces venturisporus | CDA1495 | MF802826 | - | MF802829 | - | - |
| Vicosamyces venturisporus | CDA495 | MF802827 | - | MF802830 | - | - |
| Xenocamarosporium acaciae | CPC 24755 | NR_137982 | - | NG_058163 | - | - |
| Xenocamarosporium acaciae | MFLUCC 17-2432 | MK347766 | MK347873 | MK347983 | MK360093 | - |
| Species | Strain | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1 | rpb2 | ||
| Arthopyrenia salicis | CBS 368.94 | KF443410 | AY538339 | AY538333 | KF443404 | KF443397 |
| Chromolaenomyces appendiculatus | MFLUCC 17-1455 | NR_168862 | NG_068705 | MT214394 | MT235770 | MT235806 |
| Cycasicola goaensis | MFLUCC 17-0754 | MG828885 | MG829001 | MG829112 | MG829198 | - |
| Cycasicola leucaenae | MFLUCC17-0914 | NR_163322 | MK347942 | MK347833 | MK360046 | MK434900 |
| Liua muriformis | KUMCC 18-0177 | MK433599 | MK433598 | MK433595 | MK426798 | MK426799 |
| Neoroussoella alishanense | AKW 11 FU31018 | MK503818 | MK503824 | MK503830 | MK336182 | MN037757 |
| Neoroussoella alishanense | AKW 03 FU31016 | MK503816 | MK503822 | MK503828 | MK336181 | MN037756 |
| Neoroussoella entadae | MFLUCC 18-0243 | MK347786 | MK348004 | MK347893 | MK360065 | MK434866 |
| Neoroussoella leucaenae | MFLUCC 18-1544 | MK347767 | MK347984 | MK347874 | MK360067 | MK434876 |
| Occultibambusa bambusae | MFLUCC 11-0394 | KU940124 | KU863113 | - | KU940194 | KU940171 |
| Occultibambusa bambusae | MFLUCC 13-0855 | KU940123 | KU863112 | KU872116 | KU940193 | KU940170 |
| Ohleria modesta | MGC | KX650562 | KX650562 | - | KX650533 | KX650582 |
| Ohleria modesta | OM | KX650563 | KX650563 | KX650513 | KX650534 | KX650583 |
| Parathyridaria clematidis | MFLUCC 17-2154 | MT310645 | MT214601 | MT226712 | MT394657 | MT394712 |
| Parathyridaria clematidis | MFLUCC 17-2157 | MT310644 | MT214600 | MT226711 | MT394656 | MT394711 |
| Parathyridaria clematidis | MFLUCC 17-2160 | MT310643 | MT214599 | MT226710 | MT394655 | MT394710 |
| Parathyridaria clematidis | MFLUCC 17-2185 | MT310642 | MT214598 | NG_070668 | MT394654 | MT394709 |
| Parathyridaria ellipsoidea | KNU-JJ-1829 | LC552950 | LC552952 | - | - | - |
| Parathyridaria flabelliae | MUT 4886 | KR014358 | KP671720 | KT587317 | MN605910 | MN605930 |
| Parathyridaria flabelliae | MUT 4859 | KR014355 | KP671716 | KT587315 | MN605909 | MN605929 |
| Parathyridaria percutanea | CBS 128203 | KF322117 | KF366448 | KF366450 | KF407988 | KF366453 |
| Parathyridaria percutanea | CBS 868.95 | KF322118 | KF366449 | KF366451 | KF407987 | KF366452 |
| Parathyridaria philadelphi | CBS 143432 | MH107905 | NG_063958 | - | MH108023 | - |
| Parathyridaria ramulicola | MUT 4397 | KC339235 | KF636775 | MN556311 | MN605913 | MN605933 |
| Parathyridaria ramulicola | CBS 141479 | NR_147657 | KX650565 | KX650514 | KX650536 | KX650584 |
| Parathyridaria robiniae | MUT 2452 | MG813183 | MG816491 | MN556312 | MN605903 | MN605923 |
| Parathyridaria robiniae | MUT 4893 | KM355998 | MN556328 | KM355993 | MN605904 | MN605924 |
| Parathyridaria robiniae | MFLUCC 14-1119 | KY511142 | KY511141 | - | KY549682 | - |
| Parathyridaria rosae | MFLU 17-0623 | NR_157530 | NG_059873 | - | - | - |
| Parathyridaria serratifoliae | MFLUCC 17-2210 | MT310646 | MT214602 | NG_070669 | MT394658 | MT394713 |
| Parathyridaria tyrrhenica | MUT 4966 | KR014366 | KP671740 | KT587309 | MN605911 | MN605931 |
| Parathyridaria tyrrhenica | MUT 5371 | KU314951 | MN556329 | KU314952 | MN605912 | MN605932 |
| Parathyridaria virginianae | MFLUCC 17-2163 | MT310647 | NG_073853 | NG_070670 | MT394659 | MT394714 |
| Parathyridariella dematiacea | MUT 4419 | KC339245 | KF636786 | MN556313 | MN605905 | MN605925 |
| Parathyridariella dematiacea | MUT 5310 | KU255057 | MN556330 | MN556314 | MN605907 | MN605927 |
| Parathyridariella dematiacea | MUT 5381 | KU314959 | MN556331 | KU314960 | MN605908 | MN605928 |
| Parathyridariella dematiacea | MUT 4884 | MN556317 | KP671726 | KT587329 | MN605906 | MN605926 |
| Pseudothyridariella chromolaenae | MFLUCC 17-1472 | NR_168863 | NG_068706 | MT214395 | MT235771 | MT235807 |
| Pseudothyridariella mahakoshae | NFCCI 4215 | MG020435 | MG020438 | MG020441 | MG023140 | MG020446 |
| Roussoella hysterioides | CBS 546.94 | KF443405 | KF443381 | AY642528 | KF443399 | KF443392 |
| Roussoella intermedia | CBS 170.96 | KF443407 | KF443382 | KF443390 | KF443398 | KF443394 |
| Roussoella japanensis | MAFF 239636 | KJ474829 | AB524621 | AB524480 | AB539114 | AB539101 |
| Roussoella margidorensis | MUT 5329 | KU314944 | MN556322 | MN556309 | MN605897 | MN605917 |
| Roussoella mediterranea | MUT 5306 | KU255054 | MN556323 | MN556310 | MN605898 | MN605918 |
| Roussoella padinae | MUT 5341 | KU158153 | MN556325 | KU158176 | MN605900 | MN605920 |
| Roussoella pustulans | KT 1709 | KJ474830 | AB524623 | AB524482 | AB539116 | AB539103 |
| Roussoella siamensis | MFLUCC 11-0149 | KJ474837 | KJ474845 | KU872125 | KJ474854 | KJ474861 |
| Thyridaria acaciae | CBS 138873 | KP004469 | KP004497 | - | - | - |
| Thyridaria aureobrunnea | MFLUCC 21-0090 | MZ538528 | MZ538562 | - | - | - |
| Thyridaria broussonetiae | TB | KX650567 | KX650567 | - | KX650538 | KX650585 |
| Thyridaria broussonetiae | TB1a | KX650569 | KX650569 | - | - | - |
| Thyridaria broussonetiae | TB2 | KX650570 | KX650570 | - | KX650540 | KX650587 |
| Thyridaria broussonetiae | TB1 | KX650568 | KX650568 | KX650515 | KX650539 | KX650586 |
| Thyridaria jonahhulmei | KUMCC 21-0816 | ON007041 | ON007037 | ON007046 | ON009131 | ON009135 |
| Thyridaria jonahhulmei | KUMCC 21-0817 | ON007042 | ON007038 | ON007047 | ON009132 | ON009136 |
| Thyridariella mangrovei | NFCCI 4214 | MG020436 | MG020439 | MG020442 | MG020444 | MG020447 |
| Thyridariella mangrovei | NFCCI 4213 | MG020434 | MG020437 | MG020440 | MG020443 | MG020445 |
| Torula herbarum | CBS 111855 | KF443409 | KF443386 | KF443391 | KF443403 | KF443396 |
| Torula herbarum | CBS 595.96 | KF443408 | KF443385 | KF443387 | KF443402 | KF443395 |
| Analyses | Didymosphaeriaceae | Thyridariaceae | |
|---|---|---|---|
| Number of taxa | 88 | 59 | |
| Gene regions | SSU, LSU, ITS, tef1, and rpb2 | SSU, LSU, ITS, tef1, and rpb2 | |
| Number of character positions (including gaps) | 5016 | 4529 | |
| ML optimization likelihood value | −35,672.743881 | −30,606.10565 | |
| Distinct alignment patterns in the matrix | 2249 | 1796 | |
| Number of undetermined characters or gaps (%) | 41.88% | 19.56% | |
| Estimated base frequencies | A | 0.240418 | 0.249274 |
| C | 0.253351 | 0.25578 | |
| G | 0.270784 | 0.267476 | |
| T | 0.235446 | 0.227469 | |
| Substitution rates | AC | 1.561664 | 1.486771 |
| AG | 3.248718 | 3.744601 | |
| AT | 1.433496 | 1.706836 | |
| CG | 1.323566 | 1.014483 | |
| CT | 7.428045 | 7.933665 | |
| GT | 1.0 | 1.0 | |
| Proportion of invariable sites (I) | 0.396829 | 0.505108 | |
| Gamma distribution shape parameter (α) | 0.454368 | 0.442817 | |
| Number of generated trees in BI | 11,301 | 2501 | |
| Number of trees sampled in BI after 25% were discarded as burn-in | 8476 | 1876 | |
| Final split frequency | 0.009959 | 0.009966 | |
| The total of unique site patterns | 2252 | 1798 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wanasinghe, D.N.; Mortimer, P.E. Taxonomic and Phylogenetic Insights into Novel Ascomycota from Forest Woody Litter. Biology 2022, 11, 889. https://doi.org/10.3390/biology11060889
Wanasinghe DN, Mortimer PE. Taxonomic and Phylogenetic Insights into Novel Ascomycota from Forest Woody Litter. Biology. 2022; 11(6):889. https://doi.org/10.3390/biology11060889
Chicago/Turabian StyleWanasinghe, Dhanushka N., and Peter E. Mortimer. 2022. "Taxonomic and Phylogenetic Insights into Novel Ascomycota from Forest Woody Litter" Biology 11, no. 6: 889. https://doi.org/10.3390/biology11060889
APA StyleWanasinghe, D. N., & Mortimer, P. E. (2022). Taxonomic and Phylogenetic Insights into Novel Ascomycota from Forest Woody Litter. Biology, 11(6), 889. https://doi.org/10.3390/biology11060889







