De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
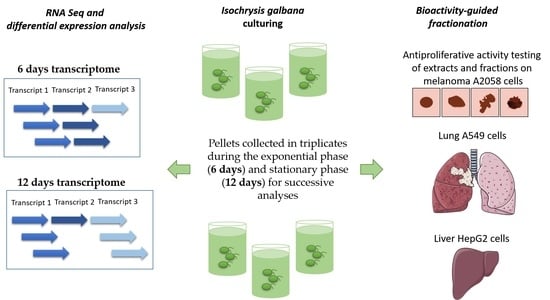
2.1. Cell Culturing and Harvesting
2.2. RNA Extraction
2.3. RNA Sequencing, Transcriptome Assembly, Annotation, Expression Quantification and Differential Expression Analysis
2.4. Chemical Extraction and Pre-Fractionation
2.5. In Vitro Antiproliferative Assay
3. Results
3.1. Transcriptome Sequencing and De Novo Assembly
3.2. Functional Annotation
3.3. Differential Expression Analysis
3.4. Bioactivity and Chemical Fractionation of I. galbana Extract
3.5. Identification of Differentially Expressed Transcripts Related to Nucleoside, Glycolipid/Phospholipid, Sterol and Triglyceride Metabolism
3.6. Differentially Expressed Genes Related to the Synthesis of Secondary Metabolites with Known Anticancer Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaspars, M.; De Pascale, D.; Andersen, J.H.; Reyes, F.; Crawford, A.D.; Ianora, A. The Marine Biodiscovery Pipeline and Ocean Medicines of Tomorrow. J. Mar. Biol. Assoc. UK 2016, 96, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Mimouni, V.; Ulmann, L.; Pasquet, V.; Mathieu, M.; Picot, L.; Bougaran, G.; Cadoret, J.P.; Morant-Manceau, A.; Schoefs, B. The Potential of Microalgae for the Production of Bioactive Molecules of Pharmaceutical Interest. Curr. Pharm. Biotechnol. 2012, 13, 2733–2750. [Google Scholar] [CrossRef] [PubMed]
- Koyande, A.K.; Chew, K.W.; Rambabu, K.; Tao, Y.; Chu, D.-T.; Show, P.-L. Microalgae: A Potential Alternative to Health Supplementation for Humans. Food Sci. Hum. Wellness 2019, 8, 16–24. [Google Scholar] [CrossRef]
- Lauritano, C.; Helland, K.; Riccio, G.; Andersen, J.H.; Ianora, A.; Hansen, E.H. Lysophosphatidylcholines and Chlorophyll-Derived Molecules from the Diatom Cylindrotheca Closterium with Anti-Inflammatory Activity. Mar. Drugs 2020, 18, 166. [Google Scholar] [CrossRef] [Green Version]
- Saide, A.; Martínez, K.A.; Ianora, A.; Lauritano, C. Unlocking the Health Potential of Microalgae as Sustainable Sources of Bioactive Compounds. Int. J. Mol. Sci. 2021, 22, 4383. [Google Scholar] [CrossRef]
- Riccio, G.; Lauritano, C. Microalgae with Immunomodulatory Activities. Mar. Drugs 2020, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Ugwu, C.U.; Aoyagi, H.; Uchiyama, H. Photobioreactors for Mass Cultivation of Algae. Bioresour. Technol. 2008, 99, 4021–4028. [Google Scholar] [CrossRef]
- Not, F.; Siano, R.; Kooistra, W.H.C.F.; Simon, N.; Vaulot, D.; Probert, I. Diversity and Ecology of Eukaryotic Marine Phytoplankton. Adv. Bot. Res. 2012, 64, 1–53. [Google Scholar] [CrossRef]
- Eikrem, W.; Medlin, L.K.; Henderiks, J.; Rokitta, S.; Rost, B.; Probert, I.; Throndsen, J.; Edvardsen, B. Haptophyta. In Handbook of the Protists, 2nd ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 893–953. ISBN 9783319281490. [Google Scholar]
- Eltanahy, E.; Torky, A. CHAPTER 1. Microalgae as Cell Factories: Food and Feed-Grade High-Value Metabolites. In Microalgal Biotechnology; Royal Society of Chemistry: London, UK, 2021; pp. 1–35. [Google Scholar]
- Parke, M. Studies on Marine Flagellates. J. Mar. Biol. Assoc. UK 1949, 28, 255–288. [Google Scholar] [CrossRef] [Green Version]
- Gouveia, L.; Coutinho, C.; Mendonca, E.; Batista, A.P.; Sousa, I.; Bandarra, N.M.; Raymundo, A. Functional Biscuits with PUFA-Omega 3 from Isochrysis Galbana. J. Sci. Food Agric. 2008, 88, 891–896. [Google Scholar] [CrossRef] [Green Version]
- Fradique, M.; Batista, A.P.; Nunes, M.C.; Gouveia, L.; Bandarra, N.M.; Raymundo, A. Isochrysis galbana and Diacronema vlkianum Biomass Incorporation in Pasta Products as PUFA’s Source. LWT—Food Sci. Technol. 2013, 50, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Matos, J.; Afonso, C.; Cardoso, C.; Serralheiro, M.L.; Bandarra, N.M. Yogurt Enriched with Isochrysis galbana: An Innovative Functional Food. Foods 2021, 10, 1458. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, H.; Guo, G.; Pu, Y.; Yan, B. The Isolation and Antioxidant Activity of Polysaccharides from the Marine Microalgae Isochrysis galbana. Carbohydr. Polym. 2014, 113, 22–31. [Google Scholar] [CrossRef]
- Sukenik, A.; Wahnon, R. Biochemical Quality of Marine Unicellular Algae with Special Emphasis on Lipid Composition. I. Isochrysis galbana. Aquaculture 1991, 97, 61–72. [Google Scholar] [CrossRef]
- De Los Reyes, C.; Ortega, M.J.; Rodríguez-Luna, A.; Talero, E.; Motilva, V.; Zubía, E. Molecular Characterization and Anti-Inflammatory Activity of Galactosylglycerides and Galactosylceramides from the Microalga Isochrysis galbana. J. Agric. Food Chem. 2016, 64, 8783–8794. [Google Scholar] [CrossRef]
- Li, M.; Ou, X.; Yang, X.; Guo, D.; Qian, X.; Xing, L.; Li, M. Isolation of a Novel C18-Δ9 Polyunsaturated Fatty Acid Specific Elongase Gene from DHA-Producing Isochrysis galbana H29 and Its Use for the Reconstitution of the Alternative Δ8 Pathway in Saccharomyces cerevisiae. Biotechnol. Lett. 2011, 33, 1823–1830. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, L.; Liu, J. Proteomic Analysis in Nitrogen-Deprived Isochrysis galbana during Lipid Accumulation. PLoS ONE 2013, 8, e82188. [Google Scholar] [CrossRef] [Green Version]
- Bustamam, M.S.A.; Pantami, H.A.; Azizan, A.; Shaari, K.; Min, C.C.; Abas, F.; Nagao, N.; Maulidiani, M.; Banerjee, S.; Sulaiman, F.; et al. Complementary Analytical Platforms of NMR Spectroscopy and LCMS Analysis in the Metabolite Profiling of Isochrysis galbana. Mar. Drugs 2021, 19, 139. [Google Scholar] [CrossRef]
- Di Lena, G.; Casini, I.; Lucarini, M.; Lombardi-Boccia, G. Carotenoid Profiling of Five Microalgae Species from Large-Scale Production. Food Res. Int. 2019, 120, 810–818. [Google Scholar] [CrossRef]
- Sadovskaya, I.; Souissi, A.; Souissi, S.; Grard, T.; Lencel, P.; Greene, C.M.; Duin, S.; Dmitrenok, P.S.; Chizhov, A.O.; Shashkov, A.S.; et al. Chemical Structure and Biological Activity of a Highly Branched (1 → 3,1 → 6)-β-D-Glucan from Isochrysis galbana. Carbohydr. Polym. 2014, 111, 139–148. [Google Scholar] [CrossRef]
- Prakash, S.; Sasikala, S.L.; Aldous, V.H.J. Isolation and Identification of MDR-Mycobacterium tuberculosis and Screening of Partially Characterised Antimycobacterial Compounds from Chosen Marine Micro Algae. Asian Pac. J. Trop. Med. 2010, 3, 655–661. [Google Scholar] [CrossRef] [Green Version]
- Nuño, K.; Villarruel-López, A.; Puebla-Pérez, A.M.; Romero-Velarde, E.; Puebla-Mora, A.G.; Ascencio, F. Effects of the Marine Microalgae Isochrysis galbana and Nannochloropsis oculata in Diabetic Rats. J. Funct. Foods 2013, 5, 106–115. [Google Scholar] [CrossRef]
- Gerecht, A.; Romano, G.; Ianora, A.; d’Ippolito, G.; Cutignano, A.; Fontana, A. Plasticity of Oxylipin Metabolism among Clones of the Marine Diatom Skeletonema marinoi (Bacillariophyceae). J. Phycol. 2011, 47, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Vidoudez, C.; Pohnert, G. Comparative Metabolomics of the Diatom Skeletonema marinoi in Different Growth Phases. Metabolomics 2012, 8, 654–669. [Google Scholar] [CrossRef]
- Ingebrigtsen, R.A.; Hansen, E.; Andersen, J.H.; Eilertsen, H.C. Light and Temperature Effects on Bioactivity in Diatoms. J. Appl. Phycol. 2016, 28, 939–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619. [Google Scholar] [CrossRef]
- Lauritano, C.; Martín, J.; De La Cruz, M.; Reyes, F.; Romano, G.; Ianora, A. First Identification of Marine Diatoms with Anti-Tuberculosis Activity. Sci. Rep. 2018, 8, 2284. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, V.; Uemura, Y.; Thanh, N.T.; Khalid, N.A.; Osman, N.; Mansor, N. Three Types of Marine Microalgae and Nannocholoropsis oculata Cultivation for Potential Source of Biomass Production. In Proceedings of the 3rd International Conference on Science & Engineering in Mathematics, Chemistry and Physics 2015 (ScieTech 2015), Bali, Indonesia, 31 January–1 February 2015; Journal of Physics: Conference Series; Institute of Physics Publishing: Bristol, UK, 2015; Volume 622, p. 012034. [Google Scholar]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and Application of a Novel SPE-Method for Bioassay-Guided Fractionation of Marine Extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef] [Green Version]
- Ribalet, F.; Wichard, T.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. Age and Nutrient Limitation Enhance Polyunsaturated Aldehyde Production in Marine Diatoms. Phytochemistry 2007, 68, 2059–2067. [Google Scholar] [CrossRef]
- Orefice, I.; Lauritano, C.; Procaccini, G.; Ianora, A.; Romano, G. Insights into Possible Cell-Death Markers in the Diatom Skeletonema marinoi in Response to Senescence and Silica Starvation. Mar. Genom. 2015, 24, 81–88. [Google Scholar] [CrossRef]
- Lauritano, C.; Orefice, I.; Procaccini, G.; Romano, G.; Ianora, A. Key Genes as Stress Indicators in the Ubiquitous Diatom Skeletonema marinoi. BMC Genom. 2015, 16, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elagoz, A.M.; Ambrosino, L.; Lauritano, C. De Novo Transcriptome of the Diatom Cylindrotheca closterium Identifies Genes Involved in the Metabolism of Anti-Inflammatory Compounds. Sci. Rep. 2020, 10, 4138. [Google Scholar] [CrossRef] [PubMed]
- BBTools User Guide—DOE Joint Genome Institute. Available online: https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/ (accessed on 28 March 2022).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A Web Server for Clustering and Comparing Biological Sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Haas, B. TransDecoder Release. 2018. Available online: https://github.com/TransDecoder/TransDecoder/releases (accessed on 28 March 2022).
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A Rapid Functional Annotation Web Server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Rau, A.; Gallopin, M.; Celeux, G.; Jaffrézic, F. Data-Based Filtering for Replicated High-Throughput Transcriptome Sequencing Experiments. Bioinformatics 2013, 29, 2146–2152. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential Expression in RNA-Seq: A Matter of Depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data Quality Aware Analysis of Differential Expression in RNA-Seq with NOISeq R/Bioc Package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Martínez, K.A.; Saide, A.; Crespo, G.; Martín, J.; Romano, G.; Reyes, F.; Lauritano, C.; Ianora, A. Promising Antiproliferative Compound From the Green Microalga Dunaliella tertiolecta Against Human Cancer Cells. Front. Mar. Sci. 2022, 9, 157. [Google Scholar] [CrossRef]
- Riccio, G.; Nuzzo, G.; Zazo, G.; Coppola, D.; Senese, G.; Romano, L.; Costantini, M.; Ruocco, N.; Bertolino, M.; Fontana, A.; et al. Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols. Mar. Drugs 2021, 19, 459. [Google Scholar] [CrossRef] [PubMed]
- Elledge, S.J.; Zhou, Z.; Allen, J.B. Ribonucleotide Reductase: Regulation, Regulation, Regulation. Trends Biochem. Sci. 1992, 17, 119–123. [Google Scholar] [CrossRef]
- Yen, C.L.E.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V. DGAT Enzymes and Triacylglycerol Biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [Green Version]
- Tamanna Ferdous, U.; Norhana Balia Yusof, Z. Algal Terpenoids: A Potential Source of Antioxidants for Cancer Therapy. In Terpenes and Terpenoids; IntechOpen: London, UK, 2021. [Google Scholar]
- Ávila-Román, J.; García-Gil, S.; Rodríguez-Luna, A.; Motilva, V.; Talero, E. Anti-Inflammatory and Anticancer Effects of Microalgal Carotenoids. Mar. Drugs 2021, 19, 531. [Google Scholar] [CrossRef]
- Jara-Gutiérrez, Á.; Baladrón, V. The Role of Prostaglandins in Different Types of Cancer. Cells 2021, 10, 1487. [Google Scholar] [CrossRef]
- Matos, J.; Cardoso, C.; Gomes, A.; Campos, A.M.; Falé, P.; Afonso, C.; Bandarra, N.M. Bioprospection of Isochrysis galbana and Its Potential as a Nutraceutical. Food Funct. 2019, 10, 7333–7342. [Google Scholar] [CrossRef]
- Chi-Cheng, Y.; Hsiao-Wei, C.; Mao-Jing, C.; Yu-Ching, C.; Shih-Chang, C.; Yueh-Hsiung, K.; Feng-Ling, Y.; Shih-Hsiung, W.; Jie, C.; Hsiao-Hui, Y.; et al. Chemical Composition and Bioactivities of the Marine Alga Isochrysis galbana from Taiwan—PubMed. Nat. Prod. Commun. 2010, 5, 1941–1944. [Google Scholar]
- Custódio, L.; Soares, F.; Pereira, H.; Barreira, L.; Vizetto-Duarte, C.; Rodrigues, M.J.; Rauter, A.P.; Alberício, F.; Varela, J. Fatty Acid Composition and Biological Activities of Isochrysis galbana T-ISO, Tetraselmis sp. and Scenedesmus sp.: Possible Application in the Pharmaceutical and Functional Food Industries. J. Appl. Phycol. 2014, 26, 151–161. [Google Scholar] [CrossRef]
- Bonfanti, C.; Cardoso, C.; Afonso, C.; Matos, J.; Garcia, T.; Tanni, S.; Bandarra, N.M. Potential of Microalga Isochrysis galbana: Bioactivity and Bioaccessibility. Algal Res. 2018, 29, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Romero, S.; Torrella, J.R.; Pagès, T.; Viscor, G.; Torres, J.L. Edible Microalgae and Their Bioactive Compounds in the Prevention and Treatment of Metabolic Alterations. Nutrients 2021, 13, 563. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Guerra, I.; Costa, M.; Silva, J.; Gouveia, L. Future Perspectives of Microalgae in the Food Industry. In Cultured Microalgae for the Food Industry; Lafarga, T., Acién, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 387–433. [Google Scholar]
- Franzolin, E.; Pontarin, G.; Rampazzo, C.; Miazzi, C.; Ferraro, P.; Palumbo, E.; Reichard, P.; Bianchi, V. The Deoxynucleotide Triphosphohydrolase SAMHD1 Is a Major Regulator of DNA Precursor Pools in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 14272–14277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meldrum, E.; Parker, P.J.; Carozzi, A. The PtdIns-PLC Superfamily and Signal Transduction. BBA—Mol. Cell Res. 1991, 1092, 49–71. [Google Scholar] [CrossRef]
- Quazi, F.; Molday, R.S. Differential Phospholipid Substrates and Directional Transport by ATP-Binding Cassette Proteins ABCA1, ABCA7, and ABCA4 and Disease-Causing Mutants. J. Biol. Chem. 2013, 288, 34414–34426. [Google Scholar] [CrossRef] [Green Version]

| GO Class | GO Description | FDR | Enrichment Score |
|---|---|---|---|
| BP | cellular protein complex disassembly | 3.78 × 10−9 | 3.54 |
| BP | sterol biosynthetic process | 3.03 × 10−9 | 3.47 |
| BP | regulation of protein localization | 1.42 × 10−7 | 3.30 |
| BP | protein localization to membrane | 3.28 × 10−9 | 3.21 |
| BP | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly | 1.87 × 10−4 | 3.10 |
| BP | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 2.73 × 10−5 | 3.07 |
| BP | negative regulation of DNA recombination | 1.30 × 10−4 | 3.03 |
| BP | acetyl-CoA metabolic process | 1.31 × 10−3 | 2.79 |
| BP | organic cyclic compound biosynthetic process | 8.64 × 10−4 | 2.75 |
| BP | carbohydrate derivative biosynthetic process | 5.61 × 10−4 | 2.72 |
| CC | recycling endosome membrane | 3.12 × 10−6 | 4.40 |
| CC | proton-transporting V-type ATPase complex | 5.60 × 10−6 | 4.05 |
| CC | AP-2 adaptor complex | 4.56 × 10−4 | 3.72 |
| CC | WASH complex | 1.81 × 10−3 | 3.54 |
| CC | eukaryotic translation initiation factor 2 complex | 1.81 × 10−3 | 3.54 |
| CC | transporter complex | 4.27 × 10−4 | 3.47 |
| CC | clathrin coat of coated pit | 1.99 × 10−5 | 3.41 |
| CC | endocytic vesicle | 1.54 × 10−5 | 3.30 |
| CC | clathrin coat of trans-Golgi network vesicle | 7.34 × 10−5 | 3.30 |
| CC | proton-transporting ATP synthase complex, catalytic core F(1) | 7.34 × 10−5 | 3.30 |
| MF | protein tag | 5.60 × 10−6 | 4.05 |
| MF | ADP-ribose diphosphatase activity | 1.23 × 10−9 | 4.01 |
| MF | chorismate mutase activity | 2.69 × 10−5 | 3.72 |
| MF | structural constituent of cell wall | 5.33 × 10−6 | 3.50 |
| MF | clathrin adaptor activity | 9.37 × 10−5 | 3.43 |
| MF | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | 1.92 × 10−12 | 3.27 |
| MF | beta-tubulin binding | 1.24 × 10−3 | 3.15 |
| MF | ATPase activator activity | 3.98 × 10−6 | 3.05 |
| MF | clathrin light chain binding | 6.16 × 10−4 | 2.97 |
| MF | ubiquitin conjugating enzyme activity | 6.16 × 10−4 | 2.97 |
| GO Class | GO Description | FDR | Enrichment Score |
|---|---|---|---|
| BP | photosystem II stabilization | 3.09 × 10−6 | 3.02 |
| BP | NADH metabolic process | 3.35 × 10−5 | 2.97 |
| BP | response to high light intensity | 1.10 × 10−4 | 2.93 |
| BP | protein localization to nucleolar rDNA repeats | 1.10 × 10−4 | 2.93 |
| BP | 7-methylguanosine mRNA capping | 1.10 × 10−4 | 2.93 |
| BP | lipid A biosynthetic process | 4.08 × 10−4 | 2.53 |
| BP | photosynthesis, light harvesting | 0.00 × 10 | 2.41 |
| BP | heterochromatin organization | 3.09 × 10−3 | 2.40 |
| BP | glycerophospholipid metabolic process | 8.20 × 10−3 | 2.31 |
| BP | cellular ketone metabolic process | 8.20 × 10−3 | 2.31 |
| CC | myelin sheath | 0.00 × 10 | 3.30 |
| CC | CORVET complex | 0.00 × 10 | 3.30 |
| CC | TAT protein transport complex | 0.00 × 10 | 3.30 |
| CC | phosphatidylinositol 3-kinase complex, class III | 0.00 × 10 | 3.30 |
| CC | ribonuclease MRP complex | 0.00 × 10 | 3.30 |
| CC | mRNA cap binding complex | 0.00 × 10 | 3.30 |
| CC | contractile actin filament bundle | 0.00 × 10 | 3.30 |
| CC | Chromocenter | 0.00 × 10 | 3.30 |
| CC | glutathione synthase complex | 0.00 × 10 | 3.30 |
| CC | sarcoplasmic reticulum | 0.00 × 10 | 3.30 |
| MF | D-cysteine desulfhydrase activity | 0.00 × 10 | 3.30 |
| MF | sodium channel regulator activity | 0.00 × 10 | 3.30 |
| MF | phosphodiesterase I activity | 0.00 × 10 | 3.30 |
| MF | isocitrate dehydrogenase (NADP+) activity | 1.17 × 10−3 | 2.82 |
| MF | glutathione-disulfide reductase activity | 1.17 × 10−3 | 2.82 |
| MF | mRNA guanylyltransferase activity | 3.77 × 10−3 | 2.75 |
| MF | palmitoyl-(protein) hydrolase activity | 1.17 × 10−2 | 2.64 |
| MF | alpha-L-arabinofuranosidase activity | 1.17 × 10−2 | 2.64 |
| MF | pyruvate carboxylase activity | 1.17 × 10−2 | 2.64 |
| MF | plastoquinol–plastocyanin reductase activity | 1.17 × 10−2 | 2.64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riccio, G.; Martinez, K.A.; Ianora, A.; Lauritano, C. De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites. Biology 2022, 11, 771. https://doi.org/10.3390/biology11050771
Riccio G, Martinez KA, Ianora A, Lauritano C. De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites. Biology. 2022; 11(5):771. https://doi.org/10.3390/biology11050771
Chicago/Turabian StyleRiccio, Gennaro, Kevin A. Martinez, Adrianna Ianora, and Chiara Lauritano. 2022. "De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites" Biology 11, no. 5: 771. https://doi.org/10.3390/biology11050771
APA StyleRiccio, G., Martinez, K. A., Ianora, A., & Lauritano, C. (2022). De Novo Transcriptome of the Flagellate Isochrysis galbana Identifies Genes Involved in the Metabolism of Antiproliferative Metabolites. Biology, 11(5), 771. https://doi.org/10.3390/biology11050771