Isolation and Characterization a Novel Catabolic Gene Cluster Involved in Chlorobenzene Degradation in Haloalkaliphilic Alcanivorax sp. HA03
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Conditions and Preparation of Cell Extracts from Alcanivorax sp. HA03
2.2. PCR Amplification of Chlorobenzene Dioxygenase Encoding Genes from HA03
2.3. Extraction of mRNA and cDNA Synthesis and RT-PCR
2.4. Amplification and Characterization of Chlorocatechol 1,2 Dioxygenase
2.5. Cloning and Transformation of Chlorobenzene Dioxygenase from HA03
2.6. Nucleotide Sequence Accession Numbers
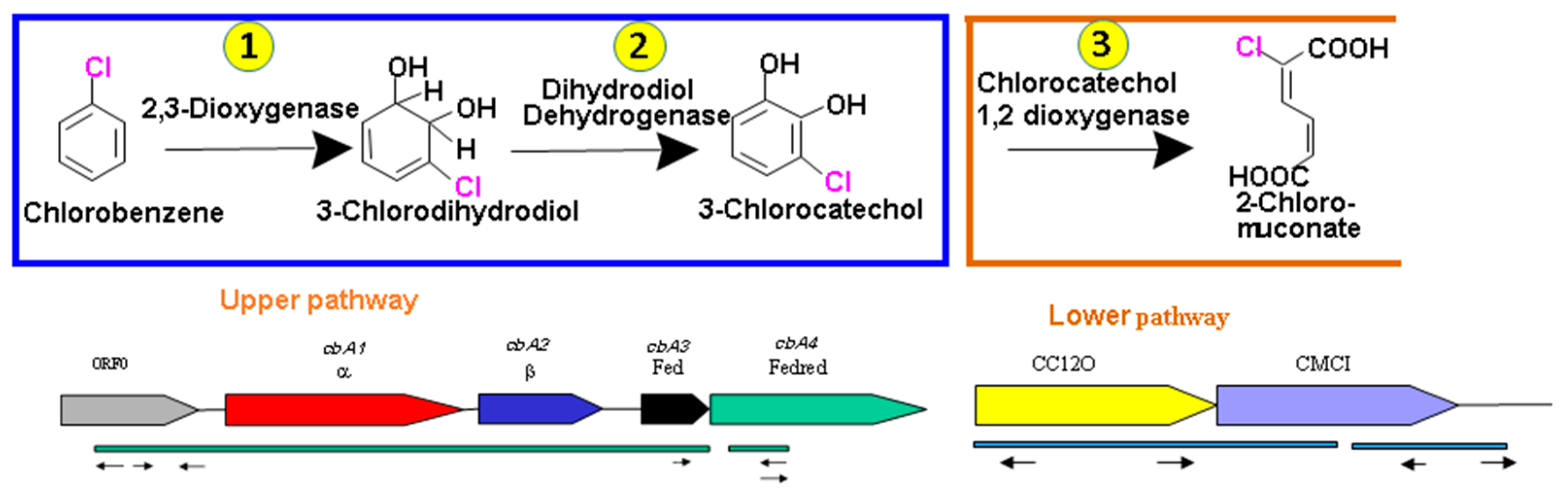
3. Results
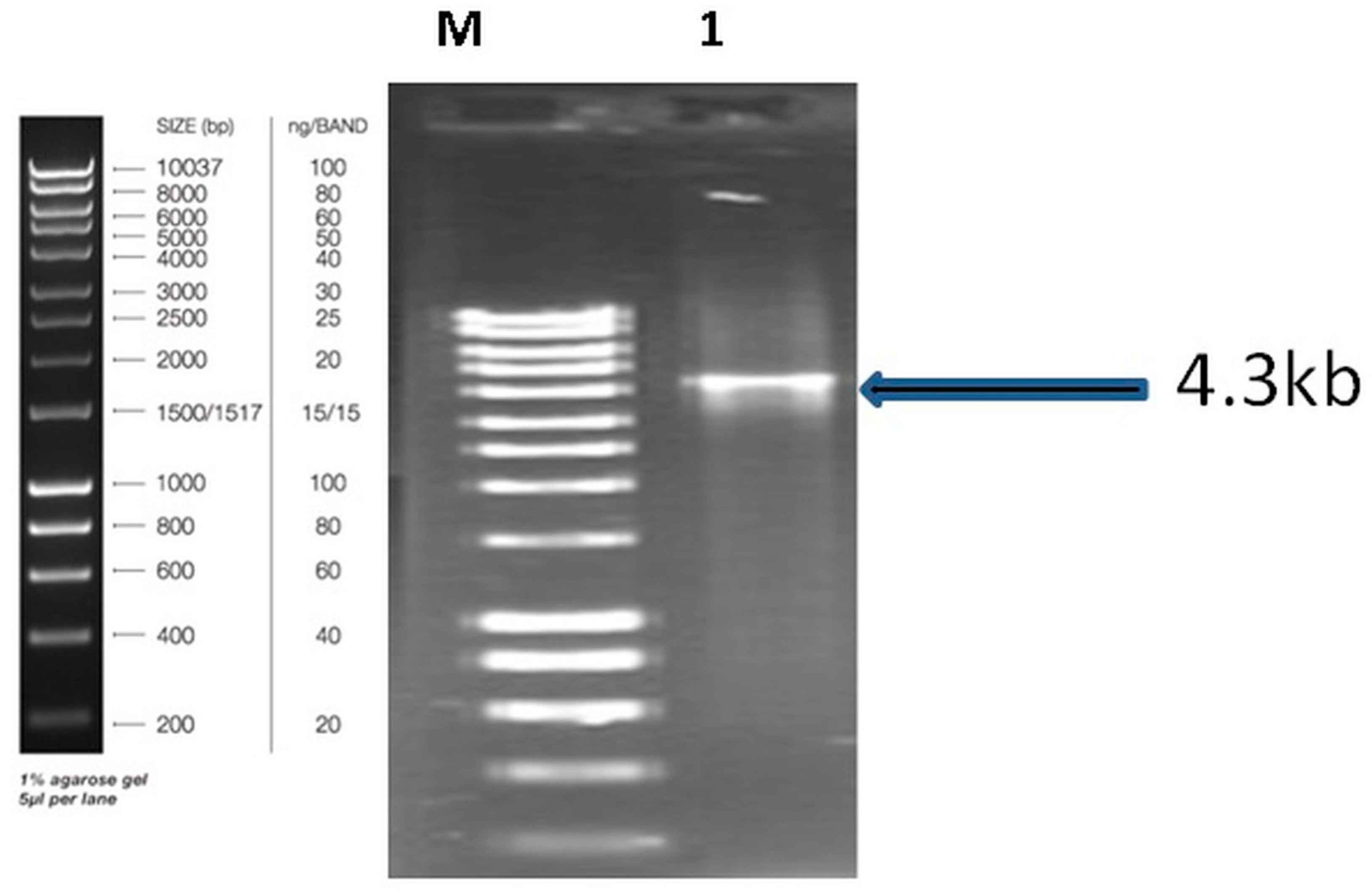
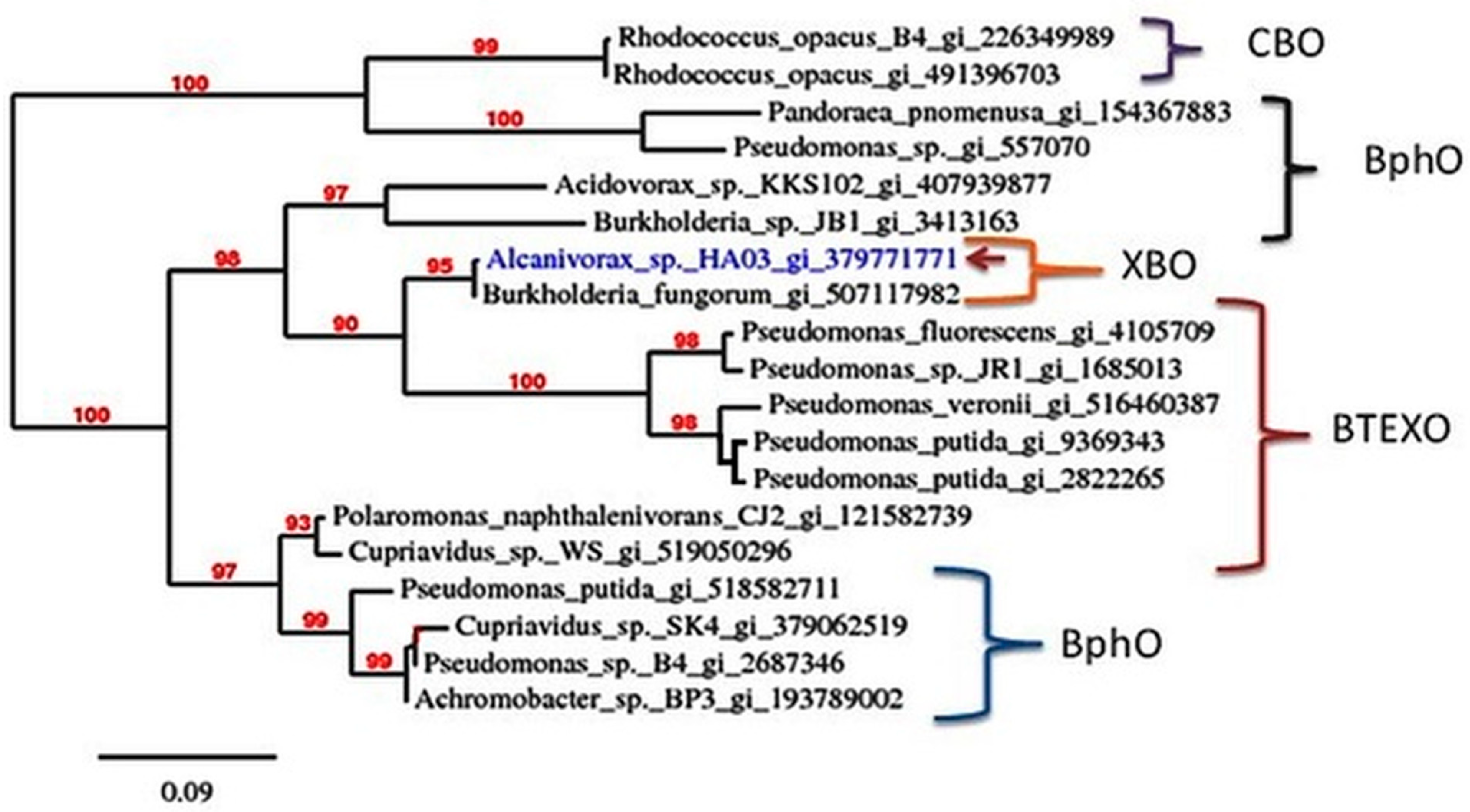
3.1. Amplification and Characterization the Initial Dioxygenase in Alcanivorax sp. HA03
3.2. Heterologous Expression of Chlorobenzene Dioxygenase Genes
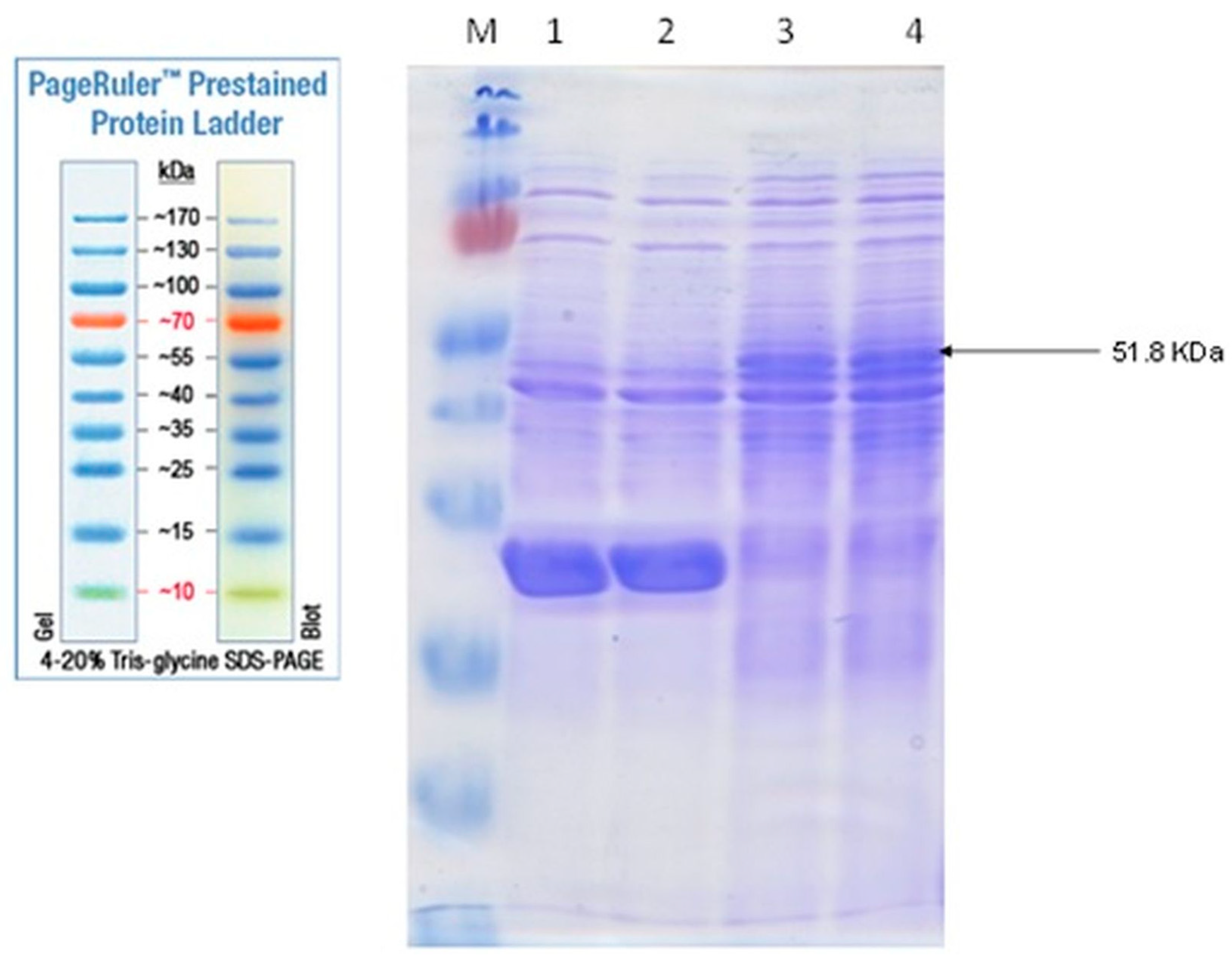
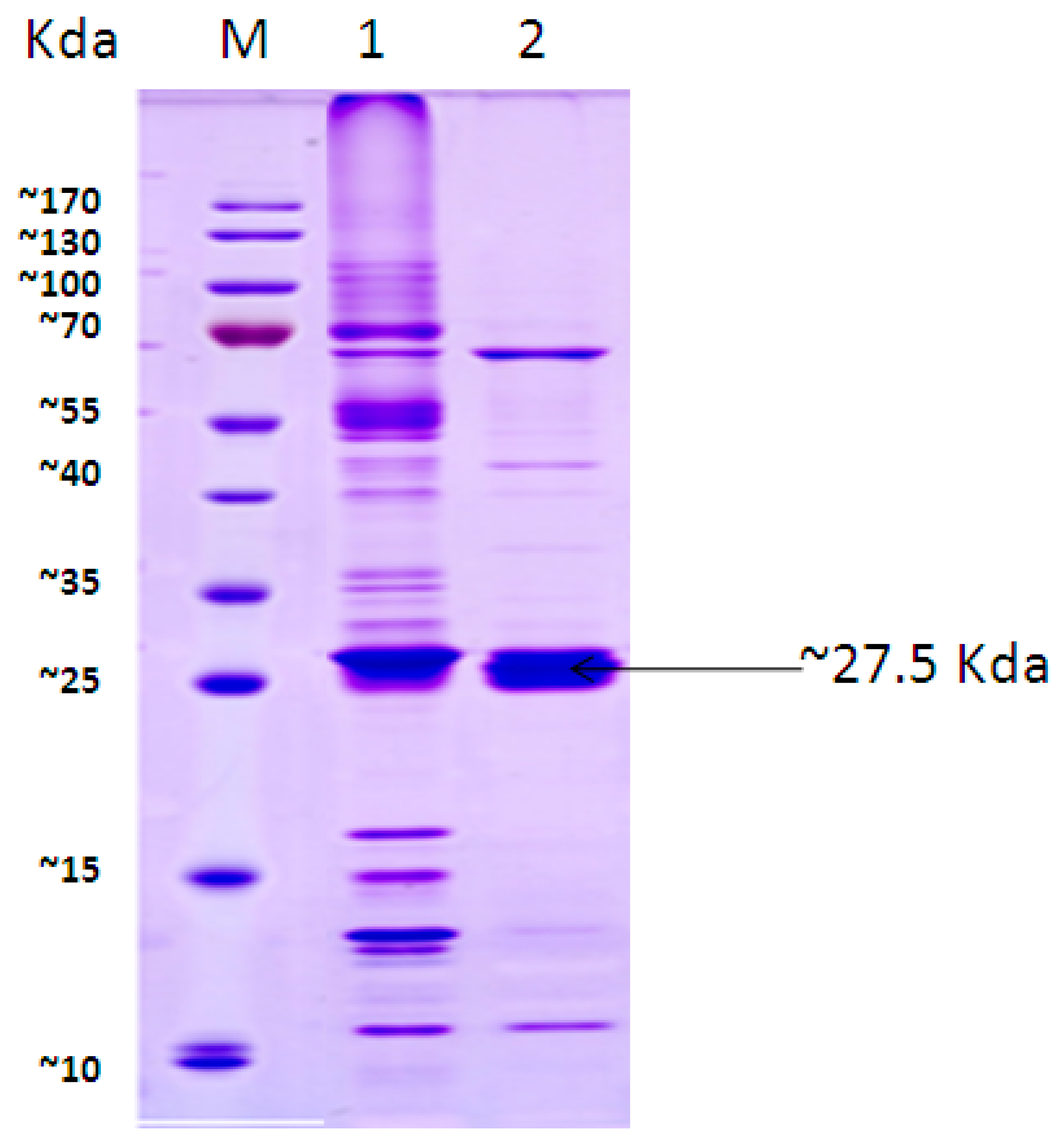
3.3. CBA Expression Analysis by SDS-PAGE
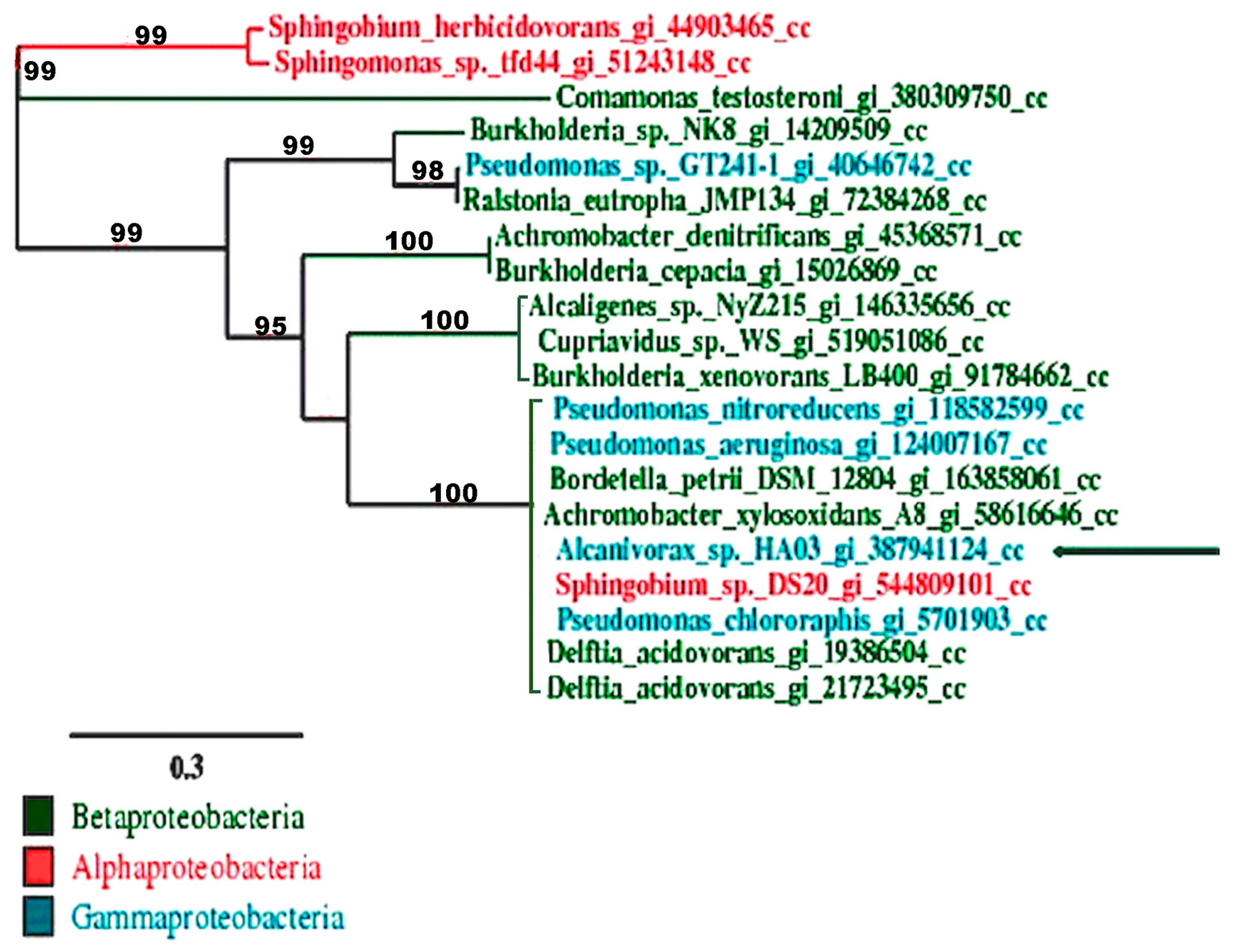
3.4. PCR Amplification and Characterization of CC12Dos in Alcanivorax sp. HA03
3.5. Heterologous Expression of CC12Dos in E. coli JM109
3.6. CC12Dos Expression Analysis by SDS-PAGE
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- U.S. EPA. Chlorpyrifos: Revised Human Health Risk Assessment for Registration Review; U.S. EPA: Washington, DC, USA, 2014.
- Kunze, M.; Zerlin, K.F.; Retzlaff, A.; Pohl, J.O.; Schmidt, E.; Janssen, D.B.; Vilchez-Vargas, R.; Pieper, D.H.; Reineke, W. Degradation of chloroaromatics by Pseudomonas putida GJ31: Assembled route for chlorobenzene degradation encoded by clusters on plasmid pKW1 and the chromosome. Microbiology 2009, 155, 4069–4083. [Google Scholar] [CrossRef] [PubMed]
- Thiel, M.; Kaschabek, S.; Groning, J.; Mau, M.; Schlömann, M. Two unusual chlorocatechol catabolic gene clusters in Sphingomonas sp. TFD44. Arch. Microbiol. 2005, 183, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Kirkok, S.K.; Kibet, J.K.; Kinyanjui, T.K.; Okanga, F.I. A review of persistent organic pollutants: Dioxins, furans, and their associated nitrogenated analogues. SN Appl. Sci. 2020, 2, 1729. [Google Scholar] [CrossRef]
- Haigler, B.E.; Pettigrew, C.A.; Spain, J.C. Biodegradation of mixtures of substituted benzenes by Pseudomonas sp. strain JS150. Appl. Environ. Microbiol. 1992, 58, 2237–2244. [Google Scholar] [CrossRef] [PubMed]
- Schlömann, M. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation 1994, 5, 301–321. [Google Scholar] [CrossRef]
- Werlen, C.; Kohler, H.P.; van der Meer, J.R. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J. Biol. Chem. 1996, 271, 4009–4016. [Google Scholar] [CrossRef]
- Van der Meer, J.R.; Werlen, C.; Nishino, S.F.; Spain, J.C. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microb. 1998, 64, 4185–4193. [Google Scholar] [CrossRef]
- Manikandan, R.; Prabhu, H.J.; Sivashanmugam, P. Biodegradation of chlorobenzene using immobilized crude extracts in packed bed column. Afr. J. Biotechnol. 2007, 6, 2259–2266. [Google Scholar]
- Vyas, T.K.; Murthy, S.R. Chlorobenzene degradation by Bacillus sp. TAS6CB: A potential candidate to remediate chlorinated hydrocarbon contaminated sites. J. Basic Microbiol. 2015, 55, 382–388. [Google Scholar] [CrossRef]
- Patel, A.; Vyas, T.K. Chlorobenzene Degradation via Ortho-Cleavage Pathway by Newly Isolated Microbacterium sp. Strain TAS1CB from a Petrochemical-Contaminated Site. Soil Sediment Contam. 2015, 24, 786–795. [Google Scholar] [CrossRef]
- Zhang, S.H.; Ying, Z.Y.; You, J.P.; Ye, J.X.; Cheng, Z.W.; Cheng, D.Z.; Chen, J.M. Superior performance and mechanism of chlorobenzene degradation by a novel bacterium. RSC Adv. 2019, 9, 15004–15012. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.H.; Luo, N.; Zhang, X.Y.; Luan, T.G.; Hu, J.M.; Wang, Z.Y.; Wu, P.C.; Chen, M.J.; Lu, J.Q. Biodegradation of benzene and its derivatives by a psychrotolerant and moderately haloalkaliphilic Planococcus sp. strain ZD22. Res. Microbiol. 2006, 157, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.A.; Nashwa, M.H.R.; Hefnawy, M.A.; Ahmad, M.A. Isolation and characterization of halophilic aromatic and chloroaromatic degrader from Wadi El-Natrun Soda lakes. Life Sci. J. 2012, 9, 1565–1570. [Google Scholar]
- Hassan, H.A.; Eldein, A.B.Z.; Rizk, N.M.H. Cloning and kinetic properties of catechol 2,3-dioxygenase from a novel alkaliphilic BTEX degrading Pseudomonas sp. HB01. Life Sci. J. 2014, 11, 376–384. [Google Scholar]
- Hassan, H.A.; Aly, A.A.; Ebeid, M.E. Cloning and expression of gene encoding meta-cleavage enzyme of BTEX degradation pathway from haloalkaliphilic Pseudomonas sp. HA10. Life Sci. J. 2014, 11, 403–411. [Google Scholar]
- Hassan, H.A.; Aly, A.A. Isolation and characterization of three novel catechol 2,3-dioxygenase from three novel haloalkaliphilic BTEX-degrading Pseudomonas strains. Int. J. Biol. Macromol. 2018, 106, 1107–1114. [Google Scholar] [CrossRef]
- Reineke, W. Aerobic and Anaerobic Biodegradation Potentials of Microorganisms. In The Handbook of Environmental Chemistry, Biodegradation and Persistence 2K Vol; Beek, B., Ed.; Springer: Heidelberg/Berlin, Germany, 2001; pp. 1–161. [Google Scholar]
- Reineke, W.; Knackmuss, H. Microbial degradation of haloaromalics. Annu. Rev. Microbiol. 1988, 42, 263–287. [Google Scholar] [CrossRef]
- Kaschabek, S.R.; Reineke, W. Maleylacetate reductase of Pseudomonas sp. strain B13: Dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch. Microbiol. 1992, 158, 412–417. [Google Scholar] [CrossRef]
- Mars, A.E.; Kasberg, T.; Kaschabek, S.R.; van Agteren, M.H.; Janssen, D.B.; Reineke, W. Microbial degradation of chloroaromatics: Use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 1997, 179, 4530–4537. [Google Scholar] [CrossRef]
- Kuhm, A.E.; Schlömann, M.; Knackmuss, H.J.; Pieper, D.H. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem. J. 1990, 266, 877–883. [Google Scholar]
- Dorn, E.; Hellwig, M.; Reineke, W.; Knackmuss, H.J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 1974, 99, 61–70. [Google Scholar] [CrossRef] [PubMed]
- McKay, D.B.; Prucha, M.; Reineke, W.; Timmis, K.N.; Pieper, D.H. Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6. J. Bacteriol. 2003, 185, 2944–2951. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Witzig, R.; Aly, H.A.; Strömpl, C.; Wray, V.; Junca, H.; Pieper, D.H. Molecular detection and diversity of novel diterpenoid dioxygenase DitA1 genes from proteobacterial strains and soil samples. Environ. Microbiol. 2007, 9, 1202–1218. [Google Scholar] [CrossRef] [PubMed]
- Aly, H.A.; Huu, N.B.; Wray, V.; Junca, H.; Pieper, D.H. Two angular dioxygenases contribute to the metabolic versatility of dibenzofuran-degrading Rhodococcus sp. Strain HA01. Appl. Environ. Microb. 2008, 74, 3812–3822. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.A.; Dawah, S.E.; El-Sheekh, M.M. Monitoring the degradation capability of novel haloalkaliphilic tributyltin chloride (TBTCl) resistant bacteria from butyltin-polluted site. Rev. Argent. Microbiol. 2019, 51, 39–46. [Google Scholar] [CrossRef]
- Monferrán, M.V.; Echenique, J.R.; Wunderlin, D.A. Degradation of chlorobenzenes by a strain of Acidovorax avenae isolated from a polluted aquifer. Chemosphere 2005, 61, 98–106. [Google Scholar] [CrossRef]
- Strunk, N.; Engesser, K.H. Degradation of fluorobenzene and its central metabolites 3-fluorocatechol and 2-fluoromuconate by Burkholderia fungorum FLU100. Appl. Microbiol. Biotechnol. 2013, 97, 5605–5614. [Google Scholar] [CrossRef]
- Dobslaw, D.; Engesser, K.H. Degradation of toluene by ortho-cleavage enzymes in Burkholderia fungorum FLU100. Microb. Biotechnol. 2015, 8, 143–154. [Google Scholar] [CrossRef]
- Chablain, P.A.; Zgoda, A.L.; Sarde, C.O.; Truffaut, N. Genetic and molecular organization of the alkylbenzene catabolism operon in the psychrotrophic strain Pseudomonas putida 01G3. Appl. Environ. Microb. 2001, 67, 453–458. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sander, P.; Wittich, R.M.; Fortnagel, P.; Wilkes, H.; Francke, W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl. Environ. Microb. 1991, 57, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Spiess, E.; Sommer, C.; Görisch, H. Degradation of 1,4-dichlorobenzene by Xanthobacter flavus 14p1. Appl. Environ. Microbiol. 1995, 61, 3884–3888. [Google Scholar] [CrossRef] [PubMed]
- Beil, S.; Timmis, K.N.; Pieper, D.H. Genetic and biochemical analyses of the tec operon suggest a route for evolution of chlorobenzene degradation genes. J. Bacteriol. 1999, 181, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Beil, S.; Mason, J.R.; Timmis, K.N.; Pieper, D.H. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 1998, 180, 5520–5528. [Google Scholar] [CrossRef] [PubMed]
- Ju, K.S.; Parales, R.E. Control of substrate specificity by active-site residues in nitrobenzene dioxygenase. Appl. Environ. Microbiol. 2006, 72, 1817–1824. [Google Scholar] [CrossRef]
- Parales, R.E.; Emig, M.D.; Lynch, N.A.; Gibson, D.T. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol. 1998, 180, 2337–2344. [Google Scholar] [CrossRef]
- Gibson, D.T.; Parales, R.E. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 2000, 11, 236–243. [Google Scholar] [CrossRef]
- Nam, J.W.; Nojiri, H.; Yoshida, T.; Habe, H.; Yamane, H.; Omori, T. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 2001, 65, 254–263. [Google Scholar] [CrossRef]
- Jiang, X.W.; Liu, H.; Xu, Y.; Wang, S.J.; Leak, D.J.; Zhou, N.Y. Genetic and biochemical analyses of chlorobenzene degradation gene clusters in Pandoraea sp. strain MCB032. Arch. Microbiol. 2009, 191, 485–492. [Google Scholar] [CrossRef]
- Hoffmann, D.; Kleinsteuber, S.; Müller, R.H.; Babel, W. A transposon encoding the complete 2,4-dichlorophenoxyacetic acid degradation pathway in the alkalitolerant strain Delftia acidovorans P4a. Microbiology 2003, 149, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Potrawfke, T.; Armengaud, J.; Wittich, R.M. Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71. J. Bacteriol. 2001, 183, 997–1011. [Google Scholar] [CrossRef]
- Potrawfke, T.; Timmis, K.N.; Wittich, R.M. Degradation of 1,2,3,4-tetrachlorobenzene by Pseudomonas chlororaphis RW71. Appl. Environ. Microbiol. 1998, 64, 3798–3806. [Google Scholar] [CrossRef] [PubMed]
- Pavlû, L.; Vosáhlová, J.; Klierová, H.; Prouza, M.; Demnerová, K.; Brenner, V. Characterization of chlorobenzoate degraders isolated from polychlorinated biphenyl-contaminated soil and sediment in the Czech Republic. J. Appl. Microbiol. 1999, 87, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Jenčová, V.; Strnad, H.; Chodora, Z.; Ulbrich, P.; Hickey, W.J.; Paces, V. Chlorocatechol catabolic enzymes from Achromobacter xylosoxidans A8. Int. Biodeterior. Biodegrad. 2004, 54, 175–181. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alghuthaymi, M.A.; Awad, A.M.; Hassan, H.A. Isolation and Characterization a Novel Catabolic Gene Cluster Involved in Chlorobenzene Degradation in Haloalkaliphilic Alcanivorax sp. HA03. Biology 2022, 11, 724. https://doi.org/10.3390/biology11050724
Alghuthaymi MA, Awad AM, Hassan HA. Isolation and Characterization a Novel Catabolic Gene Cluster Involved in Chlorobenzene Degradation in Haloalkaliphilic Alcanivorax sp. HA03. Biology. 2022; 11(5):724. https://doi.org/10.3390/biology11050724
Chicago/Turabian StyleAlghuthaymi, Mousa A., Ahmed M. Awad, and Hamdy A. Hassan. 2022. "Isolation and Characterization a Novel Catabolic Gene Cluster Involved in Chlorobenzene Degradation in Haloalkaliphilic Alcanivorax sp. HA03" Biology 11, no. 5: 724. https://doi.org/10.3390/biology11050724
APA StyleAlghuthaymi, M. A., Awad, A. M., & Hassan, H. A. (2022). Isolation and Characterization a Novel Catabolic Gene Cluster Involved in Chlorobenzene Degradation in Haloalkaliphilic Alcanivorax sp. HA03. Biology, 11(5), 724. https://doi.org/10.3390/biology11050724





