Simple Summary
Biological control of plant diseases caused by fungal pathogens using antagonistic microorganisms including Bacilli has been considered to be an effective and safe alternative to chemical fungicides. Fusarium crown rot of wheat is a serious fungal disease affecting yield and grain quality. In this study, a newly isolated strain of Bacillus subtilis YB-15 from soil of wheat rhizosphere significantly inhibited Fusarium crown rot as well as improved growth of wheat seedlings. Multiple potential biocontrol and growth-promoting attributes of Bacillus subtilis YB-15 were determined in vitro and according to the whole genome sequencing analysis. Overall, the results demonstrated that Bacillus subtilis YB-15 has great potential for practical application in controlling plant fungal diseases and improving plant growth.
Abstract
Fusarium crown rot caused by Fusarium pseudograminearum is one of the most devastating diseases of wheat worldwide causing major yield and economic losses. In this study, strain YB-15 was isolated from soil of wheat rhizosphere and classified as Bacillus subtilis by average nucleotide identity analysis. It significantly reduced Fusarium crown rot with a control efficacy of 81.50% and significantly improved the growth of wheat seedlings by increasing root and shoot fresh weight by 11.4% and 4.2%, respectively. Reduced Fusarium crown rot may have been due to direct antagonism by the production of β-1, 3-glucanase, amylase, protease and cellulase, or by the ability of B. subtilis YB-15 to induce defense-related enzyme activities of wheat seedlings, both alone and in seedlings infected with F. pseudograminearum. Improved plant growth may be related to the ability of B. subtilis YB-15 to secrete indole acetic acid and siderophores, as well as to solubilize phosphorus. In addition, the genome of strain YB-15 was determined, resulting in a complete assembled circular genome of 4,233,040 bp with GC content of 43.52% consisting of 4207 protein-encoding genes. Sequencing the B. subtilis YB-15 genome further revealed genes for encoding carbohydrate-active enzymes, biosynthesis of various secondary metabolites, nutrient acquisition, phytohormone production, chemotaxis and motility, which could explain the potential of strain YB-15 to be plant growth-promoting bacteria and biological control agent. B. subtilis YB-15 appears to be a promising biocontrol agent against Fusarium crown rot as well as for wheat growth promotion.
1. Introduction
Wheat (Triticum aestivum L.) is one of the most important food crops in the world []. However, in many wheat-producing countries, yields are significantly reduced by Fusarium crown rot predominantly caused by Fusarium graminearum, Fusarium pseudograminearum and Fusarium culmorum []. However, F. pseudograminearum is more common in drier and warmer areas []. At present, the application of chemical fungicides is the principal approach to controlling Fusarium crown rot, but long-term use of fungicides can lead to negative environmental effects and reduced effectiveness due to fungicide resistance []. More sustainable and ecologically friendly alternatives are needed for Fusarium crown rot management []. Recently, there have been a number of studies on the biocontrol of wheat Fusarium crown rot as an alternative strategy [,,,].
Bacillus species, such as Bacillus velezensis and Bacillus subtilis, have been widely studied for their strong antagonistic activities against many plant pathogens and their abilities to promote growth [,,,,]. They can protect crops against a broad range of fungal pathogens by both indirect and direct mechanisms, including secretion of antimicrobial compounds such as antibiotics and hydrolytic enzymes, competition for resources and triggering induced systemic resistance (ISR) [,]. Moreover, Bacillus species can promote plant growth through improving nutrient availability, reducing the severity of environmental stresses and altering plant growth hormone homeostasis [,,]. There have been several reports of growth promotion of wheat by Bacillus species [,,]. There have also been a few reports of Bacillus species biologically preventing F. pseudograminearum from causing crown rot on wheat and sorghum [,].
With the rapid progress of DNA sequencing technology, high-throughput sequencing has been applied to assemble and analyze many bacterial genomes. For example, the genome of B. subtilis PMB102 isolated from leaf of tomato has been sequenced, and genes related to growth promotion, including acetoin and siderophore production, and genes related to disease control, such as exopolysaccharide synthesis and cell wall-degrading enzymes, were found []. Another example is an analysis of the 13 genomes of B. subtilis strains from soil where the production of non-ribosomal peptides was different among the strains, and in vitro antagonism assays showed that plipastatin alone was enough to inhibit Fusarium spp., but both surfactin and plipastatin were required to inhibit Botrytis cinerea []. Sequencing the B. subtilis EA-CB0575 genome revealed that it has homologous genes involved in plant growth promotion, such as the release of volatile compounds, solubilizing phosphate, fixing nitrogen, and producing indole and siderophore []. Thus, whole-genome sequencing of biological control agents (BCA)s and plant growth-promoting bacteria (PGPB) is a valuable resource for exploring the characteristics responsible for biocontrol and growth promotion.
In this study, strain YB-15 was isolated from rhizospheric soil of wheat and investigated for its ability to inhibit Fusarium crown rot caused by F. pseudograminearum as well as to improve the growth of wheat seedlings. To understand its effects, five wheat defense-related enzyme activities were examined following inoculation with strain YB-15 alone, F. pseudograminearum alone and the combination of strain YB-15 and F. pseudograminearum. In addition, the genome of B. subtilis YB-15 was sequenced and assembled by Nanopore and Illumina sequencing, resulting in a complete circular genome of 4,233,040 bp with GC content of 43.52% consisting of 4207 protein-encoding genes. Sequencing the genome of strain YB-15 was used to conduct an average nucleotide identity (ANI) analysis to determine the species as well as identify a number of genes for traits commonly associated with PGPB and BCA.
2. Materials and Methods
2.1. Bacteria and Pathogens Used in this Study
Five rhizospheric soil samples were collected from a commercial wheat field in Lushan, Henan Province, China, and strain YB-15 was isolated from wheat rhizospheric soil attached to roots at 5 cm beneath the soil surface as previously described []. An isolate of F. pseudograminearum, WZ001, was obtained from the Institute of Plant Protection Research, Henan Academy of Agricultural Sciences, Zhengzhou, China.
2.2. Detection of In Vitro Plant Growth Promotion and Biocontrol Traits
IAA production was detected with strain YB-15 colonies grown in L-tryptophan nutrient broth []. Colonies of strain YB-15 were grown on Chrome Azurol S blue agar to determine siderophore production [], phosphorus agar to determine phosphorus solubilization [], β-glucan agar to determine β-glucanaseactivity [], starch agar to determine amylase activity [], skim milk agar to determine protease activity [] and carboxymethylcellulose agar to determine cellulase activity [].
2.3. Growth Promotion and Biocontrol Assay of Strain YB-15 against Fusarium Crown Rot of Wheat Seedlings
F. pseudograminearum isolate WZ001 was grown on PDA for 5 days at 28 °C, and then six 5 mm agar plugs were transferred from the growing edge to a 1000 mL flask with 200 g sterilized boiled wheat grain and sand (3:1, v/v) and incubated at 28 °C for 7 days [,]. Strain YB-15 was grown in 100 mL LB broth with 180 rpm shaking at 37 °C until an OD595 nm of 0.8. Wheat cultivar zhengmai 366 seeds were surface-sterilized in 75% ethanol (v/v) for 30 s, rinsed three times with sterile distilled water and air-dried at room temperature []. The seeds were then soaked with strain YB-15 suspension for 12 h at 28 °C. A total of 25 seeds were sown per pot (10 cm diameter,10 cm high) containing 350 g mixture of sterile soil with or without 5% inoculum (w/w), and the pots were maintained in a greenhouse at 25 °C under a 12 h light/12 h dark photoperiod. The treatments were: (1) seeds soaked with YB-15 and planted in sterile soil without pathogen inoculum; (2) seeds not soaked with YB-15 and planted in sterile soil without pathogen inoculum; (3) seeds soaked with YB-15 and planted in sterile soil with pathogen inoculum; and (4) seeds not soaked with YB-15 and planted in sterile soil with pathogen inoculum. The Fusarium crown rot disease severity was assessed using scale of 0–4 at 20 days post-planting, and disease index (DI) was calculated using DI = [(0 × S0) + (1 × S1) + (2 × S2) + (3 × S3)]/A, where S is the number of wheat seedlings for each disease class and A is total number of tested wheat seedlings []. Control efficacy (CE) was calculated using CE= [(DI of control−DI of treatment)/DI of control] ×100% []. Each treatment had 20 wheat seedlings with six replicates. At 20 days post-planting, shoot height and root length were determined manually with a ruler, and root and shoot fresh weights were weighted with an electronic analytical balance (ME203E, Mettler Toledo, Changzhou, China).
2.4. Wheat Defense-Related Enzyme Activities
After 20 days post-planting, wheat leaves were harvested into liquid nitrogen and stored at −80 °C. Firstly, 1 g leaves of wheat seedlings were ground in pre-chilled mortar, and then transferred into a new 1.5 mL Eppendorf tube with 1 mL extraction buffer. After centrifugation for 10 min at 8000× g, the supernatant was transferred to another new 1.5 mL Eppendorf tube for detecting enzyme activities. Enzyme activities were measured by using commercially available assay kits of LOX (Cat. No. BC0325), PAL (Cat. No. BC0215), CAT (Cat. No. BC0205), PPO (Cat. No. BC0195) and POD (Cat. No. BC0095) accordingto the manufacturer’s instructions (Solarbio, Beijing, China). Absorbance was recorded with a plate reader (Tecan Spark, Tecan, Männedorf, Switzerland).
2.5. DNA Preparation, Genome Sequencing, Assembly and Annotation of Strain YB-15
Strain YB-15 was grown in NB medium on a rotary shaker (QYC-200, Fuma, Shanghai, China) with shaking speed of 180 rpm at 28 °C for 18 h. Genomic DNA of strain YB-15 was extracted by using the Mini-BEST Bacterial Genomic DNA Extraction Kit Ver. 3.0 (Takara, Beijing, China) according to the manufacturer’s directions. Two separate genomic DNA libraries were constructed for the Oxford Nanopore and Illumina NovaSeq sequencing systems. Genome assembly was conducted by Unicycler v0.4.9 []. A circular map of the strain YB-15 genome was generated by CGView server []. Nanopore and Illumina sequencing reads were mapped to the genome using Minimap2 (2.17-r974-dirty) [] and BWA (0.7.17-r1198-dirty) [], respectively. The depth of genome coverage was estimated with SAMtools []. Annotation of strain YB-15 genome was performed with Prokka (1.13) [], and protein-coding, tRNA and rRNA genes were predicted using Prodigal (v2.6.3) [], Aragorn (v1.2.38) [] and RNAmmer (v1.2) [], respectively.
2.6. Molecular Identification of Strain YB-15
The genome sequences of Bacillus velezensis FZB42, Bacillus subtilis H1, Bacillus velezensis FJAT-45028, Bacillus subtilis 168, Bacillus licheniformis ATCC 14580, Bacillus altitudinis GQYP101, Bacillus licheniformis SRCM103583, Bacillus altitudinis CHB19, Bacillus pumilus ZB201701 and Bacillus pumilus SF-4 were obtained from the NCBI genome database (GenBank Accession Nos: CP000560.2, CP026662.1, CP047157.1, NC_000964.3, CP034569.1, CP040514.1, CP035404.1, CP043559.1, CP029464.1 and CP047089.1, respectively). ANI among the above genomes was analyzed with ANI calculator [].
2.7. Analysis of CAZymes and Genes Associated with Growth Promotion and Secondary Metabolites of Strain YB-15
Annotated protein-coding sequences of strain YB-15 were aligned against the carbohydrate-active enzyme (CAZy) database using dbCAN2 with the threshold of E-value1e-15 []. Signal peptide was predicted by SignalP (v4.1) []. Gene clusters for synthesis of secondary metabolite were identified by antiSMASH []. Local BLASTP was used to identify genes associated with plant growth promotion.
2.8. Statistical Analysis
Statistical analysis was performed using SPSS v21.0 software according to the one-way analysis of variance. The differences among means were determined usingDuncan’s multiple range tests with p value ≤ 0.05.
3. Results
3.1. Isolation of Strain YB-15 and Its Antagonism against F. pseudograminearum
A number of 78 bacterial strains were isolated and screened from wheat rhizospheric soil by dilution plating. Out of the 78 strains, 59 showed varying levels of antagonism against F. pseudograminearum in dual PDA culture (data not shown). Strain YB-15 was selected for further investigation because it showed the greatest antagonism against F. pseudograminearum on PDA (Figure 1A,B). Strain YB-15 colonies were opaque white with rod-shaped and Gram-positive (data not shown).
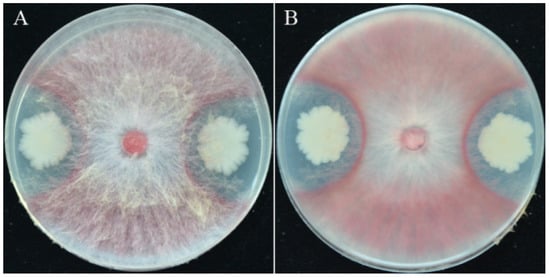
Figure 1.
Characteristics of strain YB-15: (A,B) Dual culture of F. pseudograminearum and strain YB-15 on PDA.
3.2. Plant Growth Promotion and Antifungal Traits of Strain YB-15 in Culture
Possible plant growth promotion traits of strain YB-15 detected in culture were the production of indole acetic acid (IAA) (Figure 2A), siderophore (Figure 2B) and phosphorus solubilization (Figure 2C). Potential antifungal traits detected in culture were secretion of β-1, 3-glucanase (Figure 2D), amylase (Figure 2E), protease (Figure 2F) and cellulase (Figure 2G).
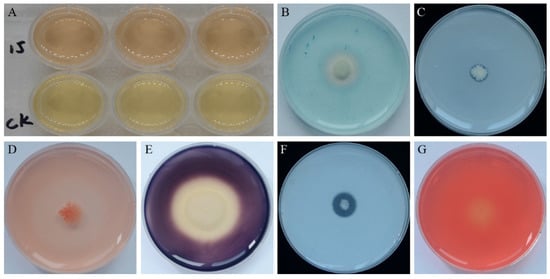
Figure 2.
PGP and Antifungal traits of strain YB-15: (A) The upper indicating IAA production by pink coloration; (B) Yellow-orange halos indicating siderophore production; (C) Phosphorus solubilization indicated by a halo zone around strain YB-15 colonies; (D) β-1, 3-glucanase activity indicated by a zone around strain YB-15 colonies; (E) Amylase activity indicated by a clear zone around strain YB-15 colonies; (F) Protease activity indicated by an obvious hydrolytic zone around strain YB-15 colonies; (G) Cellulase activity indicated by a zone of hydrolysis around strain YB-15 colonies.
3.3. Effects of Strain YB-15 on Fusarium Crown Rot of Wheat Seedlings
Typical Fusarium crown rot symptoms with brown lesions on the stems were observed in F. pseudograminearum inoculated seedlings at 20 days post-planting, but no disease symptoms were observed in seedlings with both F. pseudograminearum and strain YB-15 or control treatments not inoculated with F. pseudograminearum (Figure 3). The disease severity of wheat seedlings infected with F. pseudograminearum was 2.63 ± 0.03, whereas it was only 0.48 ± 0.02 with the combination of strain YB-15 and F. pseudograminearum. Disease incidence was 91.67 ± 1.67% infected with F. pseudograminearum, but only 15.00 ± 2.89% with the combination of strain YB-15 and F. pseudograminearum. Control efficacy was 81.50 ± 0.76% with seed treatment of strain YB-15 compared to non-treated control seeds infected with F. pseudograminearum (Table 1).

Figure 3.
Effect of strain YB-15 against Fusarium crown rot caused by F.pseudograminearum and on growth of wheat seedlings: (A,E) seeds soaked with YB-15 and planted in sterile soil without pathogen inoculum; (B,F) seeds not soaked with YB-15 and planted in sterile soil without pathogen inoculum; (C,G) seeds soaked with YB-15 and planted in sterile soil with pathogen inoculum; (D,H) seeds not soaked with YB-15 and planted in sterile soil with pathogen inoculum.

Table 1.
Disease index, disease incidence and control efficacy of B. subtilis YB-15 against Fusarium crown rot of wheat.
3.4. Growth Promotion of Wheat Seedlings by Strain YB-15
After 20 days post-planting, the shoot and root fresh weight of seedlings treated with strain YB-15 was significantly increased by 4.2% and 11.4%, respectively; although, root length and shoot height were not significantly affected (Table 2). Inoculation with F. pseudograminearum resulted in significantly lower shoot height and weight as well as root length and weight, compared to the control. Most notable was the reduction in root fresh weight by 46.7%. Inoculation with both strain YB-15 and F. pseudograminearum resulted in significantly higher shoot height and weight as well as root weight and length, compared to that of the F. pseudograminearum inoculated wheat seedlings. There was no significant difference in shoot height and root length between the non-treated control and seedlingsinoculated with both strain YB-15 and F. pseudograminearum; although, the fresh weights remained lower.

Table 2.
Effects of B. subtilis YB-15 on the growth of wheat seedlings.
3.5. Defense-Related Enzyme Activities of Wheat Seedlings
Wheat seedlings with strain YB-15 treatment exhibited significantly higher activities of PAL, POD, CAT and PPO, but not LOX, compared to non-treated control seedlings (Table 3). Inoculation with F. pseudograminearum resulted in significantly higher PAL, POD, CAT, PPO and LOX activities, compared to non-treated seedlings, but the enzyme activities were significantly higher with the combination of strain YB-15 and F. pseudograminearum compared to only F. pseudograminearum inoculation. The activities with the combination of strain YB-15 and F. pseudograminearum were also significantly higher compared to strain YB-15 alone, except for POD activity, which was significantly lower.

Table 3.
Activities of defense-related enzymes in wheat leaves.
3.6. Genome Assembly and Annotation of Strain YB-15
A total of 21,392 long reads containing 1,000,070,603 bases with a mean length of 46,749.7 bp and an N50 of 45,733 bp were obtained by Nanopore sequencing, and 13,740,276 reads containing 2,061,041,400 bases were generated by Illumina sequencing. These were used to assemble the genome of strain YB-15 with 238.54X and 486.61X genome coverage for the Nanopore and Illumina sequencing data, respectively. The completed assembled genome of strain YB-15 was deposited at GenBank with Accession number CP092631. The strain YB-15 genome was a 4,233,040 bp single circular chromosome with 43.52% GC content (Figure 4). There were 4207 protein-encoding genes, 86 tRNAs and 27 rRNAs, which were annotated (Table S1).
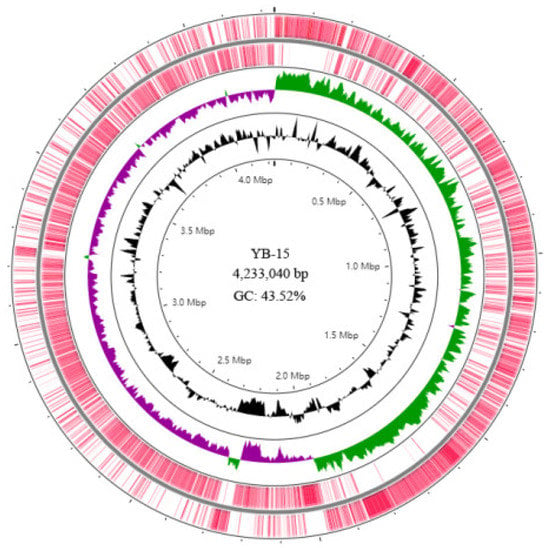
Figure 4.
Circular map of B. subtilis YB-15 genome visualized by CGView Server. The distribution of rings from outwards to inwards be: ring 1 and 2 for protein-coding genes on the forward strand and reverse strand; ring 3 for GC skew in plus (green) and GC skew in minus (purple); ring 4 for GC content (black).
3.7. Species Identification of Strain YB-15
ANI values of the genome of strain YB-15 to those of ten other Bacillus strains ranged from 71.02 to 98.76% (Figure 5). The ANI values between strain YB-15 and Bacillus subtilis strain 168 was 98.76% and between strain YB-15 and Bacillus subtilis strain H1 was 98.70%, both of which were above the thresholds of 95–96% for recognizing prokaryotic species boundaries []. Thus, strain YB-15 was identified as Bacillus subtilis.
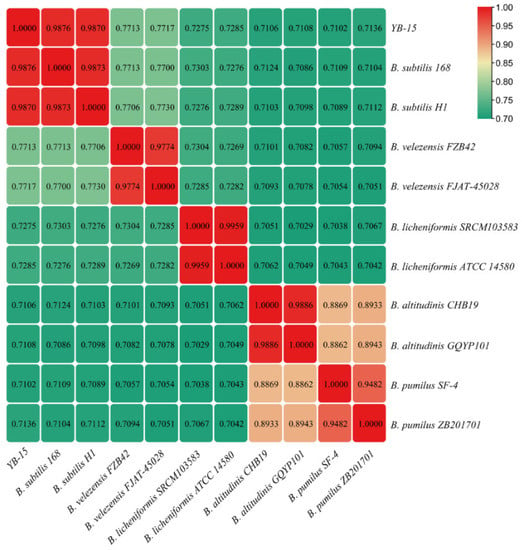
Figure 5.
Heatmap of ANIvalues based on whole-genome sequences of B. subtilis YB-15 and 10 other Bacillus species.
3.8. Predicted Genes for CAZymes
The B. subtilis YB-15 genome had 122 putative genes encoding CAZymes, including 48 glycoside hydrolases (GHs), 19 carbohydrate esterases (CEs), 7 polysaccharide lyases (PLs), 43 glycosyltransferases (GTs), 7 carbohydrate-binding modules (CBMs) and 4 auxiliary activities (AAs) (Table S2). Among those, six genes are classified to be both GHs and CBMs. There were 28 genes predicted to have signal peptides, and most belonged to the GHs (17) followed by CBMs (5), PLs (5), CEs (4) and GTs (1) with 4 of them having both GHs and CBMs.
3.9. Predicted Genes for Secondary Metabolites
The B. subtilis YB-15 genome had 11 putative gene clusters for secondary metabolites (Table 4). For antimicrobial compound synthesis, there were five predicted gene clusters with 100% similarity to clusters for the synthesis of bacillibactin, fengycin, bacillaene, subtilosin A and bacilysin. There was also one gene cluster with 82% similarity to the gene cluster for surfactin synthesis and one gene cluster with 60% similarity to the gene cluster for paenibacterin synthesis. Four other predicted gene clusters were only identified to type with no highly similar known clusters in the antiSMASH database. Two were for terpenes, one was for a type III polyketide synthase, and one was for atRNA-dependent cyclodipeptide synthase.

Table 4.
The putative gene clusters for synthesis of secondary metabolites in B. subtilis YB-15 genome analyzed by antiSMASH.
3.10. Predicted Genes for Plant Growth Promotion
In the B. subtilis YB-15 genome, there were genes possibly involved in plant growth promotion related to nutrient acquisition, phytohormone production, chemotaxis and motility (Table S3). Genes associated with nutrient acquisition were all the genes in the nasABCDEF operon for nitrate/nitrite assimilation, KtrAB and KtrCD for potassium uptake, phoP and phoR in the phoPR operon as well as phoB, phoA and phoD for phosphate assimilation and all the genes in the PstABCS operon for an ABC-type phosphate transport system (Table S3). Genes associated with phytohormone production were all the genes in the trpABCDEF operon as well as yclC for IAA biosynthesis; miaA and miaB for cytokinin biosynthesis; speA and speB for putrescine biosynthesis; speD and speE for spermidine biosynthesis; and alsS, alsD, ilvB, ilvH and bdhA for acetoin and butanediol biosynthesis. For chemotaxis, there were cheA, cheB, cheD, cheR, cheY, and cheW genes related to the two-component sensor kinase, MCP-glutamate methylesterase, protein deaminase, chemotaxis protein methyltransferase, two-component response regulator and CheA modulator chemotaxis, respectively. For motility, there were genes associated with biosynthesis and regulation of flagellum assembly. These were flgD, flgE(flgG), flgK, flgL, hag (fliC), fliD for the hook and filament; fliH, fliP, fliI, fliR, fliQ, flhB and flhA for flagellar protein secretion; flgB, flgC and fliE for the proximal rod; fliF, fliG, fliM and fliN(fliY) for the MS and C ring; flgM, flgN, fliK, fliJ, flit and fliS for other flagellar proteins; and motA and motB for the rotary motor.
4. Discussion
In this study, B. subtilis YB-15, isolated from wheat rhizospheric soil, significantly reduced Fusarium crown rotcaused by F. pseudograminearum with a control efficacy of 81.50%. B. subtilis YB-15 also increased shoot fresh weight by 13.7% and root fresh weight by 70.3% relative to F. pseudograminearum inoculated wheat seedlings. This was similar to the control efficacy of 80.33% for F. pseudograminearum crown and root rot of wheat seeds with B. subtilis strain UTBMS7 inoculation, which also significantly increased wheat stem length and root dry weight by 68 and 64%, respectively, compared to F. pseudograminearum inoculated wheat seedlings []. For F. pseudograminearum crown rot of sorghum, Bacillus velezensis N54 significantly decreased crown rot incidence by 55.6%, which was the most among the rhizobacterial isolates tested. However, it did not improve plant growth as infected plants and strain N54 had significantly lowers hoot and root weight by 14 and 35%, respectively, compared to plants infected with only F. pseudograminearum [].
In addition, B. subtilis YB-15 also promoted plant growth in the absence of infection by F. pseudograminearum. Shoot fresh weight was increased by 4.2% and root fresh weight increased by 11.4% at 20 days in this study, which was less than the increase in wheat with B. subtilis strain UTBMS7, where the strain significantly increased wheat stem and root dry weight by 117 and 107%, respectively, without F. pseudograminearum []. However, neither this study nor that of Sasani and Ahmadzadeh (2021) showed a significant increase in stem height or root length. For other studies of Bacillus species in promoting wheat growth, Chanway et al. (1988) showed a significant increase in root dry weight of 37.8% and shoot height of 2.8% but not shoot dry weight in wheat cv.Katepwa with Bacillus strain 5A1; Akinrinlola et al., (2018) showed a significant increase in shoot height of 36.5% but no significant increase in root and shoot fresh weights with Bacillus megaterium strain R181; Ku et al., (2018) showed a significant increase in shoot fresh weight of 77% and root fresh weight of 177% with Bacillus cereus strain YL6; and Rojas Padilla et al., (2020) showed a significant increase of 27% in shoot dry weight and 30% in root dry weight with B. megaterium strain TRQ8. Thus, there is considerable variation among Bacillus strains for their ability to improve wheat growth, which could be due to differences in the strains as well as in the types of wheat used in each study as Chanway et al., (1988) showed a plant genotypic effect on plant growth promotion with Bacillus isolates being able to enhance root growth of cv. Katepwa but having no effect on root growth of the parental cv. Neepawa.
The biocontrol potential of B. subtilis against plant pathogens could be due to its direct effects on a pathogen, such as by producing antimicrobial compounds and hydrolytic enzymes, or indirect effects on the pathogen by inducing host systemic resistance [,,,]. For antimicrobial compounds, B. subtilis YB-15 has genes to produce the antimicrobial compounds bacillaene, fengycin, bacillibactin, surfactin, subtilosinA, bacilysin and paenibacterin. For biocontrol agents of Fusarium species, there are many examples of them having genes for the production of such compounds, such as B. velezensis LM2303 for the cyclic lipopeptides fengycin and surfactin [], and B. subtilis SEM-2 for fengycin and surfactin as well as the polyene bacillaene, catechol-based siderophore bacillibactin, cyclic peptide subtilosin A, and dipeptide bacilysin []. A gene cluster also with 60% similarity to paenibacterin biosynthesis was found in Paenibacillus polymyxa WLY78, which was a biocontrol agent of Fusarium wilt of cucumber []. Although found in several Paenibacillus species, this appears to be the first report of this type of lipopeptide possibly produced by a Bacillus species. For hydrolytic enzymes, B. subtilis YB-15 was able to secrete β-1, 3-glucanase, amylase, protease and cellulase, which are able to degrade various cell wall components of fungal pathogens []. Furthermore, 122 putative genes encoding CAZymes were found in the B. subtilis YB-15 genome, some of which could act against fungal pathogens []. Bacillus species are well-known inducers of systemic resistance in plants [,], and B. subtilis YB-15 also appears to be able to induce systemic host resistance as indicated by seed inoculation resulting in significant increases in the activities of several defense enzymes in leaves of wheat seedlings, which may contribute to the control of Fusarium crown rot in this study.
For the promotion of plant growth, Bacillus subtilis possesses several mechanisms, such as the production of phytohormones and siderophores, increasing tolerance to abiotic stresses and improved nutrient acquisition [,,,]. Production of IAA by B. subtilis YB-15 could increase cell elongation and production of cytokinin could increase cell division, both of which result in increased size of plant roots and shoots []. Improved plant growth and root development could be due to the production of the polyamines putrescine and spermidine that can act by regulating expansin and ethylene levels, and the production of the related C4 compounds acetoin and butanediol that can act through altering auxin and cytokinin homeostasis []. Production of siderophores by B. subtilis YB-15 can chelate iron thus promoting plant growth by making iron more available []. Other genes in the B. subtilis YB-15 genome that can directly promote growth through enhanced nutrient availability were for nitrate/nitrite assimilation, potassium uptake and phosphate assimilation and transport []. In addition, genes for flagellar motility and chemotaxis in the B. subtilis YB-15 genome indicate that they can likely identify root exudates and migrate to the roots, which is common among effective PGPB and BCA [].
5. Conclusions
In summary, B. subtilis YB-15 is a promising BCA exhibiting significant biocontrol effects against Fusarium crown rot caused by F. pseudograminearum, which could be due to the production of hydrolytic enzymes, antimicrobial compounds and inducing host systemic resistance. It is also a promising PGPB demonstrating growth promotion of wheat seedlings both with and without F. pseudograminearum infection, which could be due to the potential of strain YB-15 to produce phytohormone and siderophores and improve nutrient acquisition for wheat seedlings. While in vitro and in planta tests can reveal common mechanisms employed by BCA and PGPB, genome sequencing can rapidly determine a much wider range of molecular mechanisms underlying BCA and PGPB-mediated biocontrol and growth promotion.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11050778/s1, Table S1: Genome annotation of B. subtilis YB-15; Table S2: CAZymes identified in the B. subtilis YB-15 genome. Table S3: Genes associated with plant growth promotion identified in the strain YB-15 genome.
Author Contributions
Q.W., L.Y. and W.X. conceived the research and designed the experiments. W.X. prepared the manuscript draft. Q.Y., X.X., J.Z., X.D., R.S. and C.W. performed the experiments and analyzed the data. P.H.G., Q.W., M.X. and L.Y. critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Outstanding Youth Science and Technology Fund of Henan Academy of Agricultural Sciences (2022YQ06), the Key Research and Development and Promotion of Special Scientific and Technological Projects in Henan Province (212102110459), the Special Project for Science and Technology Innovation Team of Henan Academy of Agricultural Sciences (2022TD25) and the Central Government Guiding Local Projects (2020 [44]).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The completed assembled genome of strain YB-15 was deposited at GenBank with Accession No. CP092631.
Conflicts of Interest
All the authors declare that there was no potential conflict of commercial or financial interest.
References
- Alomari, D.Z.; Eggert, K.; von Wiren, N.; Alqudah, A.M.; Polley, A.; Plieske, J.; Ganal, M.W.; Pillen, K.; Roder, M.S. Identifying Candidate Genes for Enhancing Grain Zn Concentration in Wheat. Front. Plant Sci. 2018, 9, 1313. [Google Scholar] [CrossRef] [PubMed]
- Moya, E. Fusarium crown rot disease: Biology, interactions, management and function as a possible sensor of global climate change. Cienc. E Investig. Agrar. Rev. Latinoam. Cienc. Agric. 2013, 40, 235–252. [Google Scholar]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolopoulou-Stamati, P.; Maipas, S.; Kotampasi, C.; Stamatis, P.; Hens, L. Chemical Pesticides and Human Health: The Urgent Need for a New Concept in Agriculture. Front. Public Health 2016, 4, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardar, M.F.; Abbas, T.; Naveed, M.; Siddique, S.; Mustafa, A.; Abbasi, B.; Khan, K. Biopesticides: Importance and Challenges; Apple Academic Press: Palm Bay, FL, USA, 2021; pp. 129–151. [Google Scholar]
- Dehghanpour-Farashah, S.; Taheri, P.; Falahati-Rastegar, M. Effect of polyamines and nitric oxide in Piriformospora indica-induced resistance and basal immunity of wheat against Fusarium pseudograminearum. Biol. Control. 2019, 136, 104006. [Google Scholar] [CrossRef]
- Kim, Y.T.; Monkhung, S.; Lee, Y.S.; Kim, K.Y. Effects of Lysobacter antibioticus HS124, an effective biocontrol agent against Fusarium graminearum, on crown rot disease and growth promotion of wheat. Can. J. Microbiol. 2019, 65, 904–912. [Google Scholar] [CrossRef]
- Winter, M.; Samuels, P.L.; Otto-Hanson, L.K.; Dill-Macky, R.; Kinkel, L.L. Biological control of Fusarium crown and root rot of wheat by Streptomyces isolates—It’s complicated. Phytobiomes J. 2019, 3, 52–60. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, C.A.; Roper, M.M.; Myers, C.A.; Thatcher, L.F. Developing Actinobacterial Endophytes as Biocontrol Products for Fusarium pseudograminearum in Wheat. Front. Bioeng. Biotechnol. 2021, 9, 691770. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, R.C.; Maheshwari, D.K. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol. Res. 2012, 167, 493–499. [Google Scholar] [CrossRef]
- Miljakovic, D.; Marinkovic, J.; Balesevic-Tubic, S. The Significance of Bacillus spp. in Disease Suppression and Growth Promotion of Field and Vegetable Crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef]
- Blake, C.; Christensen, M.N.; Kovacs, A.T. Molecular Aspects of Plant Growth Promotion and Protection by Bacillus subtilis. Mol. Plant Microbe Interact. 2021, 34, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Naveed, M.; Mustafa, A.; Abbas, A. The Good, the Bad, and the Ugly of Rhizosphere Microbiome. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 253–290. [Google Scholar]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, Identification, and Complete Genome Assembly of an Endophytic Bacillus velezensis YB-130, Potential Biocontrol Agent Against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef] [PubMed]
- Saeed, Q.; Xiukang, W.; Haider, F.U.; Kučerik, J.; Mumtaz, M.Z.; Holatko, J.; Naseem, M.; Kintl, A.; Ejaz, M.; Naveed, M.; et al. Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated Sites: A Comprehensive Review of Effects and Mechanisms. Int. J. Mol. Sci. 2021, 22, 10529. [Google Scholar] [CrossRef] [PubMed]
- Chanway, C.P.; Nelson, L.M.; Holl, F.B. Cultivar-specific growth promotion of spring wheat (Triticum aestivum L.) by coexistent Bacillus species. Can. J. Microbiol. 1988, 34, 925–929. [Google Scholar] [CrossRef]
- Ku, Y.; Xu, G.; Tian, X.; Xie, H.; Yang, X.; Cao, C.; Chen, Y. Root colonization and growth promotion of soybean, wheat and Chinese cabbage by Bacillus cereus YL6. PLoS ONE 2018, 13, e0200181. [Google Scholar] [CrossRef] [Green Version]
- Rojas Padilla, J.; Chaparro Encinas, L.A.; Robles Montoya, R.I.; de los Santos Villalobos, S. Growth promotion on wheat (Triticum turgidum L. subsp. durum) by co-inoculation of native Bacillus strains isolated from the Yaqui Valley, Mexico. Nova Scientia 2020, 12, 1–27. [Google Scholar] [CrossRef]
- Sasani, m.; Ahmadzadeh, M. Evaluation of antagonistic effect of several strain of Bacillus bacteria on control of crown and root rot of wheat with Fusarium pseudograminearum. Biol. Control Pests Plant Dis. 2021, 9, 115–126. [Google Scholar] [CrossRef]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Hassen, A.I.; Labuschagne, N. Rhizobacteria-induced systemic resilience in Sorghum bicolor (L.) moench against Fusarium pseudograminearum crown rot under drought stress conditions. Biol. Control 2020, 151, 104395. [Google Scholar] [CrossRef]
- Wu, J.J.; Chou, H.P.; Huang, J.W.; Deng, W.L. Genomic and biochemical characterization of antifungal compounds produced by Bacillus subtilis PMB102 against Alternaria brassicicola. Microbiol. Res. 2021, 251, 126815. [Google Scholar] [CrossRef]
- Kiesewalter, H.T.; Lozano-Andrade, C.N.; Wibowo, M.; Strube, M.L.; Maroti, G.; Snyder, D.; Jorgensen, T.S.; Larsen, T.O.; Cooper, V.S.; Weber, T.; et al. Genomic and Chemical Diversity of Bacillus subtilis Secondary Metabolites against Plant Pathogenic Fungi. mSystems 2021, 6, e00770-20. [Google Scholar] [CrossRef]
- Franco-Sierra, N.D.; Posada, L.F.; Santa-Maria, G.; Romero-Tabarez, M.; Villegas-Escobar, V.; Alvarez, J.C. Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genom. 2020, 20, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Quan, X.; Xue, B.; Goodwin, P.H.; Lu, S.; Wang, J.; Du, W.; Wu, C. Isolation and identification of Bacillus subtilis strain YB-05 and its antifungal substances showing antagonism against Gaeumannomyces graminis var. tritici. Biol. Control 2015, 85, 52–58. [Google Scholar] [CrossRef]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, S. Identification and Characterization of the Phosphate-Solubilizing Bacterium Pantoea sp. S32 in Reclamation Soil in Shanxi, China. Front. Microbiol. 2019, 10, 2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef] [Green Version]
- Al-Naamani, L.S.H.; Dobretsov, S.; Al-Sabahi, J.; Soussi, B. Identification and characterization of two amylase producing bacteria Cellulosimicrobium sp. and Demequina sp. isolated from marine organisms. J. Agric. Mar. Sci. [JAMS] 2015, 20, 8–15. [Google Scholar]
- Kazanas, N. Proteolytic activity of microorganisms isolated from freshwater fish. Appl. Microbiol. 1968, 16, 128–132. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.M.; Li, Y.H.; Ding, S.L.; Yuan, H.X.; Riley, I.T.; Li, H.L. Biocontrol of cereal cyst nematode by Streptomyces anulatus isolate S07. Australas. Plant Pathol. 2016, 45, 57–64. [Google Scholar] [CrossRef]
- Xu, W.; Zhao, F.; Deng, X.; Goodwin, P.; Xia, M.; Zhang, J.; Sun, R.; Liang, J.; Wu, C.; Yang, L. First Report of Collar Canker and Dieback of Camellia sinensis caused by Fusarium solani Species Complex in Henan, China. Plant Dis. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Xu, W.; Xu, L.; Deng, X.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Sun, R.; Pan, Y.; Wu, C.; et al. Biological Control of Take-All and Growth Promotion in Wheat by Pseudomonas chlororaphis YB-10. Pathogens 2021, 10, 903. [Google Scholar] [CrossRef]
- Ullah, H.; Yasmin, H.; Mumtaz, S.; Jabeen, Z.; Naz, R.; Nosheen, A.; Hassan, M.N. Multitrait Pseudomonas spp. Isolated from Monocropped Wheat (Triticum aestivum) Suppress Fusarium Root and Crown Rot. Phytopathology 2020, 110, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2004, 21, 537–539. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Ali, S.; Shahid, M.A.; Mustafa, A.; Sayyed, R.Z.; Curá, J.A. Insights into the Interactions among Roots, Rhizosphere, and Rhizobacteria for Improving Plant Growth and Tolerance to Abiotic Stresses: A Review. Cells 2021, 10, 1551. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, V. Genome wide underpinning of antagonistic and plant beneficial attributes of Bacillus sp. SBA12. Genomics 2020, 112, 2894–2902. [Google Scholar] [CrossRef]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Liao, S.; Wei, J.; Xing, D.; Xiao, Y.; Yang, Q. Isolation of Bacillus subtilis strain SEM-2 from silkworm excrement and characterisation of its antagonistic effect against Fusarium spp. Can. J. Microbiol. 2020, 66, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, S. Fusaricidin Produced by Paenibacillus polymyxa WLY78 Induces Systemic Resistance against Fusarium Wilt of Cucumber. Int. J. Mol. Sci. 2019, 20, 5240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadhav, H.; Sayyed, R. Hydrolytic enzymes of rhizospheric microbes in crop protection. MOJ Cell Sci. Rep. 2016, 3, 135–136. [Google Scholar]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants—With special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W.; Ryu, C.M.; Zhang, S. Induced Systemic Resistance and Promotion of Plant Growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Scavino, A.F.; Pedraza, R.O. The role of siderophores in plant growth-promoting bacteria. In Bacteria in Agrobiology: Crop Productivity; Springer: New York, NY, USA, 2013; pp. 265–285. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).