Low-Salt Diet Reduces Anti-CTLA4 Mediated Systemic Immune-Related Adverse Events while Retaining Therapeutic Efficacy against Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Murine Breast Tumor Model
2.2. CD4+T Cell Isolation
2.3. Real Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.4. Western Blot
2.5. Luminex
2.6. Histochemical Staining
2.7. Sodium Magnetic Resonance Imaging
2.8. Electrolyte and Tissue Osmolarity Measurements
2.9. Enzyme Linked Immunosorbent Assay (ELISA)
2.10. Caspase 1 Activity Assay
2.11. Statistical Analysis
3. Results
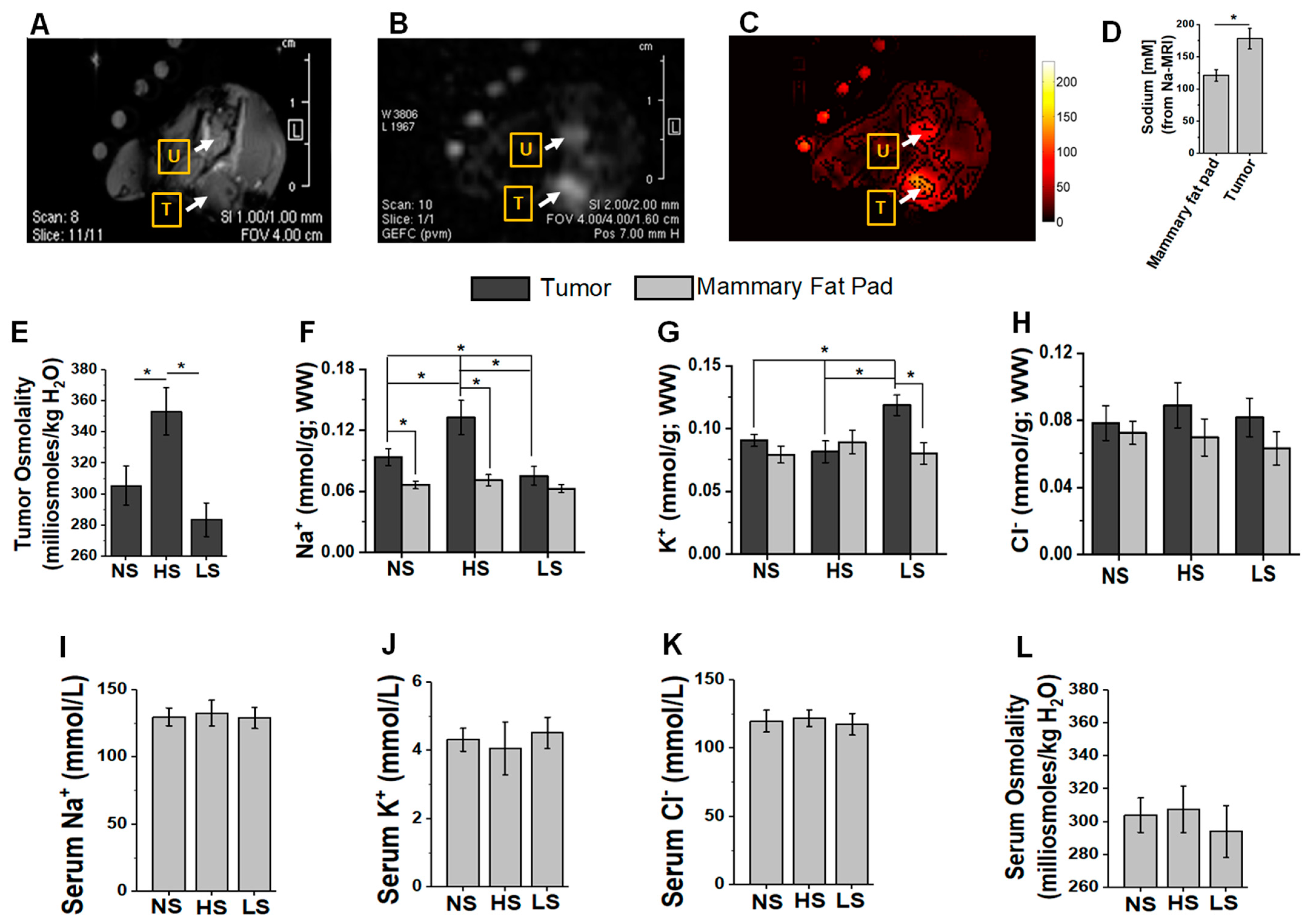
3.1. Tumor and Plasma Electrolyte Changes following Salt Modified Diet
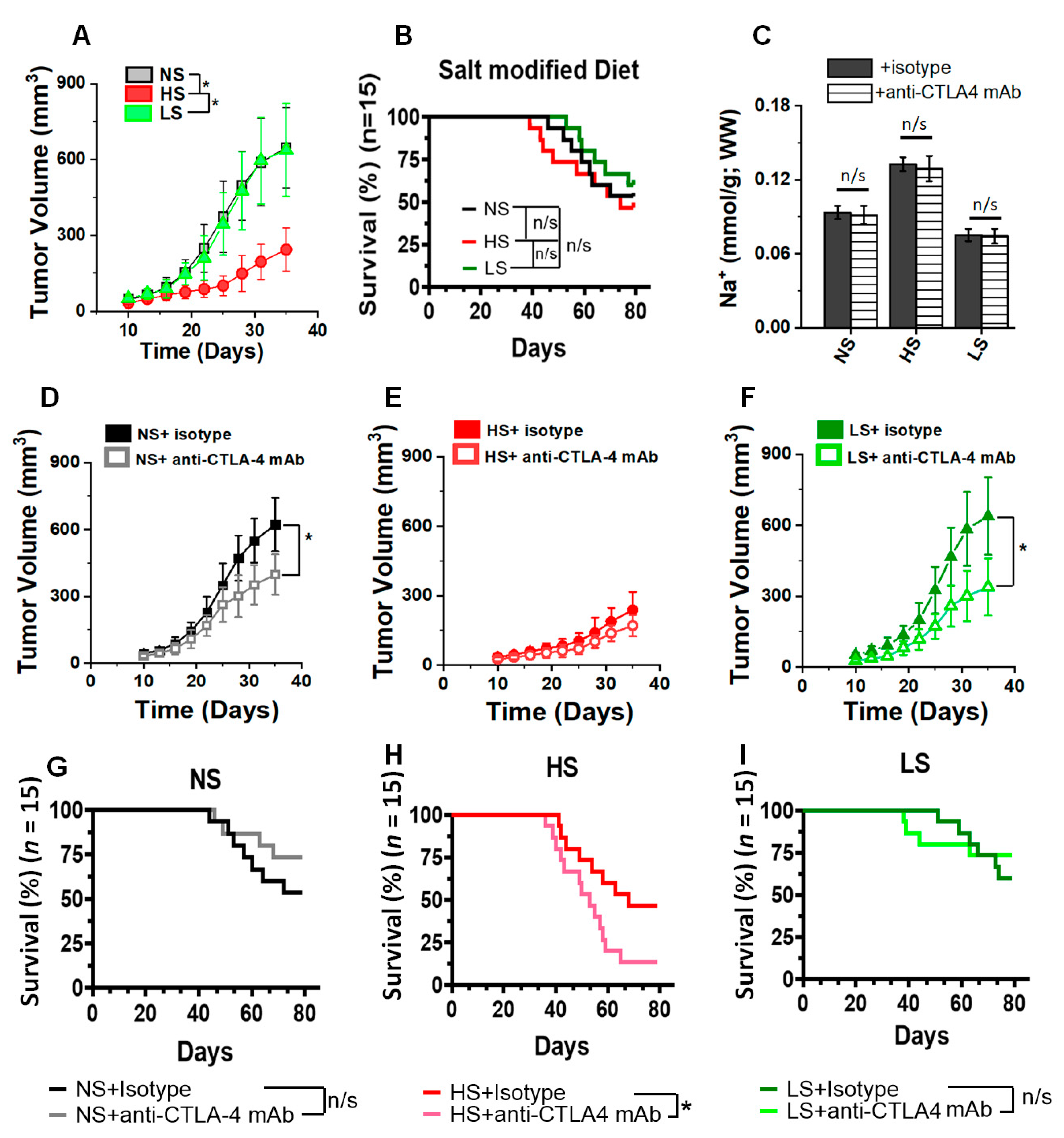
3.2. Anti-CTLA4 mAb Treatment in Low-Salt-Diet Cohort Enhanced the Survival
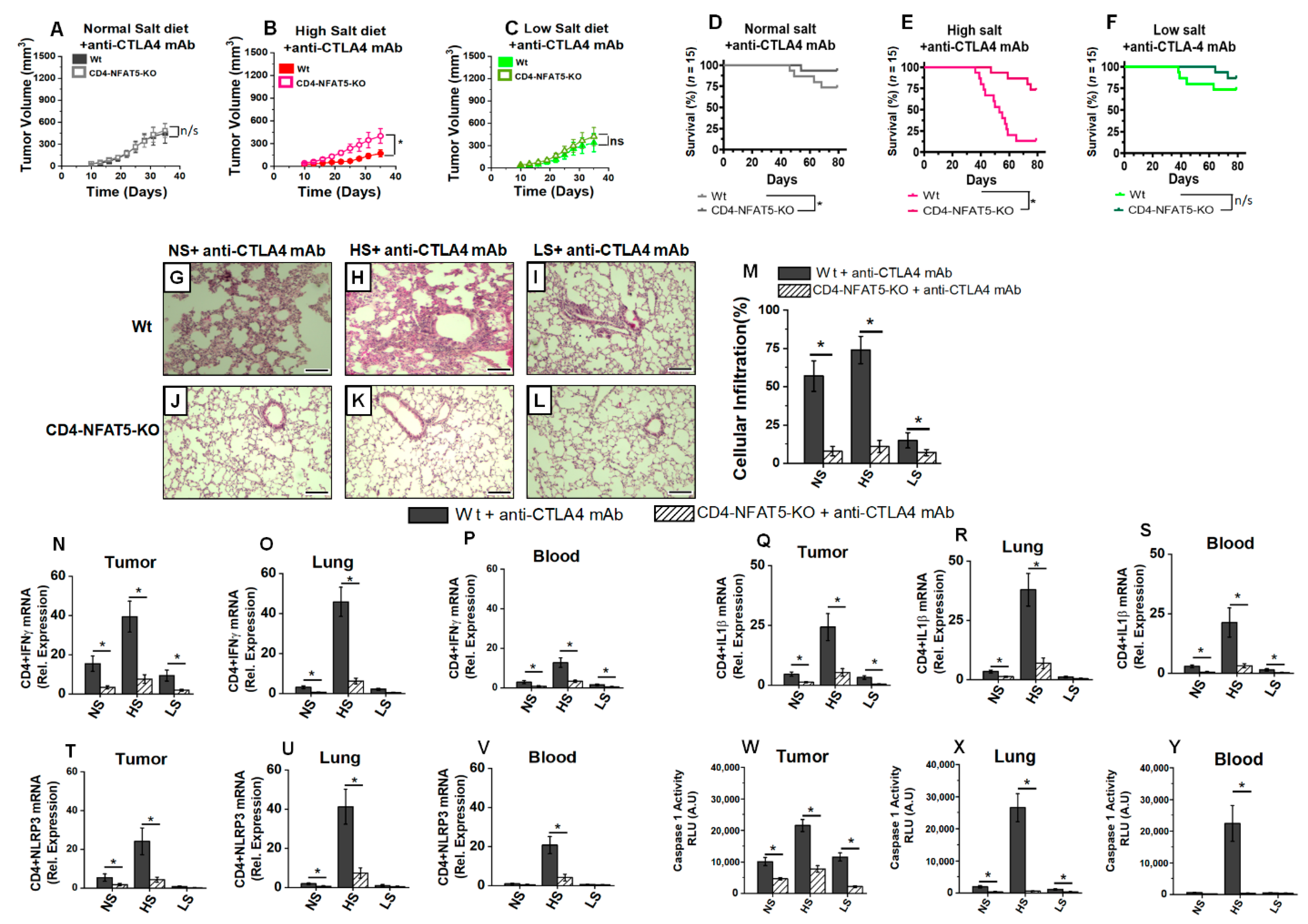
3.3. Low-Salt Diet Reduced Anti-CTLA4-Mediated Inflammatory Lung Infiltration
3.4. Low-Salt Diet Reduced Peripheral Inflammatory Cytokine Response and Alveolar Infiltration with Inflammatory CD4+T Cells following Anti-CTLA4 Therapy
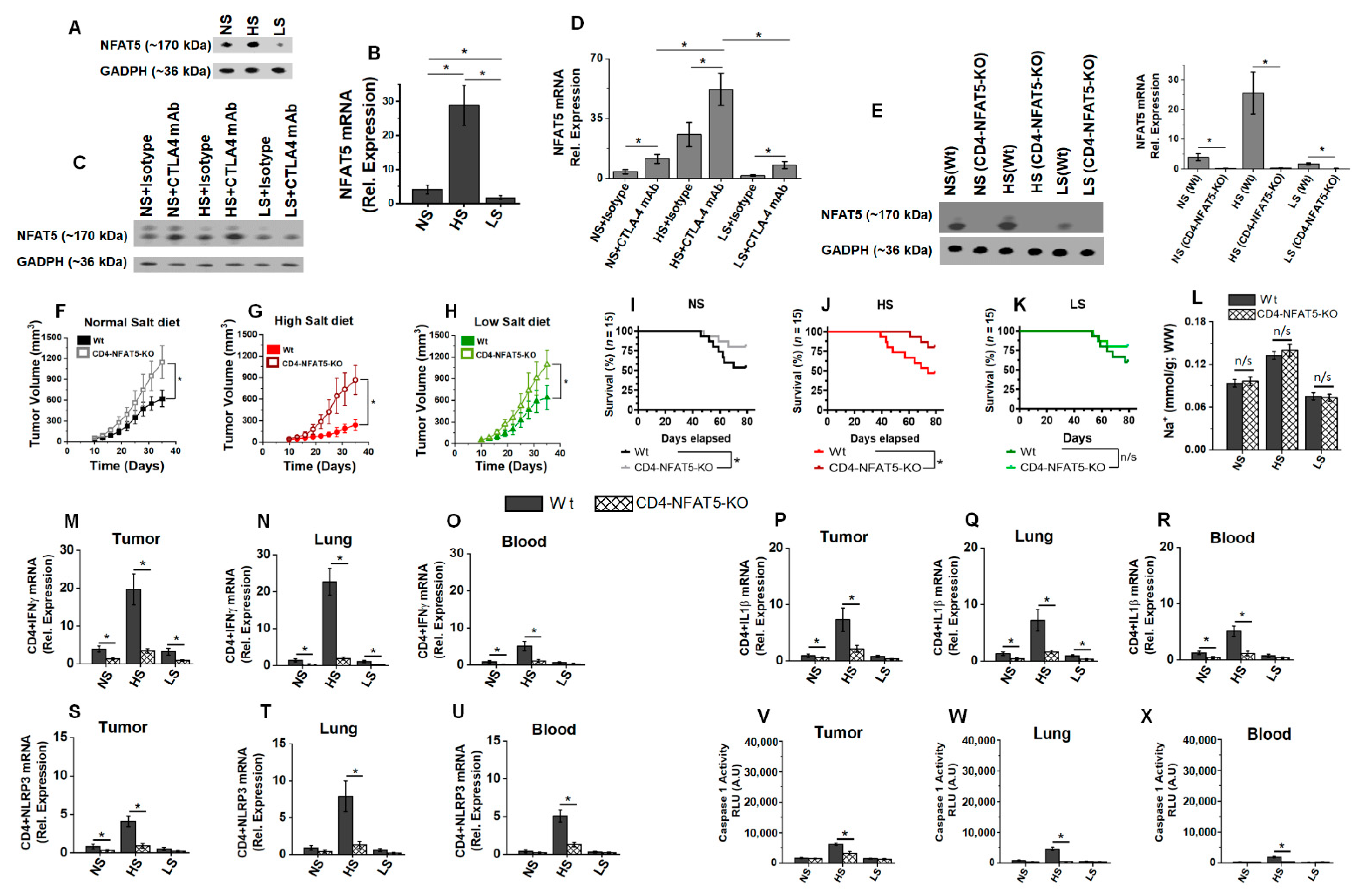
3.5. Low-Salt Diet Downregulates NLRP3-Mediated Inflammasome Complex Leading to Diminished irAE
3.6. Osmosensitive Transcription Factor, NFAT5, Is a Critical Upstream Regulator in Anti-CTLA4 Induced irAE following High-Salt Diet
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat Commun. 2020, 11, 3801. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures (2022); American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Thomas, R.; Al-Khadairi, G.; Decock, J. Immune Checkpoint Inhibitors in Triple Negative Breast Cancer Treatment: Promising Future Prospects. Front. Oncol. 2021, 10, 600573. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M. Tumor mutation burden for predicting immune checkpoint blockade response: The more, the better. J. Immunother. Cancer 2022, 10, e003087. [Google Scholar] [CrossRef] [PubMed]
- Sivapiragasam, A.; Ashok Kumar, P.; Sokol, E.S.; Albacker, L.A.; Killian, J.K.; Ramkissoon, S.H.; Huang, R.S.P.; Severson, E.A.; Brown, C.A.; Danziger, N.; et al. Predictive Biomarkers for Im mune Checkpoint Inhibitors in Metastatic Breast Cancer. Cancer Med. 2021, 10, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zou, X.; Zheng, S.; Tang, H.; Zhang, L.; Liu, P.; Xie, X. Efficacy and predictive factors of immune checkpoint inhibitors in metastatic breast cancer: A systematic review and meta-analysis. Ther. Adv. Med. Oncol. 2020, 12, 1758835920940928. [Google Scholar] [CrossRef]
- Gaynor, N.; Crown, J.; Collins, D.M. Immune checkpoint inhibitors: Key trials and an emerging role in breast cancer. Semin. Cancer Biol. 2022, 79, 44–57. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Wang, J.; Chen, P.; Su, M.; Zhong, G.; Zhang, S.; Gou, D. Integrative Modeling of Multiomics Data for Predicting Tumor Mutation Burden in Patients with Lung Cancer. Biomed. Res. Int. 2022, 2022, 2698190. [Google Scholar] [CrossRef]
- Johnson, D.B.; Nebhan, C.A.; Moslehi, J.J.; Balko, J.M. Immune-checkpoint inhibitors: Long-term implications of toxicity. Nat. Rev. Clin. Oncol. 2022, 19, 254–267. [Google Scholar] [CrossRef]
- Martins, F.; Sofiya, L.; Sykiotis, G.P.; Lamine, F.; Maillard, M.; Fraga, M.; Shabafrouz, K.; Ribi, C.; Cairoli, A.; Guex-Crosier, Y.; et al. Adverse effects of immune-checkpoint inhibitors: Epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 2019, 16, 563–580. [Google Scholar] [CrossRef]
- Zhong, L.; Altan, M.; Shannon, V.R.; Sheshadri, A. Immune-Related Adverse Events: Pneumonitis. Adv. Exp. Med. Biol. 2020, 1244, 255–269. [Google Scholar] [PubMed]
- Conroy, M.; Naidoo, J. Immune-related adverse events and the balancing act of immunotherapy. Nat. Commun. 2022, 13, 392. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.R.; Kanneganti, T.D. NLRP3 inflammasome in cancer and metabolic diseases. Nat. Immunol. 2021, 22, 550–559. [Google Scholar] [CrossRef]
- Moossavi, M.; Parsamanesh, N.; Bahrami, A.; Atkin, S.L.; Sahebkar, A. Role of the NLRP3 inflammasome in cancer. Mol. Cancer. 2018, 17, 158. [Google Scholar] [CrossRef]
- Anderson, R.; Theron, A.J.; Rapoport, B.L. Immunopathogenesis of Immune Checkpoint Inhibitor-Related Adverse Events: Roles of the Intestinal Microbiome and Th17 Cells. Front. Immunol. 2019, 10, 2254. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Hamarsheh, S.; Zeiser, R. NLRP3 Inflammasome Activation in Cancer: A Double-Edged Sword. Front. Immunol. 2020, 11, 144. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Wu, C.; Yosef, N.; Thalhamer, T.; Zhu, C.; Xiao, S.; Kishi, Y.; Regev, A.; Kuchroo, V.K. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 2013, 496, 513–517. [Google Scholar] [CrossRef]
- Hernandez, A.L.; Kitz, A.; Wu, C.; Lowther, D.E.; Rodriguez, D.M.; Vudattu, N.; Deng, S.; Herold, K.C.; Kuchroo, V.K.; Kleinewietfeld, M.; et al. Sodium chloride inhibits the suppressive function of FOXP3+ regulatory T cells. J. Clin. Investig. 2015, 125, 4212–4222. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Ji, W.J.; Yuan, F.; Guo, Z.Z.; Li, Y.X.; Dong, Y.; Ma, Y.Q.; Zhou, X.; Li, Y.M. Th17/Treg Imbalance Induced by Dietary Salt Variation Indicates Inflammation of Target Organs in Humans. Sci. Rep. 2016, 6, 26767. [Google Scholar] [CrossRef] [PubMed]
- Willebrand, R.; Hamad, I.; Van Zeebroeck, L.; Kiss, M.; Bruderek, K.; Geuzens, A.; Swinnen, D.; Corte-Real, B.F.; Marko, L.; Lebegge, E.; et al. High Salt Inhibits Tumor Growth by Enhancing Anti-tumor Immunity. Front. Immunol. 2019, 10, 1141. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Xu, J.; Mu, R.; Li, Q.; Lv, D.L.; Huang, Z.; Zhang, J.; Wang, C.; Dong, L. High-salt diet inhibits tumour growth in mice via regulating myeloid-derived suppressor cell differentiation. Nat. Commun. 2020, 11, 1732. [Google Scholar] [CrossRef]
- Rizvi, Z.A.; Dalal, R.; Sadhu, S.; Kumar, Y.; Kumar, S.; Gupta, S.K.; Tripathy, M.R.; Rathore, D.K.; Awasthi, A. High-salt diet mediates interplay between NK cells and gut microbiota to induce potent tumor immunity. Sci. Adv. 2021, 7, eabg5016. [Google Scholar] [CrossRef]
- Scrivo, R.; Perricone, C.; Altobelli, A.; Castellani, C.; Tinti, L.; Conti, F.; Valesini, G. Dietary Habits Bursting into the Complex Pathogenesis of Autoimmune Diseases: The Emerging Role of Salt from Experimental and Clinical Studies. Nutrients 2019, 11, 1013. [Google Scholar] [CrossRef]
- Allu, A.S.; Tiriveedhi, V. Cancer Salt Nostalgia. Cells 2021, 10, 1285. [Google Scholar] [CrossRef]
- Amara, S.; Tiriveedhi, V. Inflammatory role of high salt level in tumor microenvironment (Review). Int. J. Oncol. 2019, 50, 1477–1481. [Google Scholar] [CrossRef]
- Wiig, H.; Schroder, A.; Neuhofer, W.; Jantsch, J.; Kopp, C.; Karlsen, T.V.; Boschmann, M.; Goss, J.; Bry, M.; Rakova, N.; et al. Immune cells control skin lymphatic electrolyte homeostasis and blood pressure. J. Clin. Investig. 2013, 123, 2803–2815. [Google Scholar] [CrossRef]
- Berga-Bolanos, R.; Alberdi, M.; Buxade, M.; Aramburu, J.; Lopez-Rodriguez, C. NFAT5 induction by the pre-T-cell receptor serves as a selective survival signal in T-lymphocyte development. Proc. Natl. Acad. Sci. USA 2013, 110, 16091–16096. [Google Scholar] [CrossRef]
- Go, W.Y.; Liu, X.; Roti, M.A.; Liu, F.; Ho, S.N. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. USA 2004, 101, 10673–10678. [Google Scholar] [CrossRef] [PubMed]
- Tiriveedhi, V.; Ivy, M.T.; Myles, E.L.; Zent, R.; Rathmell, J.C.; Titze, J. Ex Vivo High Salt Activated Tumor-Primed CD4+T Lymphocytes Exert a Potent Anti-Cancer Response. Cancers 2021, 13, 1690. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.K.; Medzhitov, R. Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat. Commun. 2015, 6, 6931. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Zha, S.; Shen, X.; Zhao, Y.; Li, L.; Yang, L.; Lei, M.; Liu, W. NFAT5 mediates hypertonic stress-induced atherosclerosis via activating NLRP3 inflammasome in endothelium. Cell Commun. Signal. 2019, 17, 102. [Google Scholar] [CrossRef]
- Babaer, D.; Zheng, M.; Ivy, M.T.; Zent, R.; Tiriveedhi, V. Methylselenol producing selenocompounds enhance the efficiency of mammaglobin-A peptide vaccination against breast cancer cells. Oncol. Lett. 2019, 18, 6891–6898. [Google Scholar] [CrossRef]
- Amara, S.; Majors, C.; Roy, B.; Hill, S.; Rose, K.L.; Myles, E.L.; Tiriveedhi, V. Critical role of SIK3 in mediating high salt and IL-17 synergy leading to breast cancer cell proliferation. PLoS ONE 2017, 12, e0180097. [Google Scholar] [CrossRef]
- Babaer, D.; Amara, S.; McAdory, B.S.; Johnson, O.; Myles, E.L.; Zent, R.; Rathmell, J.C.; Tiriveedhi, V. Oligodeoxynucleotides ODN 2006 and M362 Exert Potent Adjuvant Effect through TLR-9/-6 Synergy to Exaggerate Mammaglobin-A Peptide Specific Cytotoxic CD8+T Lymphocyte Responses against Breast Cancer Cells. Cancers 2019, 11, 672. [Google Scholar] [CrossRef]
- Tiriveedhi, V.; Takenaka, M.; Ramachandran, S.; Gelman, A.E.; Subramanian, V.; Patterson, G.A.; Mohanakumar, T. T regulatory cells play a significant role in modulating MHC class I antibody-induced obliterative airway disease. Am. J. Transplant. 2012, 12, 2663–2674. [Google Scholar] [CrossRef][Green Version]
- Ouwerkerk, R.; Jacobs, M.A.; Macura, K.J.; Wolff, A.C.; Stearns, V.; Mezban, S.D.; Khouri, N.F.; Bluemke, D.A.; Bottomley, P.A. Elevated tissue sodium concentration in malignant breast lesions detected with non-invasive 23Na, MRI. Breast Cancer Res. Treat. 2007, 106, 151–160. [Google Scholar] [CrossRef]
- Zaric, O.; Pinker, K.; Zbyn, S.; Strasser, B.; Robinson, S.; Minarikova, L.; Gruber, S.; Farr, A.; Singer, C.; Helbich, T.H.; et al. Quantitative Sodium MR Imaging at 7 T: Initial Results and Comparison with Diffusion-weighted Imaging in Patients with Breast Tumors. Radiology 2016, 280, 39–48. [Google Scholar] [CrossRef]
- Rahbar, H.; Partridge, S.C. Multiparametric MR Imaging of Breast Cancer. Magn. Reason. Imaging Clin. N. Am. 2016, 24, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Jo, S.G.; Kim, D.J.; Park, J.H. NLRP3 inflammasome mediates interleukin-1beta production in immune cells in response to Acinetobacter baumannii and contributes to pulmonary inflammation in mice. Immunology 2017, 150, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Amara, S.; Alotaibi, D.; Tiriveedhi, V. NFAT5/STAT3 interaction mediates synergism of high salt with IL-17 towards induction of VEGF-A expression in breast cancer cells. Oncol. Lett. 2016, 12, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Arnett, D.K.; Blumenthal, R.S.; Albert, M.A.; Buroker, A.B.; Goldberger, Z.D.; Hahn, E.J.; Himmelfarb, C.D.; Khera, A.; Lloyd-Jones, D.; McEvoy, J.W.; et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019, 74, 1376–1414. [Google Scholar] [CrossRef]
- Mente, A.; O‘Donnell, M.; Rangarajan, S.; Dagenais, G.; Lear, S.; McQueen, M.; Diaz, R.; Avezum, A.; Lopez-Jaramillo, P.; Lanas, F.; et al. Associations of urinary sodium excretion with cardiovascular events in individuals with and without hypertension: A pooled analysis of data from four studies. Lancet 2016, 388, 465–475. [Google Scholar] [CrossRef]
- Na, S.Y.; Janakiraman, M.; Leliavski, A.; Krishnamoorthy, G. High-salt diet suppresses autoimmune demyelination by regulating the blood-brain barrier permeability. Proc. Natl. Acad. Sci. USA 2021, 118, e2025944118. [Google Scholar] [CrossRef]
- Grillo, A.; Salvi, L.; Coruzzi, P.; Salvi, P.; Parati, G. Sodium Intake and Hypertension. Nutrients 2019, 11, 1970. [Google Scholar] [CrossRef]
- He, F.J.; MacGregor, G.A. Role of salt intake in prevention of cardiovascular disease: Controversies and challenges. Nat. Rev. Cardiol. 2018, 15, 371–377. [Google Scholar] [CrossRef]
- Wu, B.; Yang, D.; Yang, S.; Zhang, G. Dietary Salt Intake and Gastric Cancer Risk: A Systematic Review and Meta-Analysis. Front. Nutr. 2021, 8, 801228. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Radin, J.N.; Loh, J.T.; Zhang, F.; Washington, M.K.; Peek, R.M., Jr.; Algood, H.M.; Cover, T.L. High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis. Infect. Immun. 2013, 81, 2258–2267. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Fox, J.G.; Gonda, T.; Worthley, D.L.; Muthupalani, S.; Wang, T.C. Mouse models of gastric cancer. Cancers 2013, 5, 92–130. [Google Scholar] [CrossRef] [PubMed]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Amara, S.; Ivy, M.T.; Myles, E.L.; Tiriveedhi, V. Sodium channel gammaENaC mediates IL-17 synergized high salt induced inflammatory stress in breast cancer cells. Cell Immunol. 2016, 302, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Amara, S.; Zheng, M.; Tiriveedhi, V. Oleanolic Acid Inhibits High Salt-Induced Exaggeration of Warburg-like Metabolism in Breast Cancer Cells. Cell Biochem. Biophys. 2016, 74, 427–434. [Google Scholar] [CrossRef]
- Alotaibi, D.; Amara, S.; Johnson, T.L.; Tiriveedhi, V. Potential anticancer effect of prostratin through SIK3 inhibition. Oncol. Lett. 2018, 15, 3252–3258. [Google Scholar] [CrossRef]
- Steenbrugge, J.; Vander Elst, N.; Demeyere, K.; De Wever, O.; Sanders, N.N.; Van Den Broeck, W.; Dirix, L.; Van Laere, S.; Meyer, E. Comparative Profiling of Metastatic 4T1- vs. Non-metastatic Py230-Based Mammary Tumors in an Intraductal Model for Triple-Negative Breast Cancer. Front. Immunol. 2019, 10, 2928. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Wang, D.Y.; Salem, J.E.; Cohen, J.V.; Chandra, S.; Menzer, C.; Ye, F.; Zhao, S.; Das, S.; Beckermann, K.E.; Ha, L.; et al. Fatal Toxic Effects Associated With Immune Checkpoint Inhibitors: A Systematic Review and Meta-analysis. JAMA Oncol. 2018, 4, 1721–1728. [Google Scholar] [CrossRef]
- Albandar, H.J.; Fuqua, J.; Albandar, J.M.; Safi, S.; Merrill, S.A.; Ma, P.C. Immune-Related Adverse Events (irAE) in Cancer Immune Checkpoint Inhibitors (ICI) and Survival Outcomes Correlation: To Rechallenge or Not? Cancers 2021, 13, 989. [Google Scholar] [CrossRef]
- Patrinely, J.R., Jr.; Johnson, R.; Lawless, A.R.; Bhave, P.; Sawyers, A.; Dimitrova, M.; Yeoh, H.L.; Palmeri, M.; Ye, F.; Fan, R.; et al. Chronic Immune-Related Adverse Events Following Adjuvant Anti-PD-1 Therapy for High-risk Resected Melanoma. JAMA Oncol. 2021, 7, 744–748. [Google Scholar] [CrossRef]
- Dearden, H.; Au, L.; Wang, D.Y.; Zimmer, L.; Eroglu, Z.; Smith, J.L.; Cuvietto, M.; Khoo, C.; Atkinson, V.; Lo, S.; et al. Hyperacute toxicity with combination ipilimumab and anti-PD1 immunotherapy. Eur. J. Cancer. 2021, 153, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Alilou, S.; Mirzaei, H.R. Adverse Events Following Administration of Anti-CTLA4 Antibody Ipilimumab. Front. Oncol. 2021, 11, 624780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, S.; Xiao, Y.; Zhang, W.; Wu, S.; Qin, T.; Yue, Y.; Qian, W.; Li, L. NLRP3 Inflammasome and Inflammatory Diseases. Oxid. Med. Cell. Longev. 2020, 2020, 4063562. [Google Scholar] [CrossRef] [PubMed]
- Pollack, M.H.; Betof, A.; Dearden, H.; Rapazzo, K.; Valentine, I.; Brohl, A.S.; Ancell, K.K.; Long, G.V.; Menzies, A.M.; Eroglu, Z.; et al. Safety of resuming anti-PD-1 in patients with immune-related adverse events (irAEs) during combined anti-CTLA-4 and anti-PD1 in metastatic melanoma. Ann. Oncol. 2018, 29, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Spiers, L.; Coupe, N.; Payne, M. Toxicities associated with checkpoint inhibitors-an overview. Rheumatology 2019, 58, vii7–vii16. [Google Scholar] [CrossRef]
- Dimitriou, F.; Staeger, R.; Ak, M.; Maissen, M.; Kudura, K.; Barysch, M.J.; Levesque, M.P.; Cheng, P.F.; Dummer, R.; Mangana, J. Frequency, Treatment and Outcome of Immune-Related Toxicities in Patients with Immune-Checkpoint Inhibitors for Advanced Melanoma: Results from an Institutional Database Analysis. Cancers 2021, 13, 2931. [Google Scholar] [CrossRef]
- Deshpande, R.P.; Sharma, S.; Watabe, K. The Confounders of Cancer Immunotherapy: Roles of Lifestyle, Metabolic Disorders and Sociological Factors. Cancers 2020, 12, 2983. [Google Scholar] [CrossRef]



| Regular Diet | High Salt | Low Salt | |
|---|---|---|---|
| Effector Cytokines | |||
| IL-1β | 427 ± 111 | 1453 ± 256 * | 203 ± 89 *,$ |
| TNF-α | 511 ± 93 | 1818 ± 279 * | 367 ± 83 *,$ |
| IFN-γ | 342 ± 77 | 1507 ± 301 * | 312 ± 84 * |
| IL-17A | 27 ± 4 | 79 ± 13 * | n.d. |
| Anti-inflammatory Cytokines | |||
| IL-4 | n.d. | n.d. | n.d. |
| IL-10 | 3659 ± 394 | 1018 ± 238 * | 5138 ± 651 *,$ |
| Immunostimulatory Cytokines | |||
| Eotaxin | 34 ± 12 | 49 ± 17 | 33 ± 18 |
| IL-1α | n.d. | 7.8 ± 3.6 | n.d. |
| IL-2 | 6.2 ± 4.9 | 6.6 ± 3.1 | 5.9 ± 2.7 |
| IL-5 | n.d. | n.d. | n.d. |
| IL-6 | 216 ± 68 | 582 ± 91 * | 114 ± 73 $ |
| IL-12(p40) | 673 ± 116 | 803 ± 132 | 754 ± 82 |
| IL-12(p70) | 256 ± 59 | 298 ± 102 | 272 ± 73 |
| IL-13 | 12.1 ± 2.7 | 39.4 ± 8.3 * | 11.9 ± 3.7 |
| Chemoattractants | |||
| RANTES | 1042 ± 287 | 2365 ± 397 * | 1246 ± 231 $ |
| MIP-1α | 127 ± 37 | 496 ± 82 * | 94 ± 21 $ |
| MIP-1β | 214 ± 47 | 726 ± 54 * | 376 ± 97 $ |
| MCP-1 | 184 ± 39 | 212 ± 42 | 206 ± 58 |
| KC | 17 ± 4 | 15 ± 5 | 22 ± 7 |
| Growth Factors/Cell differentiation factors | |||
| IL-9 | n.d. | n.d. | n.d. |
| IL-3 | 2673 ± 538 | 3982 ± 537 * | 698 ± 269 *,$ |
| GM-CSF | 22 ± 7 | 38 ± 9 | 32 ± 9 |
| KC | 17 ± 4 | 15 ± 5 | 22 ± 7 |
| G-CSF | 513 ± 97 | 583 ± 104 | 638 ± 96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khandekar, D.; Dahunsi, D.O.; Manzanera Esteve, I.V.; Reid, S.; Rathmell, J.C.; Titze, J.; Tiriveedhi, V. Low-Salt Diet Reduces Anti-CTLA4 Mediated Systemic Immune-Related Adverse Events while Retaining Therapeutic Efficacy against Breast Cancer. Biology 2022, 11, 810. https://doi.org/10.3390/biology11060810
Khandekar D, Dahunsi DO, Manzanera Esteve IV, Reid S, Rathmell JC, Titze J, Tiriveedhi V. Low-Salt Diet Reduces Anti-CTLA4 Mediated Systemic Immune-Related Adverse Events while Retaining Therapeutic Efficacy against Breast Cancer. Biology. 2022; 11(6):810. https://doi.org/10.3390/biology11060810
Chicago/Turabian StyleKhandekar, Durga, Debolanle O. Dahunsi, Isaac V. Manzanera Esteve, Sonya Reid, Jeffrey C. Rathmell, Jens Titze, and Venkataswarup Tiriveedhi. 2022. "Low-Salt Diet Reduces Anti-CTLA4 Mediated Systemic Immune-Related Adverse Events while Retaining Therapeutic Efficacy against Breast Cancer" Biology 11, no. 6: 810. https://doi.org/10.3390/biology11060810
APA StyleKhandekar, D., Dahunsi, D. O., Manzanera Esteve, I. V., Reid, S., Rathmell, J. C., Titze, J., & Tiriveedhi, V. (2022). Low-Salt Diet Reduces Anti-CTLA4 Mediated Systemic Immune-Related Adverse Events while Retaining Therapeutic Efficacy against Breast Cancer. Biology, 11(6), 810. https://doi.org/10.3390/biology11060810







