Comprehensive Transcriptome Analysis of Follicles from Two Stages of the Estrus Cycle of Two Breeds Reveals the Roles of Long Intergenic Non-Coding RNAs in Gilts
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Isolation and Sequencing
2.3. Quality Control, Mapping, and Transcriptome Assembly
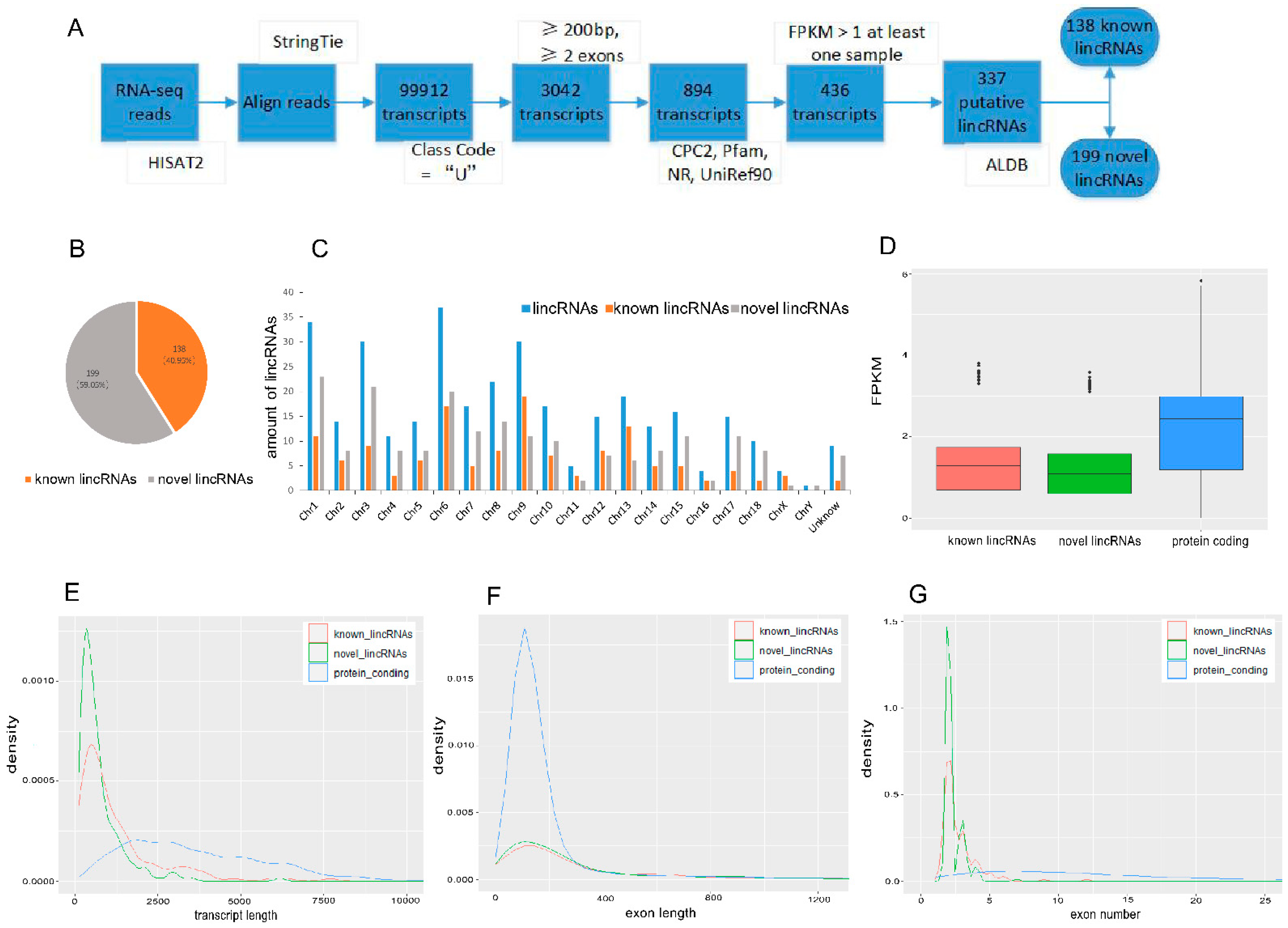
2.4. LincRNAs Identification and Characterization
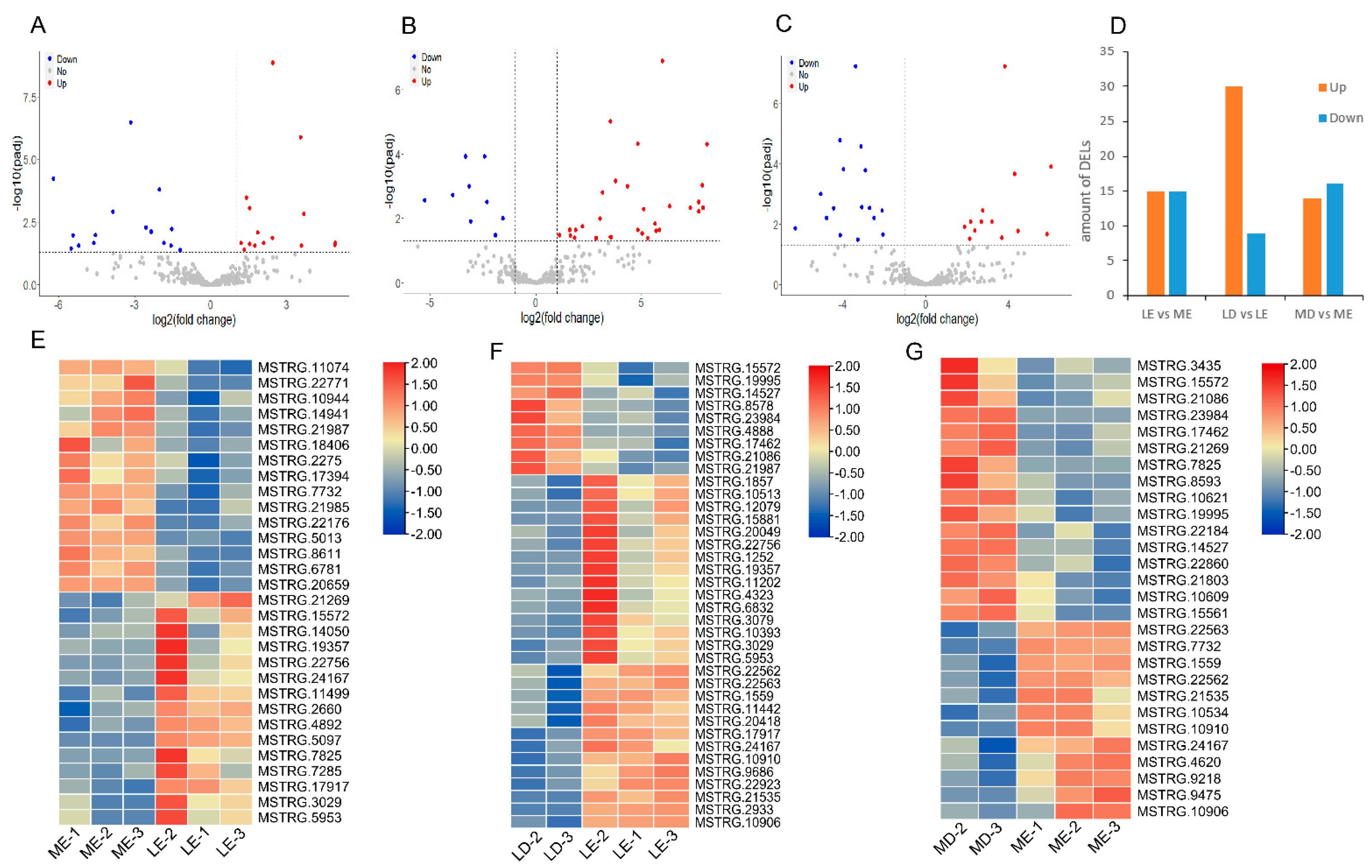
2.5. Analysis of Differential Expression
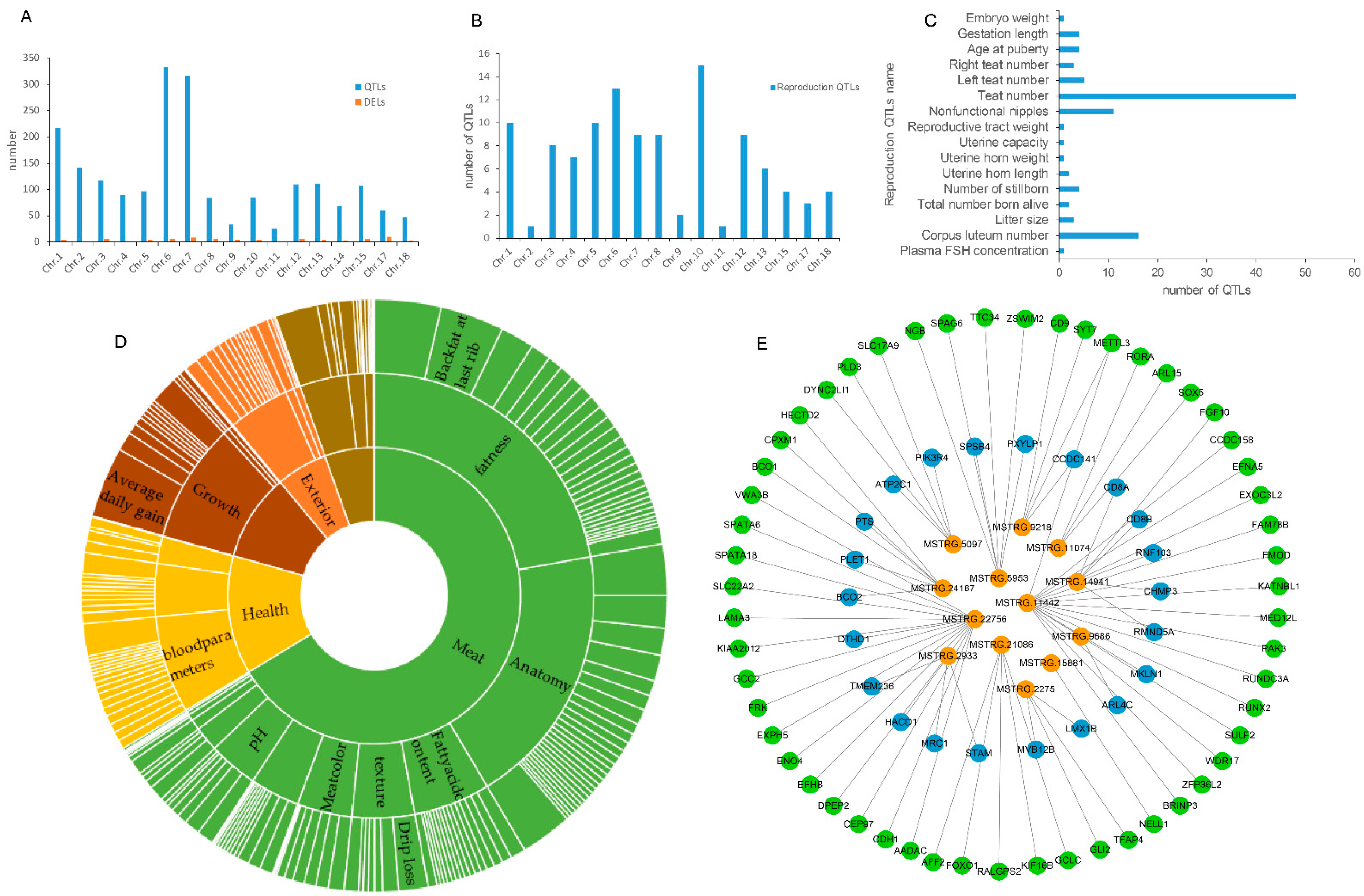
2.6. Functional Prediction of DELs by QTL Analysis
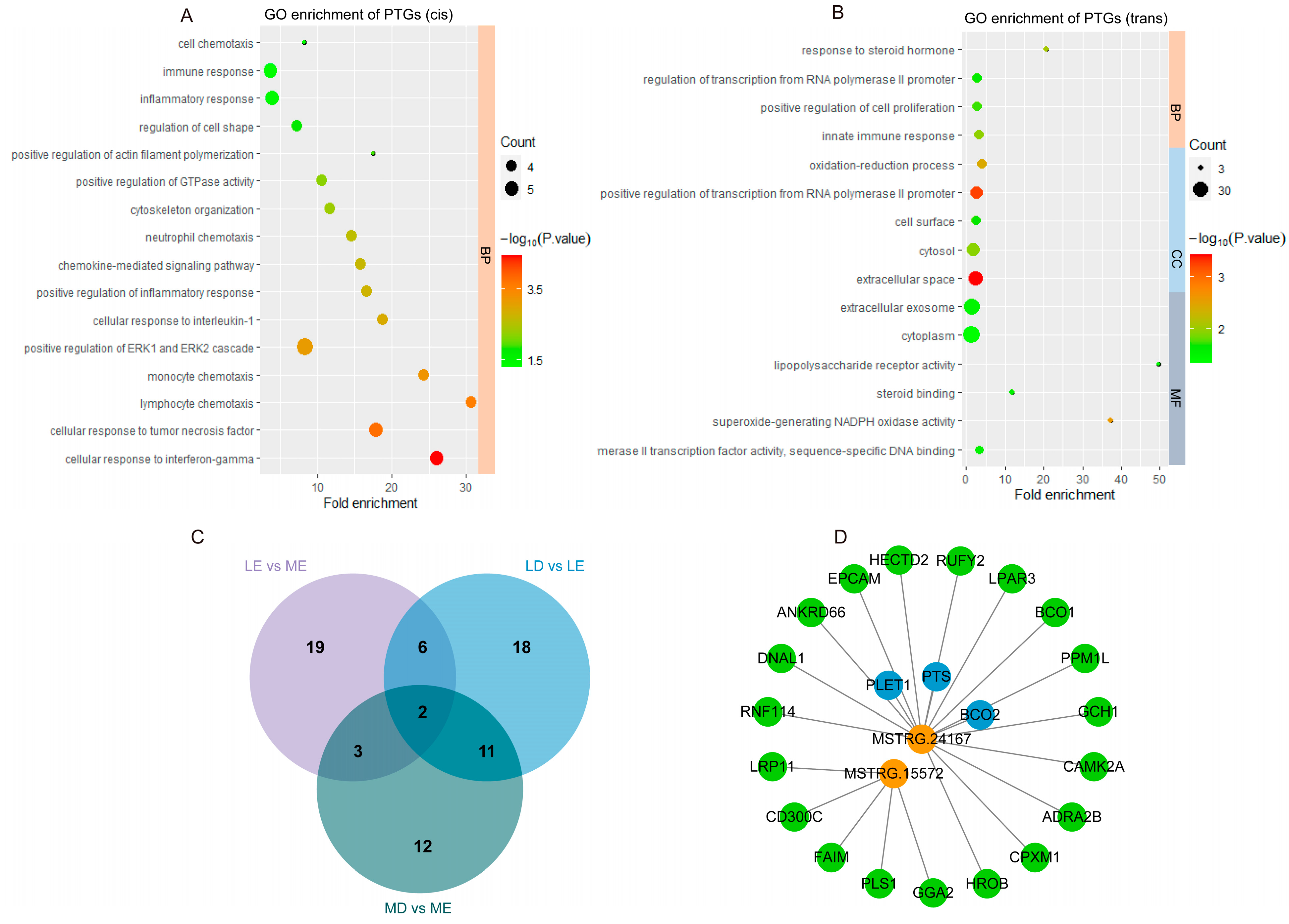
2.7. Target Genes Prediction and Function Enrichment Analysis
2.8. Construction of ceRNA Network Related to Follicular Development and Estrus Expression
2.9. RT-qPCR Verification
3. Results
3.1. Identification and Characterization of lincRNAs
3.2. Analysis of Differentially Expressed LincRNAs (DELs)
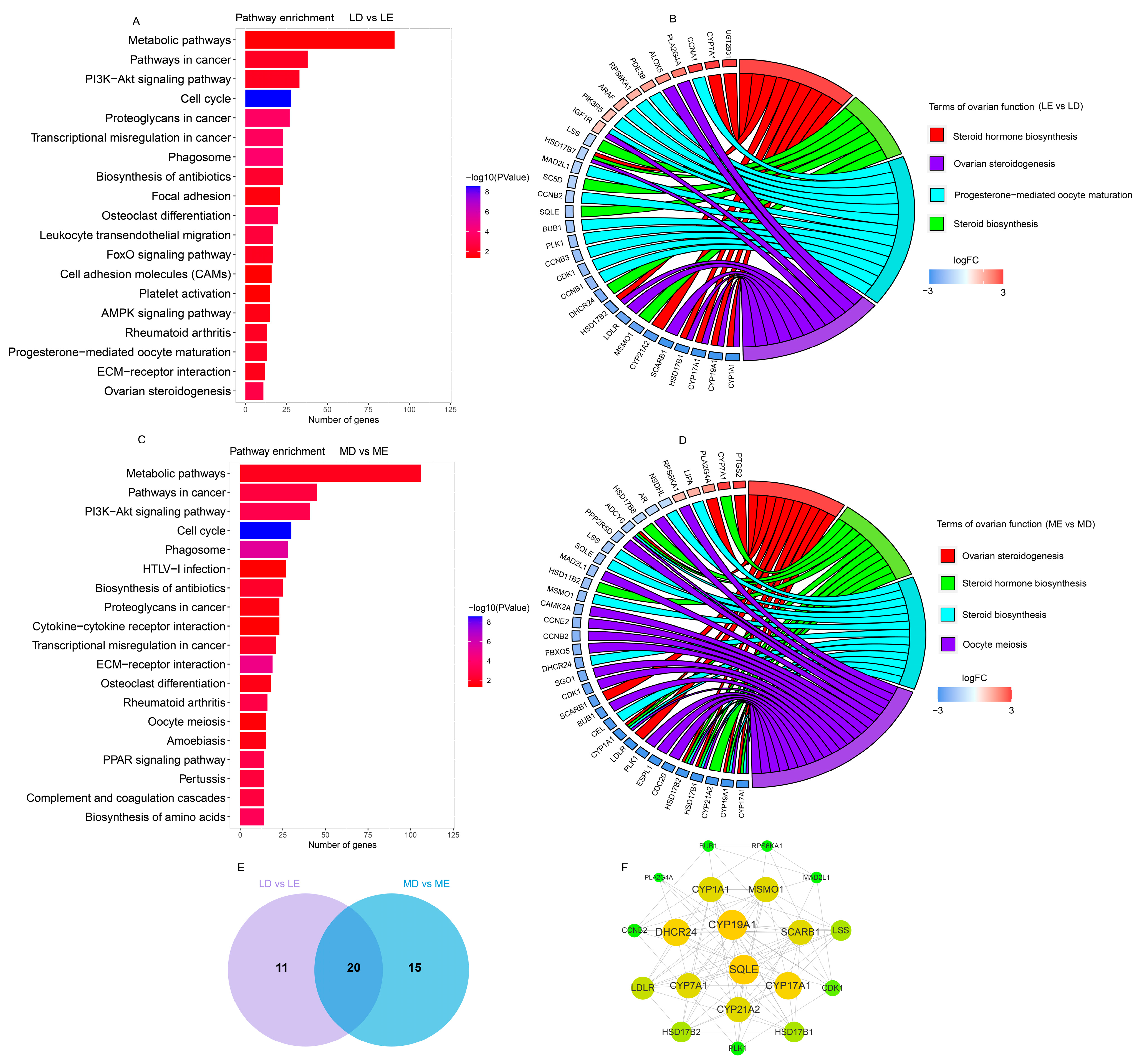
3.3. Target Gene Prediction and Function Analysis of lincRNAs
3.4. Analysis of DELs by QTLs
3.5. Analysis of DELs by Differentially Expressed Protein-Coding Genes (DEGs)
3.6. Construction of the lincRNA-miRNA-mRNA Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, L.G.; King, G.J. Reproductive performance of gilts bred on first versus third estrus. J. Anim. Sci. 1981, 53, 19–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sterning, M. Oestrous symptoms in primiparous sows. 2. Factors influencing the duration and intensity of external oestrous symptoms. Anim. Reprod. Sci. 1995, 40, 165–174. [Google Scholar] [CrossRef]
- Lents, C.A.; Lindo, A.N.; Hileman, S.M.; Nonneman, D.J. Physiological and genomic insight into neuroendocrine regulation of puberty in gilts. Domest. Anim. Endocrinol. 2020, 73, 106446. [Google Scholar] [CrossRef] [PubMed]
- Jahn, L.; Schuepbach-Regula, G.; Nathues, H.; Grahofer, A. Effect of 1,25-Dihydroxyvitamin D3-Glycosides on the Farrowing Process and Piglet Vitality in a Free Farrowing System. Animals 2022, 12, 611. [Google Scholar] [CrossRef]
- Knox, R.V. Physiology and Endocrinology Symposium: Factors influencing follicle development in gilts and sows and management strategies used to regulate growth for control of estrus and ovulation1. J. Anim. Sci. 2019, 97, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Zavy, M.T.; Geisert, R.D.; Buchanan, D.S.; Norton, S.A. Estrogen-induced pseudopregnancy in gilts: Its use in estrus synchronization and subsequent influence on litter response. Theriogenology 1988, 30, 721–732. [Google Scholar] [CrossRef]
- Kuehn, L.A.; Nonneman, D.J.; Klindt, J.M.; Wise, T.H. Genetic relationships of body composition, serum leptin, and age at puberty in gilts. J. Anim. Sci. 2009, 87, 477. [Google Scholar] [CrossRef]
- Knauer, M. Genetics of gilt estrous behavior. Diss. Theses-Gradworks 2009, 105, 716–721. Available online: http://www.lib.ncsu.edu/resolver/1840.16/4554 (accessed on 5 August 2021).
- Huang, L. Identification and differential expression of microRNAs in the ovaries of pigs (Sus scrofa) with high and low litter sizes. Anim. Genet. 2016, 47, 543–551. [Google Scholar] [CrossRef]
- Hossner, K. Hormonal Regulation of Farm Animal Growth; CABI Publishing: London, UK, 2005; p. 240. [Google Scholar] [CrossRef]
- Rena, M.; Ai, W.; Noriyuki, T.; Hiroshi, Y.; Naobumi, T.; Yoshio, S.; Tadahiro, O.; Hidenori, T.; Keiko, T.; Kenichi, I. Identification and Characterization of Novel Genotoxic Stress-Inducible Nuclear Long Noncoding RNAs in Mammalian Cells. PLoS ONE 2012, 7, e34949. [Google Scholar] [CrossRef]
- Zou, C.; Long, L.; Cheng, X.; Li, C.; Fu, Y.; Fang, C.; Li, C. Identification and Functional Analysis of Long Intergenic Non-coding RNAs Underlying Intramuscular Fat Content in Pigs. Front. Genet. 2018, 9, 102. [Google Scholar] [CrossRef]
- Chen, G.; Cheng, X.; Shi, G.; Zou, C.; Chen, L.; Li, J.; Li, M.; Fang, C.; Li, C. Transcriptome Analysis Reveals the Effect of Long Intergenic Noncoding RNAs on Pig Muscle Growth and Fat Deposition. BioMed Res. Int. 2019, 2019, 2951427. [Google Scholar] [CrossRef] [PubMed]
- Deniz, E.; Erman, B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genom. 2017, 17, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, G.; Chen, G.; Li, J.; Li, M.; Zou, C.; Fang, C.; Li, C. Transcriptome Analysis Suggests the Roles of Long Intergenic Non-coding RNAs in the Growth Performance of Weaned Piglets. Front. Genet. 2019, 10, 196. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2017, 19, 143–157. [Google Scholar] [CrossRef]
- Yan, P.; Luo, S.; Lu, J.Y.; Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming—ScienceDirect. Curr. Opin. Genet. Dev. 2017, 46, 170–178. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Li, M.; Zhang, N.; Zhang, W.; Wei, H.; Li, B. Comprehensive analysis of differentially expressed circRNAs and ceRNA regulatory network in porcine skeletal muscle. BMC Genom. 2021, 22, 320. [Google Scholar] [CrossRef]
- Tan, S.; Zhou, Y.; Zhao, H.; Wu, J.; Li, H. Comprehensive transcriptome analysis of hypothalamus reveals genes associated with disorders of sex development in pigs. J. Steroid Biochem. Mol. Biol. 2021, 210, 105875. [Google Scholar] [CrossRef]
- Swarr, D.T.; Herriges, M.; Li, S.; Morley, M.; Fernandes, S.; Sridharan, A.; Zhou, S.; Garcia, B.A.; Stewart, K.; Morrisey, E.E. The long noncoding RNA Falcor regulates Foxa2 expression to maintain lung epithelial homeostasis and promote regeneration. Genes Dev. 2019, 33, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, L.; Koganti, P.P.; Wang, L.; Hand, J.M.; Ma, H.; Yao, J. Identification and Functional Prediction of Large Intergenic Noncoding RNAs (lincRNAs) in Rainbow Trout (Oncorhynchus mykiss). Mar. Biotechnol. 2016, 18, 271–282. [Google Scholar] [CrossRef]
- Luo, H.; Sun, S.; Ping, L.; Bu, D.; Cao, H.; Zhao, Y.; Zhou, H. Comprehensive Characterization of 10,571 Mouse Large Intergenic Noncoding RNAs from Whole Transcriptome Sequencing. PLoS ONE 2013, 8, e70835. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.T.; Zheng, B.; Cao, S.Q.; Ding, J.C.; Hu, G.S.; Liu, W.; Chen, C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol. Cancer 2022, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Sang, L.J.; Ju, H.Q.; Liu, G.P.; Tian, T.; Ma, G.L.; Lu, Y.X.; Liu, Z.X.; Pan, R.L.; Li, R.H.; Piao, H.L.; et al. LncRNA CamK-A Regulates Ca(2+)-Signaling-Mediated Tumor Microenvironment Remodeling. Mol. Cell 2018, 72, 601. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Z.; Liu, Q.; Xie, S.; Li, M.; Wang, Y.; Li, C. Analysis of long intergenic non-coding RNAs transcriptomic profiling in skeletal muscle growth during porcine embryonic development. Sci. Rep. 2021, 11, 15240. [Google Scholar] [CrossRef]
- Zou, C.; Li, S.; Deng, L.; Guan, Y.; Chen, D.; Yuan, X.; Xia, T.; He, X.; Shan, Y.; Li, C. Transcriptome Analysis Reveals Long Intergenic Noncoding RNAs Contributed to Growth and Meat Quality Differences between Yorkshire and Wannanhua Pig. Genes 2017, 8, 203. [Google Scholar] [CrossRef]
- Li, J.; Gao, Z.; Wang, X.; Liu, H.; Zhang, Y.; Liu, Z. Identification and functional analysis of long intergenic noncoding RNA genes in porcine pre-implantation embryonic development. Sci. Rep. 2016, 6, 38333. [Google Scholar] [CrossRef]
- Zhao, W.; Mu, Y.; Ma, L.; Wang, C.; Tang, Z.; Yang, S.; Zhou, R.; Hu, X.; Li, M.H.; Li, K. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 2015, 5, 8957. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, Z.G.; Li, Y.J.; Zhao, X.L.; Li, K. Genetic variation analysis within and among Chinese indigenous swine populations using microsatellite markers. Anim. Genet. 2015, 33, 422–427. [Google Scholar] [CrossRef]
- Bazer, F.W.; Thatcher, W.W.; Martinat-Botte, F.; Terqui, M. Sexual maturation and morphological development of the reproductive tract in large white and prolific Chinese Meishan pigs. J. Reprod. Fertil. 1988, 83, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Liang, T.; Zhou, B. Genetic differences in oestrous signs and oestrogen metabolism-related genes between Chinese Mi and European Landrace-Large White pigs. Reprod. Domest. Anim. 2017, 52, 696–700. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Enfield, F. A characterization of Chinese breeds of swine using cluster analysis. J. Anim. Breed. Genet. 1989, 106, 379–388. [Google Scholar] [CrossRef]
- Gao, S.; Tao, R.; Tong, X.; Xu, Q.; Zhao, J.; Guo, Y.; Schinckel, A.P.; Zhou, B. Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts. Biomolecules 2020, 10, 1545. [Google Scholar] [CrossRef]
- Chu, Q.; Zhou, B.; Xu, F.; Chen, R.; Shen, C.; Liang, T.; Li, Y.; Schinckel, A.P. Genome-wide differential mRNA expression profiles in follicles of two breeds and at two stages of estrus cycle of gilts. Sci. Rep. 2017, 7, 5052. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef]
- Pirooznia, M.; Perkins, E.J.; Deng, Y. Batch Blast Extractor: An automated blastx parser application. BMC Genom. 2008, 9, S10. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, D.; Zhang, T.; Duan, A.; Zhang, J.; He, C. Transcriptomic and functional analyses unveil the role of long non-coding RNAs in anthocyanin biosynthesis during sea buckthorn fruit ripening. DNA Res. 2018, 25, 465–476. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zheng, X.; Feng, W.; Wang, H.; Kang, H.; Ning, C.; Du, H.; Yu, Y.; Li, B.; Zhao, Y.; et al. Profiling long noncoding RNA of multi-tissue transcriptome enhances porcine noncoding genome annotation. Epigenomics 2018, 10, 301–320. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Bo, X.; Li, T.; Ma, L.; Zhai, T.; Huang, T. Systematic Analysis of Long Non-Coding RNAs and mRNAs in the Ovaries of Duroc Pigs During Different Follicular Stages Using RNA Sequencing. Int. J. Mol. Sci. 2018, 19, 1722. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Pasquinelli, A.E. NON-CODING RNA MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef]
- Tang, L.T.; Ran, X.Q.; Ning, M.; Zhang, F.P.; Xi, N.; Yi-Qi, R.; Yi, F.L.; Sheng, L.; Wang, J.F. Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology 2018, 119, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Y.; Li, S.; Li, K.; Tang, Z. Analysis and comparison of long non-coding RNAs expressed in the ovaries of Meishan and Yorkshire pigs. Anim. Genet. 2019, 50, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Liu, Y.; Xie, S.; Ma, L.P.; Zhao, Z.C.; Gong, H.B.; Sun, Y.S.; Huang, T. Transcriptome analysis reveals that long noncoding RNAs contribute to developmental differences between medium-sized ovarian follicles of Meishan and Duroc sows. Sci. Rep. 2021, 11, 22510. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.B.; Zhou, X.L.; Pei, Y.; Wang, H.; He, K.; Zhao, A.Y. Identification of Differentially Expressed Genes in Porcine Ovaries at Proestrus and Estrus Stages Using RNA-Seq Technique. BioMed Res. Int. 2018, 2018, 22510. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Guigó, R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Li, Q.; Huang, Z.; Zhao, W.; Li, M.; Li, C. Transcriptome Analysis Reveals Long Intergenic Non-Coding RNAs Contributed to Intramuscular Fat Content Differences between Yorkshire and Wei Pigs. Int. J. Mol. Sci. 2020, 21, 1732. [Google Scholar] [CrossRef]
- Zang, X.; Gu, T.; Wang, W.; Zhou, C.; Ding, Y.; Gu, S.; Xu, Z.; Xie, Y.; Li, Z.; Cai, G.; et al. Integrated Insight into the Molecular Mechanisms of Spontaneous Abortion during Early Pregnancy in Pigs. Int. J. Mol. Sci. 2021, 22, 6644. [Google Scholar] [CrossRef]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282–1291. [Google Scholar] [CrossRef]
- Dode, M.; Graves, C. Involvement of steroid hormones on in vitro maturation of pig oocytes. Theriogenology 2002, 57, 811–821. [Google Scholar] [CrossRef]
- Barfield, R.; Glaser, J.; Rubin, B.; Etgen, A. Behavioral effects of progestin in the brain. Psychoneuroendocrinology 1984, 9, 217–231. [Google Scholar] [CrossRef]
- Sugiura, T.; Akiyoshi, S.; Inoue, F.; Yanagawa, Y.; Moriyoshi, M.; Tajima, M.; Katagiri, S. Relationship between bovine endometrial thickness and plasma progesterone and estradiol concentrations in natural and induced estrus. J. Reprod. Develop. 2018, 64, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Christenson, L.K.; Gunewardena, S.; Hong, X.; Spitschak, M.; Baufeld, A.; Vanselow, J. Research resource: Preovulatory LH surge effects on follicular theca and granulosa transcriptomes. Mol. Endocrinol. 2013, 27, 1153–1171. [Google Scholar] [CrossRef]
- Teng, L.; Hong, L.; Liu, R.; Chen, R.; Li, X.; Yu, M. Cellular Localization and Regulation of Expression of the PLET1 Gene in Porcine Placenta. Int. J. Mol. Sci. 2016, 17, 2048. [Google Scholar] [CrossRef]
- Zhao, S.H.; Simmons, D.G.; Cross, J.C.; Scheetz, T.E.; Tuggle, C.K. PLET1 (C11orf34), a highly expressed and processed novel gene in pig and mouse placenta, is transcribed but poorly spliced in human. Genomics 2004, 84, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Abdoli, R.; Mirhoseini, S.Z.; Hossein-Zadeh, N.G.; Zamani, P.; Gondro, C. Genome-wide association study of four composite reproductive traits in Iranian fat-tailed sheep. Reprod. Fertil. Dev. 2019, 31, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.R.; Swann, K. Molecular triggers of egg activation at fertilization in mammals. Reproduction 2016, 152, 41–51. [Google Scholar] [CrossRef]
- Rovani, M.T.; Gasperin, B.G.; Ilha, G.F.; Ferreira, R.; Bohrer, R.C.; Duggavathi, R.; Bordignon, V.; Gonçalves, P.B.D. Expression and molecular consequences of inhibition of estrogen receptors in granulosa cells of bovine follicles. J. Ovarian Res. 2014, 7, 96. [Google Scholar] [CrossRef]
- Li, G.; Qin, X.; Cui, S.; Li, Y.; Gong, J. Upregulated microRNAb alleviates ovarian cancer through inhitbition of the PI3K/Akt pathway by targeting LPAR3. J. Cell. Physiol. 2019, 234, 22331–22342. [Google Scholar] [CrossRef]
- Hou, Q.; Wu, J.; Zhao, Y.; Wang, X.; Li, J.D. Genotypic and phenotypic spectra of CCDC141 variants in a Chinese cohort with congenital hypogonadotropic hypogonadism. Eur. J. Endocrinol. 2020, 183, 245–254. [Google Scholar] [CrossRef]
- Meng, L.; Teerds, K.; Tao, J.; Wei, H.; Jaklofsky, M.; Zhao, Z.; Liang, Y.; Li, L.; Wang, C.C.; Zhang, S. Characteristics of Circular RNA Expression Profiles of Porcine Granulosa Cells in Healthy and Atretic Antral Follicles. Int. J. Mol. Sci. 2020, 21, 5217. [Google Scholar] [CrossRef] [PubMed]
- Desbois-Mouthon, C.; Cadoret, A.; Eggelpoël, M.-J.B.-V.; Bertrand, F.; Cherqui, G.; Perret, C.; Capeau, J. Insulin and IGF-1 stimulate the β-catenin pathway through two signalling cascades involving GSK-3β inhibition and Ras activation. Oncogene 2001, 20, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Nimz, M.; Spitschak, M.; Furbass, R.; Vanselow, J. The Pre-Ovulatory Luteinizing Hormone Surge Is Followed by Down-Regulation of CYP19A1, HSD3B1, and CYP17A1 and Chromatin Condensation of the Corresponding Promoters in Bovine Follicles. Mol. Reprod. Dev. 2010, 77, 1040–1048. [Google Scholar] [CrossRef]
- Sekar, N.; Veldhuis, J.D. Concerted transcriptional activation of the low density lipoprotein receptor gene by insulin and luteinizing hormone in cultured porcine granulosa-luteal cells: Possible convergence of protein kinase a, phosphatidylinositol 3-kinase, and mitogen-activated protein kinase signaling pathways. Endocrinology 2001, 142, 2921–2928. [Google Scholar] [CrossRef][Green Version]
- Lu, X.Y.; Shi, X.J.; Hu, A.; Wang, J.Q.; Ding, Y.; Jiang, W.; Sun, M.; Zhao, X.; Luo, J.; Qi, W.; et al. Feeding induces cholesterol biosynthesis via the mTORC1-USP20-HMGCR axis. Nature 2020, 588, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Mesmin, B.; Pipalia, N.H.; Lund, F.W.; Ramlall, T.F.; Sokolov, A.; Eliezer, D.; Maxfield, F.R. STARD4 abundance regulates sterol transport and sensing. Mol. Biol. Cell 2011, 22, 4004–4015. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Stocco, D.M.; Clark, B.J. Current knowledge on the acute regulation of steroidogenesis. Biol. Reprod. 2018, 99, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.L.; Liu, L.; Wang, N.; Chen, Z.J.; Zhang, C. The function of high-density lipoprotein and low-density lipoprotein in the maintenance of mouse ovarian steroid balance. Biol. Reprod. 2017, 97, 862–872. [Google Scholar] [CrossRef]
- Chotiner, J.Y.; Wolgemuth, D.J.; Wang, P.J. Functions of cyclins and CDKs in mammalian gametogenesisdagger. Biol. Reprod. 2019, 101, 591–601. [Google Scholar] [CrossRef]
- Li, J.; Dong, F.; Ouyang, Y.C.; Sun, Q.Y.; Qian, W.P. Overexpression of cyclin A1 promotes meiotic resumption but induces premature chromosome separation in mouse oocyte. J. Cell. Physiol. 2020, 235, 7136–7145. [Google Scholar] [CrossRef]
- Tahir, M.S.; Porto-Neto, L.R.; Gondro, C.; Shittu, O.B.; Fortes, M. Meta-Analysis of Heifer Traits Identified Reproductive Pathways in Bos indicus Cattle. Genes 2021, 12, 768. [Google Scholar] [CrossRef] [PubMed]
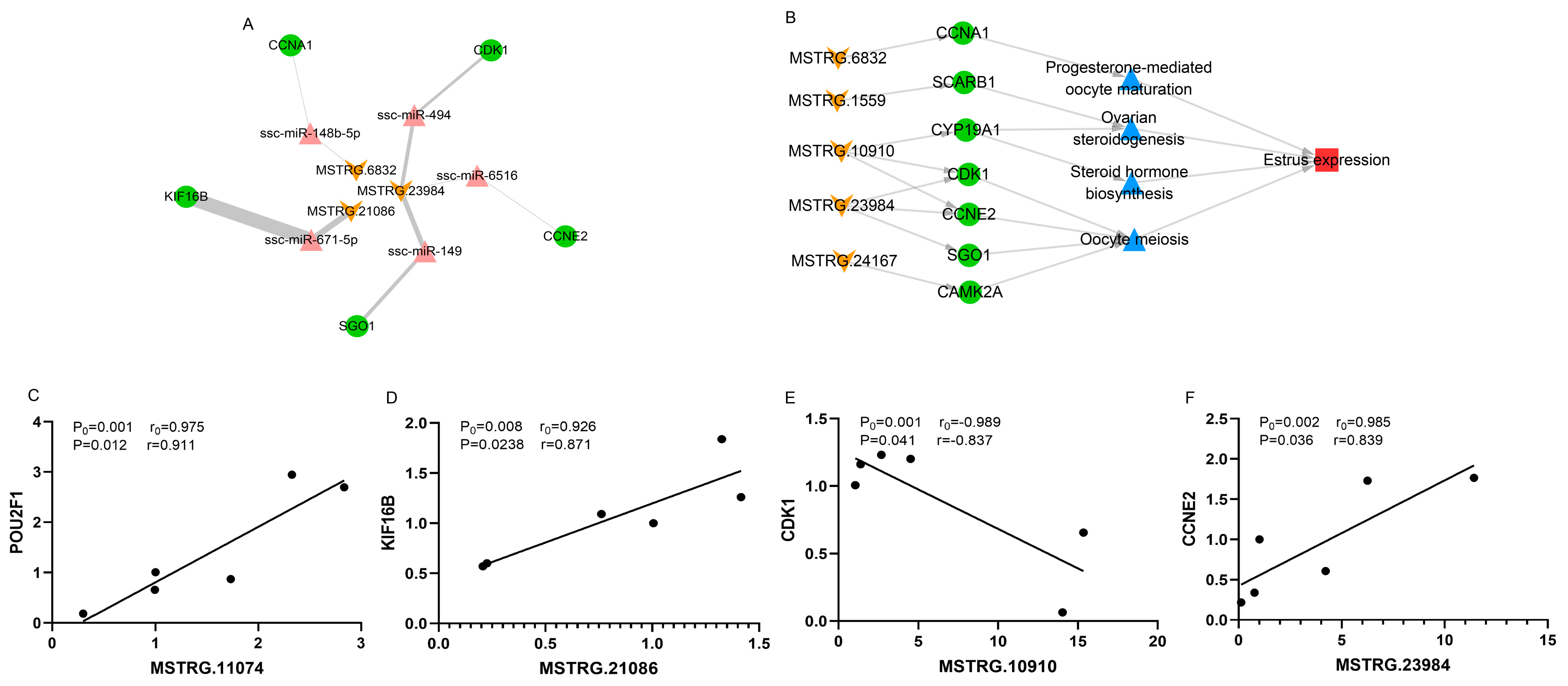
| Comparison | DELs | PTGs | Desription | KEGG Pathway | r | p |
|---|---|---|---|---|---|---|
| MD vs. ME | MSTRG.10910 | CCNE2 | cyclin E2 | Oocyte meiosis | −0.96 | <0.01 |
| MSTRG.23984 | CCNE2 | 0.97 | <0.01 | |||
| MSTRG.1559 | SCARB1 | scavenger receptor class B member 1 | Ovarian steroidogenesis | −0.95 | <0.01 | |
| MSTRG.10910 | CDK1 | cyclin dependent kinase 1 | Oocyte meiosis | −0.96 | <0.01 | |
| MSTRG.23984 | CDK1 | 0.96 | <0.01 | |||
| MSTRG.23984 | SGO1 | shugoshin 1 | Oocyte meiosis | 0.96 | <0.01 | |
| MSTRG.24167 | CAMK2A | calcium/calmodulin dependent protein kinase II alpha | Oocyte meiosis | −0.96 | <0.01 | |
| MSTRG.10910 | CYP19A1 | cytochrome P450 19A1 | Steroid hormone biosynthesis, Ovarian steroidogenesis | −0.96 | <0.01 | |
| LD vs. LE | MSTRG.6832 | CCNA1 | cyclin A1 | Progesterone-mediated oocyte maturation | 0.95 | <0.01 |
| MSTRG.1559 | SCARB1 | scavenger receptor class B member 1 | Ovarian steroidogenesis | −0.95 | <0.01 | |
| MSTRG.10910 | CDK1 | cyclin dependent kinase 1 | Progesterone-mediated oocyte maturation | −0.96 | <0.01 | |
| MSTRG.23984 | CDK1 | 0.96 | <0.01 | |||
| MSTRG.23984 | SGO1 | shugoshin 1 | Oocyte meiosis | 0.96 | <0.01 | |
| MSTRG.23984 | CCNE2 | cyclin E2 | Oocyte meiosis | 0.97 | <0.01 | |
| MSTRG.10910 | CYP19A1 | cytochrome P450 19A1 | Steroid hormone biosynthesis, Ovarian steroidogenesis | −0.96 | <0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Xu, Q.; Zhao, J.; Guo, Y.; Zhang, C.; Chao, X.; Cheng, M.; Schinckel, A.P.; Zhou, B. Comprehensive Transcriptome Analysis of Follicles from Two Stages of the Estrus Cycle of Two Breeds Reveals the Roles of Long Intergenic Non-Coding RNAs in Gilts. Biology 2022, 11, 716. https://doi.org/10.3390/biology11050716
Liu M, Xu Q, Zhao J, Guo Y, Zhang C, Chao X, Cheng M, Schinckel AP, Zhou B. Comprehensive Transcriptome Analysis of Follicles from Two Stages of the Estrus Cycle of Two Breeds Reveals the Roles of Long Intergenic Non-Coding RNAs in Gilts. Biology. 2022; 11(5):716. https://doi.org/10.3390/biology11050716
Chicago/Turabian StyleLiu, Mingzheng, Qinglei Xu, Jing Zhao, Yanli Guo, Chunlei Zhang, Xiaohuan Chao, Meng Cheng, Allan P. Schinckel, and Bo Zhou. 2022. "Comprehensive Transcriptome Analysis of Follicles from Two Stages of the Estrus Cycle of Two Breeds Reveals the Roles of Long Intergenic Non-Coding RNAs in Gilts" Biology 11, no. 5: 716. https://doi.org/10.3390/biology11050716
APA StyleLiu, M., Xu, Q., Zhao, J., Guo, Y., Zhang, C., Chao, X., Cheng, M., Schinckel, A. P., & Zhou, B. (2022). Comprehensive Transcriptome Analysis of Follicles from Two Stages of the Estrus Cycle of Two Breeds Reveals the Roles of Long Intergenic Non-Coding RNAs in Gilts. Biology, 11(5), 716. https://doi.org/10.3390/biology11050716







