Impact of Cultivation Substrate and Microbial Community on Improving Mushroom Productivity: A Review
Simple Summary
Abstract
1. Introduction
2. Mushroom Cultivation
2.1. Sources and Composition of Substrates
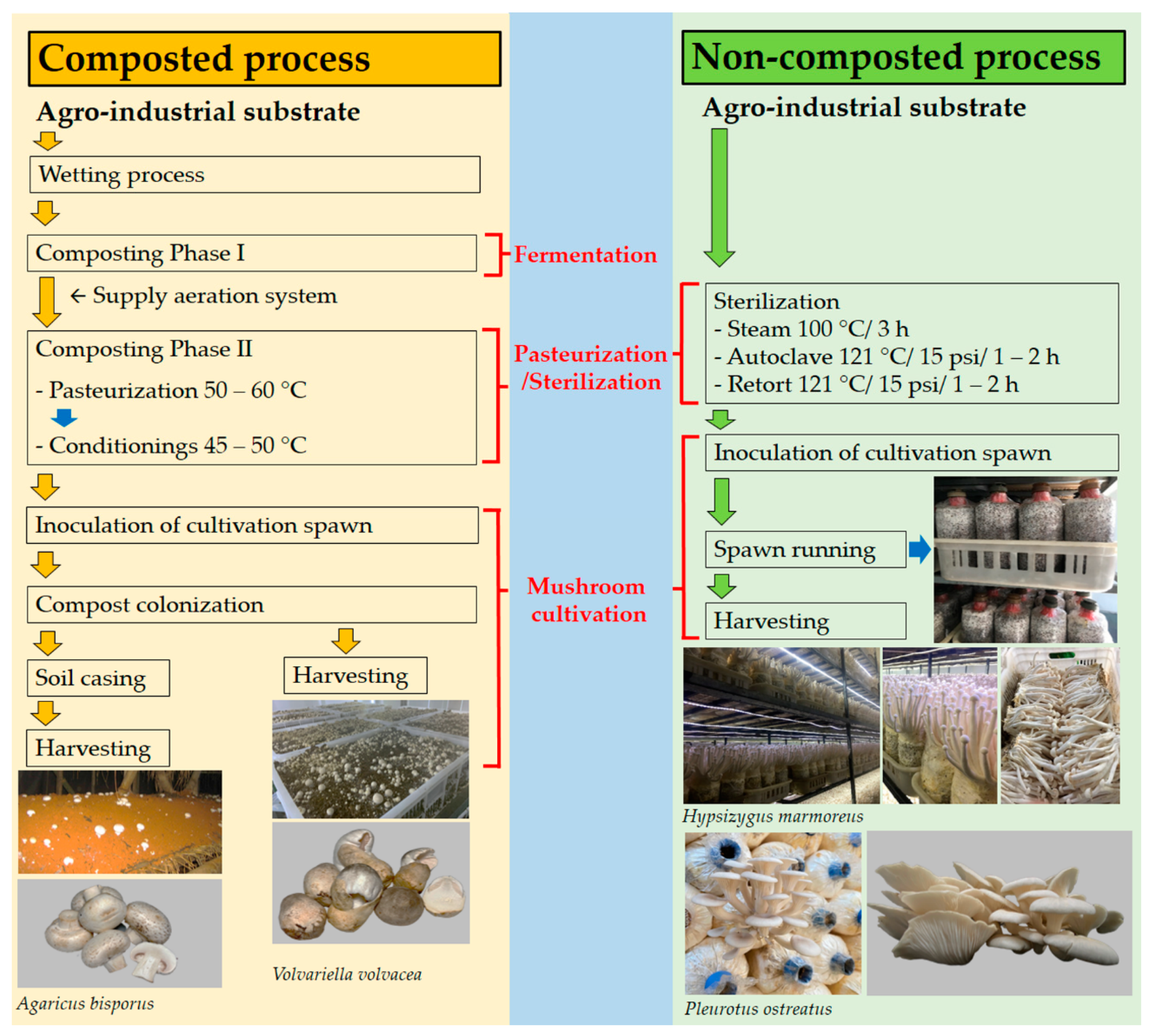
2.2. Methods for Mushroom Cultivation
2.2.1. Cultivation on Non-Composted Substrates
2.2.2. Cultivation on Composted Substrates
2.3. Enzymes Involved in Substrate Utilization
3. Microbial Community for Mushroom Cultivation
3.1. Methods Used for Analysis of Microbial Communities
3.1.1. Culture-Dependent Methods
Plate Culture Method
Biolog Microplate Method
Fingerprinting Based on Biochemical Components
3.1.2. Culture-Independent Methods
Denaturing Gradient Gel Electrophoresis (DGGE)
Amplified Ribosomal DNA Restriction Analysis (ARDRA)
Terminal Restriction Polymorphism (T-RFLP)
High-Throughput Sequencing
Quantitative Polymerase Chain Reaction (qPCR)
3.2. Microbial Community Influence on Mushroom Cultivation
| Substrate Types | Dominant Bacteria | Properties Related to the Cultivation | Method of Analysis | Mushroom | References | |
|---|---|---|---|---|---|---|
| Phylum | Class/Order/Genus | |||||
| Wheat straw-based compost | Proteobacteria | Pseudoxanthomonas | Cellulose-degrading consortium | DGGE and T-RFLP analysis | A. bisporus | [12] |
| Actinobacteria | Thermobifida Thermomonospora | |||||
| Corncob compost (Early stage) | Firmicutes | Carnobacterium | - | Metagenomic sequencing | P. ostreatus | [15] |
| Proteobacteria | Pseudomonas Stenotrophomonas | |||||
| Bacteroidetes | Sphingobacterium | |||||
| Actinobacteria | Glutamicibacter | |||||
| Corncob compost (Thermophilic stage) | Firmicutes | Aerococcus | - | |||
| Bacillus | ||||||
| Desemzia | ||||||
| Lysinibacillus | ||||||
| Enterococcus | ||||||
| Proteobacteria | Acinetobacter | |||||
| Actinobacteria | Corynebacterium | |||||
| Naturally occurring soil and mushroom | Acidobacteria | - | - | DNA sequencing | C. militaris | [16] |
| Actinobacteria | ||||||
| Bacteroidetes Proteobacteria | ||||||
| Soil in fruiting body | Firmicutes | - | - | 16 rRNA amplicons, Illumina MiSeq sequencing | O. highlandensis | [17] |
| Verrucomicrobia | ||||||
| Deltaproteobacteria | ||||||
| Proteobacteria | ||||||
| Peach sawdust-based compost | Firmicutes | - | - | Metagenomic 16S rRNA sequencing | Oyster mushroom | [80] |
| Actinobacteria | ||||||
| Proteobacteria | ||||||
| Compost | Thermodesulfobacteria | Thermodesulfobacterium | Sulfur-reducing properties | DNA and cDNA sequencing | A. bisporus | [92] |
| Proteobacteria | Pseudoxanthomonas | |||||
| Actinobacteria | - | |||||
| Firmicutes | - | |||||
| Natural composting samples (Mesophilic stage) | Actinobacteria | Actinomycetales Bacillales Clostridiales | - | DNA sequence with Roche/454 technology | - | [135] |
| Firmicutes | Rhodococcus Lactobacillus Thermobifida | - | ||||
| Actinobacteria | Amycolatopsis | Potential for lignin degradation | ||||
| Maize straw compost | Firmicutes | Sporosarcina Bacillus Staphylococcus | - | Illumina MiSeq sequencing | - | [151] |
| Proteobacteria | Pseudomonas Ochrobactrum | - | ||||
| Bacteroidetes | - | - | ||||
| Actinobacteria | Cellulosimicrobium | Produce lignocellulose hydrolytic enzymes | ||||
| Sugarcane processing | Firmicutes | Bacillales Lactobacillales Clostridiales | Fermentation | PhyloChip microarray | - | [152] |
| Proteobacteria | - | - | ||||
| Bacteroidetes | - | - | ||||
| Wood chips and sawdust compost | Actinobacteria | Brevibacterium | Degrade cellulose | Real-time PCR and DGGE | - | [153] |
| Micrococcineae | Degrade lignocellulosic materials | |||||
| Cellulomonas | Produce cellulases and hemicellulases | |||||
| Composting from waste management system | Actinobacteria | - | - | DNA sequencing | - | [154] |
| Bacteroidetes | - | - | ||||
| Firmicutes | Bacillus | Produce proteases | ||||
| Clostridium | Degrade cellulose and lignin | |||||
| Lactobacillus | Related to low pH | |||||
| Thermoactinomyces | - | |||||
| Proteobacteria | Acetobacter | Related to low pH | ||||
| Deinococcus-Thermus | - | - | ||||
4. Volatile Organic Compounds Involved in the Microbial Community and Mushroom Cultivation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Data. Available online: http://www.fao.org/faostat/en/#home (accessed on 20 May 2020).
- Carrasco, J.; Zied, D.C.; Pardo, J.E.; Preston, G.M.; Pardo-Giménez, A. Supplementation in mushroom crops and its impact on yield and quality. AMB Express 2018, 8, e146. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Thai, M. Compost bacteria and fungi that influence growth and development of Agaricus bisporus and other commercial mushrooms. Appl. Microbiol. Biotechnol. 2018, 102, 1639–1650. [Google Scholar] [CrossRef]
- Kumla, J.; Suwannarach, N.; Sujarit, K.; Penkhrue, W.; Kakumyan, P.; Jatuwong, K.; Vadthanarat, S.; Lumyong, S. Cultivation of mushrooms and their lignocellulolytic enzyme production through the utilization of agro-industrial waste. Molecules 2020, 25, 2811. [Google Scholar] [CrossRef]
- Kumar, H.; Bhardwaj, K.; Sharma, R.; Nepovimova, E.; Cruz-Martins, N.; Dhanjal, D.S.; Singh, R.; Chopra, C.; Verma, R.; Abd-Elsalam, K.A.; et al. Potential usage of edible mushrooms and their residues to retrieve valuable supplies for industrial applications. J. Fungi 2021, 7, 427. [Google Scholar] [CrossRef]
- Taylor, J.W.; Ellison, C.E. Mushrooms: Morphological complexity in the fungi. Proc. Natl. Acad. Sci. USA 2010, 107, 11655–11656. [Google Scholar] [CrossRef]
- Kabel, M.A.; Jurak, E.; Mäkelä, M.R.; de Vries, R.P. Occurrence and function of enzymes for lignocellulose degradation in commercial Agaricus bisporus cultivation. Appl. Microbiol. Biotechnol. 2017, 101, 4363–4369. [Google Scholar] [CrossRef]
- Kerrigan, R.W.; Challen, M.P.; Burton, K.S. Agaricus bisporus genome sequence: A commentary. Fungal Genet. Biol. 2013, 55, 2–5. [Google Scholar] [CrossRef]
- Vieira, F.R.; Pecchia, J.A. Bacterial community patterns in the Agaricus bisporus cultivation system, from compost raw materials to mushroom caps. Microb. Ecol. 2021, 1–13. [Google Scholar] [CrossRef]
- Carrasco, J.; Preston, G.M. Growing edible mushrooms: A conversation between bacteria and fungi. Environ. Microbiol. 2020, 22, 858–887. [Google Scholar] [CrossRef]
- Tan, H.; Hohler, A.; Mian, R.; Liu, T.; Zhang, Q.; Zhang, B.; Jiang, L.; Wang, Y.; Xie, L.; Tang, J.; et al. Multi-omic analyses of exogenous nutrient bag decomposition by the black morel Morchella importuna reveal sustained carbon acquisition and transferring. Environ. Microbiol. 2019, 21, 3909–3926. [Google Scholar] [CrossRef]
- Székely, A.J.; Sipos, R.; Berta, B.; Vajna, B.; Hajdú, C.; Márialigeti, K. DGGE and T-RFLP Analysis of Bacterial Succession during Mushroom Compost Production and Sequence-aided T-RFLP Profile of Mature Compost. Microb. Ecol. 2009, 57, 522–533. [Google Scholar] [CrossRef]
- Carrasco, J.; Tello, M.L.; de Toro, M.; Tkacz, A.; Poole, P.; Perez-Clavijo, M.; Preston, G. Casing microbiome dynamics during button mushroom cultivation: Implications for dry and wet bubble diseases. Microbiology 2019, 165, 611–624. [Google Scholar] [CrossRef]
- Vajna, B.; Nagy, A.; Sajben, E.; Manczinger, L.; Szijártó, N.; Kádár, Z.; Bordás, D.; Márialigeti, K. Microbial community structure changes during oyster mushroom substrate preparation. Appl. Microbiol. Biotechnol. 2010, 86, 367–375. [Google Scholar] [CrossRef]
- Kong, W.; Sun, B.; Zhang, J.; Zhang, Y.; Gu, L.; Bao, L.; Liu, S. Metagenomic analysis revealed the succession of microbiota and metabolic function in corncob composting for preparation of cultivation medium for Pleurotus ostreatus. Bioresour. Technol. 2020, 306, e123156. [Google Scholar] [CrossRef]
- Zhang, X.M.; Tang, D.X.; Li, Q.Q.; Wang, Y.B.; Xu, Z.H.; Li, W.J.; Yu, H. Complex microbial communities inhabiting natural Cordyceps militaris and the habitat soil and their predicted functions. Antonie Leeuwenhoek. 2021, 114, 465–477. [Google Scholar] [CrossRef]
- Li, C.; Tang, D.; Wang, Y.; Fan, Q.; Zhang, X.; Cui, X.; Yu, H. Endogenous bacteria inhabiting the Ophiocordyceps highlandensis during fruiting body development. BMC Microbiol. 2021, 21, e178. [Google Scholar] [CrossRef]
- Anwar, Z.; Gulfraz, M.; Irshad, M. Agro-industrial lignocellulosic biomass a key to unlock the future bioenergy: A brief review. J. Radiat. Res. Appl. Sci. 2014, 7, 163–173. [Google Scholar] [CrossRef]
- da Silva, L.L. Adding value to agro-Industrial wastes. Ind. Chem. 2016, 2, e103. [Google Scholar] [CrossRef]
- Siwulski, M.; Rzymski, P.; Budka, A.; Kalač, P.; Budzyńska, S.; Dawidowicz, L.; Hajduk, E.; Kozak, L.; Budzulak, J.; Sobieralski, K.; et al. The effect of different substrates on the growth of six cultivated mushroom species and composition of macro and trace elements in their fruiting bodies. Eur. Food. Res. Technol. 2019, 245, 419–431. [Google Scholar] [CrossRef]
- Hassan, F.R.H. Cultivation of the monkey head mushroom (Hericium erinaceus) in Egypt. J. Appl. Sci. Res. 2007, 3, 1229–1233. [Google Scholar]
- Atila, F. Lignocellulosic and proximate based compositional changes in substrates during cultivation of Hericium erinaceus mushroom. Sci. Hortic. 2019, 258, e108779. [Google Scholar] [CrossRef]
- Biswas, M.K.; Layak, M. Techniques for increasing the biological efficiency of paddy straw mushroom (Volvariella volvacea) in eastern India. Food Sci. Technol. 2014, 2, 52–57. [Google Scholar] [CrossRef]
- Haq, I.U.; Khan, M.A.; Khan, S.A.; Ahmad, M. Biochemical analysis of fruiting bodies of Volvariella volvacea strain Vv pk, grown on six different substrates. Soil Environ. 2011, 30, 146–150. [Google Scholar]
- Triyono, S.; Haryanto, A.; Telaumbanua, M.; Lumbanraja, D.J.; To, F. Cultivation of straw mushroom (Volvariella volvacea) on oil palm empty fruit bunch growth medium. Int. J. Recycl. Org. Waste Agric. 2019, 8, 381–392. [Google Scholar] [CrossRef]
- de Carvalho, C.S.M.; Sales-Campos, C.; de Carvalho, L.P.; Minhoni, M.T.A.; Saad, A.L.M.; Alquati, G.P.; de Andrade, M.C.N. Cultivation and bromatological analysis of medicinal mushroom Ganoderma lucidum (Curt.: Fr.) P. Karst cultivated in agricultural waste. Afr. J. Biotechnol. 2015, 14, 412–418. [Google Scholar]
- Roy, S.; Jahan, M.A.A.; Das, K.K.; Munshi, S.K.; Noor, R. Artificial cultivation of Ganoderma lucidum (Reishi medicinal mushroom) using different sawdusts as substrates. Am. J. Biocci. 2015, 3, 178–182. [Google Scholar] [CrossRef]
- Liang, C.H.; Wu, C.Y.; Lu, P.L.; Kuo, Y.C.; Liang, Z.C. Biological efficiency and nutritional value of the culinary-medicinal mushroom Auricularia cultivated on a sawdust basal substrate supplement with different proportions of grass plants. Saudi. J. Biol. Sci. 2019, 26, 263–269. [Google Scholar] [CrossRef]
- Razak, D.L.A.; Abdullah, N.; Johari, N.M.K.; Sabaratnam, V. Comparative study of mycelia growth and sporophore yield of Auricularia polytricha (Mont.) Sacc on selected palm oil wastes as fruiting substrate. Appl. Microbiol. Biotechnol. 2013, 97, 3207–3213. [Google Scholar] [CrossRef]
- Gao, S.; Huang, Z.; Feng, X.; Bian, Y.; Huang, W.; Lui, Y. Bioconversion of rice straw agroresidues by Lentinula edodes and evaluation of non-volatile taste compounds in mushrooms. Sci. Rep. 2020, 10, e1814. [Google Scholar] [CrossRef]
- Gaitán-Hernández, R.; Cortés, N.; Mata, G. Improvement of yield of the edible and medicinal mushroom Lentinula edodes on wheat straw by use of supplemented spawn. Braz. J. Microbiol. 2014, 45, 467–474. [Google Scholar] [CrossRef]
- Salmones, D.; Mata, G.; Ramos, L.M.; Waliszewski, K.N. Cultivation of shiitake mushroom, Lentinula edodes, in several lignocellulosic materials originating from the subtropics. Agronomie 1999, 19, 13–19. [Google Scholar] [CrossRef]
- Xie, C.; Yan, L.; Gong, W.; Zhu, Z.; Tan, S.; Chen, D.; Hu, Z.; Peng, Y. Effects of Different Substrates on Lignocellulosic Enzyme Expression, Enzyme Activity, Substrate Utilization and Biological Efficiency of Pleurotus eryngii. Cell Physiol. Biochem. 2016, 39, 1479–1494. [Google Scholar] [CrossRef] [PubMed]
- Sardar, H.; Ali, M.A.; Anjum, M.A.; Nawaz, F.; Hussain, S.; Naz, S.; Karimi, S.M. Agro-industrial residues influence mineral elements accumulation and nutritional composition of king oyster mushroom (Pleurotus eryngii). Sci. Hortic. 2017, 225, 327–334. [Google Scholar] [CrossRef]
- Adjapong, A.O.; Ansah, K.D.; Angfaarabung, F.; Sintim, H.O. Maize residue as a viable substrate for farm scale cultivation of oyster mushroom (Pleurotus ostreatus). Adv. Agric. 2015, 2015, e213251. [Google Scholar] [CrossRef]
- Koutrotsios, G.; Mountzouris, K.C.; Chatzipavlidis, I.; Zervakis, G.I. Bioconversion of lignocellulosic residues by Agrocybe cylindracea and Pleurotus ostreatus mushroom fungi–assessment of their effect on the final product and spent substrate properties. Food Chem. 2014, 161, 127–135. [Google Scholar] [CrossRef]
- Obodai, M.; Cleland-Okine, J.; Vowotor, K.A. Comparative study on the growth and yield of Pleurotus ostreatus. J. Ind. Microbiol. Biotechnol. 2003, 30, 146–149. [Google Scholar] [CrossRef]
- Pardo-Giménez, A.; Catalán, L.; Carrasco, J.; Álvarez-Ortí, M.; Zied, D.; Pardo, J. Effect of supplementing crop substrate with defatted pistachio meal on Agaricus bisporus and Pleurotus ostreatus production. J. Sci. Food Agric. 2016, 96, 3838–3845. [Google Scholar] [CrossRef]
- Hoa, H.T.; Wang, C.L.; Wang, C.H. The effects of different substrates on the growth, yield, and nutritional composition of two oyster mushrooms (Pleurotus ostreatus and Pleurotus cystidiosus). Mycobiology 2015, 43, 423–434. [Google Scholar] [CrossRef]
- Abou Fayssal, S.; El Sebaaly, Z.; Alsanad, M.A.; Najjar, R.; Böhme, M.; Yordanova, M.H.; Sassine, Y.N. Combined effect of olive pruning residues and spent coffee grounds on Pleurotus ostreatus production, composition, and nutritional value. PLoS ONE 2021, 16, e0255794. [Google Scholar]
- Kleofas, V.; Sommer, L.; Fraatz, M.A.; Zorn, H.; Rühl, M. Fruiting body production and aroma profile analysis of Agrocybe aegerita cultivated on different substrates. Nat. Resour. 2014, 5, 233–240. [Google Scholar]
- Harith, N.; Abdullah, N.; Sabaratnam, V. Cultivation of Flammulina velutipes mushroom using various agro-residues as a fruiting substrate. Pesq. Agropec. Bras. 2014, 49, 181–188. [Google Scholar] [CrossRef]
- Patil, S.S. Productivity and proximate content of Pleurotus sajor-caju. Biosci. Discov. 2013, 4, 169–172. [Google Scholar]
- Pardo-Giménez, A.; Pardo, J.E.; Dias, E.S.; Rinker, D.L.; Caitano, C.E.C.; Zied, D.C. Optimization of cultivation techniques improves the agronomic behavior of Agaricus subrufescens. Sci. Rep. 2020, 10, e8154. [Google Scholar] [CrossRef] [PubMed]
- Toker, H.; Baysal, E.; Yigibasi, O.N.; Colak, M.; Perker, H.; Simsek, H.; Yilmaz, F. Cultivation of Agaricus bisporus on wheat straw and waste tea leaves based composts using poplar leaves as activator material. Afr. J. Biotechnol. 2007, 6, 204–212. [Google Scholar]
- de Andrade, M.C.N.; Zied, D.C.; de Almeida Minhoni, M.T.; Filho, J.K. Yield of four Agaricus bisporus strains in three compost formulations and chemical composition analyses of the mushrooms. Braz. J. Microbiol. 2008, 39, 593–598. [Google Scholar] [CrossRef]
- Zied, D.C.; Junior, W.G.V.; Soares, D.M.M.; Stevani, C.V.; Dias, E.S.; Iossi, M.R.; Pardo-Giménez, A. Overview of four Agaricus subrufescens strains used in the last 15 years in Brazil and other countries and current potential materials for the future. Mycol. Prog. 2021, 20, 953–966. [Google Scholar] [CrossRef]
- Win, T.T.; Ohga, S. Study on the cultivation of Agaricus blazei (Almond mushroom) grown on compost mixed with selected agro-residues. Adv. Microbiol. 2018, 8, 778–789. [Google Scholar] [CrossRef][Green Version]
- Wisitrassameewong, K.; Karunarathna, S.C.; Thongklang, N.; Zhao, R.; Callac, P.; Moukha, S.; Fe´randon, C.; Chukeatirote, E.; Hyde, K.D. Agaricus subrufescens: A review. Saudi J. Biol. Sci. 2012, 19, 131–146. [Google Scholar] [CrossRef]
- Yella, V.K.; Chadrapati, A.; Kuri, A.; Miglani, I.; Andrews, A.A.; Singh, S. Cultivation technology and spawn production of Volvariella volvacea: Paddy straw mushroom. Pharm. Innov. J. 2021, 10, 1184–1190. [Google Scholar]
- Alam, N.; Singha, S.M. Effects of composition, age and sterilization techniques of mother culture on the growth and yield of Volvariella volvacea (bull.) Singer. Bangladesh J. Bot. 2020, 49, 387–393. [Google Scholar] [CrossRef]
- Zhai, F.H.; Han, J.R. Decomposition of asparagus old stalks by Pleurotus spp. under mushroom growing conditions. Sci. Hortic. 2018, 231, 11–14. [Google Scholar] [CrossRef]
- Chang, S.; Miles, P.G. Mushrooms: Cultivation, Nutritional Value, Medicinal Effect and Environmental Impact, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Bellettini, M.B.; Fiorda, F.A.; Maieves, H.A.; Teixeira, G.L.; Ávila, S.; Hornung, P.S.; Júnior, A.M.; Ribani, R.H. Factors affecting mushroom Pleurotus spp. Saudi J. Biol. Sci. 2016, 26, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, J.L.R.; Gomes, V.F.F.; Filho, J.K.; de Almeida Minhoni, M.T.; de Andrade, M.C.N. Cultivation of Agaricus blazei in the environmental protection area of the Baturite´ region under three types of casing soils. Acta. Sci. Agron. 2008, 30, 513–517. [Google Scholar]
- Souza, T.P.; Marques, S.C.; da Silveira e Santos, D.M.; Dias, E.S. Analysis of thermophilic fungal populations during phase II of composting for the cultivation of Agaricus subrufescens. World J. Microbiol. Biotechnol. 2014, 30, 2419–2425. [Google Scholar] [CrossRef]
- Scrase, R. Cultivating mushrooms: Making composted and non-composted substrates. Mycologist 1996, 10, 52–55. [Google Scholar] [CrossRef]
- Wang, L.; Mao, J.; Zhao, H.; Li, M.; Wei, Q.; Zhou, Y.; Shao, H. Comparison of characterization and microbial communities in rice straw- and wheat straw-based compost for Agaricus bisporus production. J. Ind. Microbiol. Biotechnol. 2016, 43, 1249–1260. [Google Scholar] [CrossRef]
- Cueva, M.B.R.; Hernáadez, A.; Niňo-Ruiz, Z. Influence of C/N ratio on productivity and the protein contents of Pleurotus ostreatus grown in differents residue mixtures. Rev. FCA Uncuyo. 2017, 49, 331–334. [Google Scholar]
- Ragunathan, R.; Swaminathan, K. Nutritional status of Pleurotus spp. grown on various agro-wastes. Food Chem. 2003, 80, 371–375. [Google Scholar] [CrossRef]
- Garcha, H.S.; Khanna, P.K.; Sodhi, H.S.; Dhanda, S.; Sidhu, A.; Phutela, R.P. Nutrient supplementation for Agaricus bisporus (Lange) Sing. Cultivation. Develop. Crop Sci. 1987, 10, 101–108. [Google Scholar]
- Shekhar, H.S.; Kilpatrick, M. Mushroom (Agaricus bisporus) compost quality factor for predicting potential yield of fruiting bodies. Can. J. Microbiol. 2000, 46, 515–519. [Google Scholar]
- Oei, P. Mushroom Cultivation, 3rd ed.; Backhuys Publishers: Leiden, The Netherlands, 2003; 429p. [Google Scholar]
- Lisiecka, J.; Sobieralski, K.; Siwulski, M.; Jasinska, A. Almond mushroom Agaricus brasiliensis (Wasser et al.)–properties and culture condition. Acta Sci. Pol. Hortorum Cultus 2013, 12, 27–40. [Google Scholar]
- Kopytowski, F.J.; Minhoni, M.T.A. C/N ratio on yield of Agaricus blazei Murrill ss. Heinemann. Mush. Sci. 2004, 16, 213–220. [Google Scholar]
- Shi, L.; Chen, D.; Xu, C.; Ren, A.; Yu, H.; Zhao, M. Highly-efficient liposome-mediated transformation system for the basidiomycetous fungus Flammulina velutipes. J. Gen. Appl. Microbiol. 2017, 63, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.; Yang, F.C. Reusing soy residue for the solid-state fermentation of Ganoderma lucidum. Bioresur. Technol. 2004, 91, 105–109. [Google Scholar] [CrossRef]
- Poppe, J.; Höfte, M. Twenty wastes for twenty cultivated mushroom. Mush. Sci. 1995, 14, 171–179. [Google Scholar]
- Chang, S.T.; Milles, P.G. Edible Mushroom and Their Cultivation; CRC Press: Florida, FL, USA, 1989; 345p. [Google Scholar]
- Chang-Ho, Y.; Ho, T.M. Effect of nitrogen amendment on the growth of Volvariella volvacea. Mush. Sci. 1979, 10, 619–625. [Google Scholar]
- Kaul, T.; Khurana, M.; Kachroo, J. Chemical composition of cereal straw of the Kashmir valley. Mush. Sci. 1981, 11, 19–25. [Google Scholar]
- Grimm, D.; Wösten, H.A.B. Mushroom cultivation in the circular economy. Appl. Microbiol. Biotechnol. 2018, 102, 7795–7803. [Google Scholar] [CrossRef]
- Royse, D.J.; Baars, J.; Tan, Q. Current Overview of Mushroom Production in the World. In Edible and Medicinal Mushrooms: Technology and Applications; Zied, D.C., Pardo-Gimenez, A., Eds.; Wiley-Blackwell: West Sussex, UK, 2007; pp. 5–13. [Google Scholar]
- Moonmoon, M.; Shelly, N.J.; Khan, M.A.; Uddin, M.N.; Hossain, K.; Tania, M.; Ahmed, S. Effects of different levels of wheat bran, rice bran and maize powder supplementation with saw dust on the production of shiitake mushroom (Lentinus edodes (Berk.) Singer). Saudi. J. Biol. Sci. 2011, 18, 323–328. [Google Scholar] [CrossRef]
- Cai, Y.J.; Buswell, J.A.; Chang, S.T. Production of cellulases and hemicellulases by the straw mushroom, Volvariella volvacea. Mycol. Res. 1994, 98, 1019–1024. [Google Scholar] [CrossRef]
- An, Q.; Wu, X.J.; Dai, Y.C. Comparative genomics of 40 edible and medicinal mushrooms provide an insight into the evolution of lignocellulose decomposition mechanisms. 3 Biotech 2019, 9, e157. [Google Scholar] [CrossRef] [PubMed]
- Gramss, G.; Bergmann, H. Role of plants in the vegetative and reproductive growth of saprobic basidiomycetous ground fungi. Microb. Ecol. 2008, 56, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Mahari, W.A.W.; Peng, W.; Nam, W.L.; Yang, H.; Lee, X.Y.; Lee, Y.K.; Lee, Y.K.; Liew, R.K.; Ma, N.L.; Mohammad, A.; et al. A review on valorization of oyster mushroom and waste generated in the mushroom cultivation industry. J. Hazard Mater. 2020, 400, e123156. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Sakamoto, M.; Yamada, M.; Hara, H.; Mat Taib, S.; Rezania, S.; Md Din, F.M.; Mohd Hanaf, F.H. Cultivation of oyster mushroom (Pleurotus ostrreatus) on fermented moso bamboo sawdust. J. King Saud. Univ. Sci. 2019, 31, 490–494. [Google Scholar] [CrossRef]
- Guo, Y.X.; Chen, Q.J.; Qin, Y.; Yang, Y.R.; Yang, Q.Z.; Wang, Y.X.; Cheng, Z.; Cao, Z.; Zhang, G.Q. Succession of the microbial communities and function prediction during short-term peach sawdust-based composting. Bioresour. Technol. 2021, 332, 125079. [Google Scholar] [CrossRef]
- Noble, R.; Dobrovin-Pennington, A.; Hobbs, P.J.; Pederby, J.; Rodger, A. Volatile C8 compounds and pseudomonads influence primordium formation of Agaricus bisporus. Mycologia 2009, 101, 583–591. [Google Scholar] [CrossRef]
- Straatsma, G.; Gerrits, J.P.G.; Thissen, J.T.N.M.; Amsing, J.G.M.; Loeffen, H.; Van Griensven, L.J.L.D. Adjustment of the composting process for mushroom cultivation based on initial substrate composition. Bioresour. Technol. 2000, 72, 67–74. [Google Scholar] [CrossRef]
- Zhao, Z.T.; Liu, H.Q.; Wang, C.F.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, e27. [Google Scholar] [CrossRef]
- Ospina-Giraldo, M.D.; Grifth, J.G.; Laird, E.W.; Mingora, C. The CAZyome of Phytophthora spp.: A comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genom. 2010, 11, e525. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef]
- Elisashvili, V.; Chichua, D.; Kachlishvili, E.; Tsiklauri, N.; Khardziani, T. Lignocellulolytic enzyme activity during growth and fruiting of the edible and medicinal mushroom Pleurotus ostreatus (Jacq.: Fr.) Kumm. (Agaricomycetideae). Int. J. Med. Mushrooms 2003, 5, 193–198. [Google Scholar] [CrossRef]
- Xing, Z.; Cheng, J.; Tan, Q.; Pan, Y. Effect of nutritional parameters on laccase production by the culinary and medicinal mushroom, Grifola frondosa. World J. Microbiol. Biotechnol. 2006, 22, 1215–1224. [Google Scholar] [CrossRef]
- Chen, S.C.; Ma, D.; Ge, W.; Buswell, J.A. Induction of laccase activity in the edible straw mushroom, Volvariella volvacea. FEMS Microbiol. Lett. 2003, 218, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-L.; Wei, J.-K.; Wang, Q.-H.; Yang, R.; Gao, X.-J.; Sang, Y.-X.; Cai, P.-P.; Zhang, G.-Q.; Chen, Q.-J. Lignocellulose utilization and bacterial communities of millet straw based mushroom (Agaricus bisporus) production. Sci. Rep. 2019, 9, 1151. [Google Scholar] [CrossRef]
- Montoya, S.; Orrego, C.E.; Levin, L. Growth, fruiting and lignocellulolytic enzyme production by the edible mushroom Grifola frondosa (maitake). World J. Microbiol. Biotechnol. 2012, 28, 1533–1541. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, Y.; Yang, S.; Zhang, W.; Xu, M.; Ma, A.; Zhuang, G.; Chen, G.; Liu, W. Diversity and dynamics of the microbial community on decomposing wheat straw during mushroom compost production. Bioresour. Technol. 2014, 170, 183–195. [Google Scholar] [CrossRef]
- Mcgee, C.F.; Byrne, H.; Irvine, A.; Wilson, J. Diversity and dynamics of the DNA and cDNA-derived bacterial compost communities throughout the Agaricus bisporus mushroom cropping process. Ann. Microbiol. 2017, 67, 751–761. [Google Scholar] [CrossRef]
- Agrawal, P.K.; Agrawal, S.; Shrivastava, R. Modern molecular approaches for analyzing microbial diversity from mushroom compost ecosystem. Biotech 2015, 5, 853–866. [Google Scholar] [CrossRef]
- Salar, R.K.; Aneja, K.R. Significance of thermophilic fungi in mushroom compost preparation: Effect on growth and yield of Agaricus bisporus (Lange) Sing. J. Agric. Technol. 2007, 3, 241–253. [Google Scholar]
- Asker, D. High throughput screening and profiling of high-value carotenoids from a wide diversity of bacteria in surface seawater. Food Chem. 2018, 261, 103–111. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Agarwal, P.K.; Johri, B.N. Evaluation of in situ functional activity of casing soils during growth cycle of mushroom (Agaricus bisporus (Lange) Imbach) employing community level physiological profiles (CLPPs). Indian J. Microbiol. 2010, 50, 19–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, C.L.; Zeng, G.M.; Huang, D.L.; Hu, S.; Zhao, M.H.; Lai, C.; Huang, C.; Wei, Z.; Li, N.J. Effect of ligninolytic enzymes on lignin degradation and carbon utilization during lignocellulosic waste composting. Process Biochem. 2011, 46, 1515–1520. [Google Scholar] [CrossRef]
- Zhao, M.; Yin, C.; Tao, Y.; Li, C.; Fang, S. Diversity of soil microbial community identified by Biolog method and the associated soil characteristics on reclaimed Scirpus mariqueter wetlands. SN Appl. Sci. 2019, 1, e1408. [Google Scholar] [CrossRef]
- Farnet, A.M.; Qasemian, L.; Peter-Valence, F.; Ruaudel, F.; Savoie, J.M.; Ferré, E. Capacity for colonization and degradation of horse manure and wheat-straw-based compost by different strains of Agaricus subrufescens during the first two weeks of cultivation. Bioresour. Technol. 2013, 131, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Farnet, A.M.; Qasemian, L.; Peter-Valence, F.; Ruaudel, F.; Savoie, J.M.; Roussos, S.; Gaime-Perraud, I.; Ziarelli, F.; Ferré, É. Do spawn storage conditions influence the colonization capacity of a wheat-straw-based substrate by Agaricus subrufescens? C R Biol. 2014, 337, 443–450. [Google Scholar] [CrossRef]
- Fan, F.; Zhang, B.; Morrill, P.L. Phospholipid fatty acid (PLFA) analysis for profiling microbial communities in offshore produced water. Mar. Pollut. Bull. 2017, 122, 194–206. [Google Scholar] [CrossRef]
- Vos, A.M.; Heijboer, A.; Boschker, H.T.S.; Bonnet, B.; Lugones, L.G.; Wösten, H.A.B. Microbial biomass in compost during colonization of Agaricus bisporus. AMB Express 2017, 7, e12. [Google Scholar] [CrossRef]
- Song, T.; Cai, W.; Jin, Q.; Feng, W.; Fan, L.; Shen, Y.; Fang, H.; Tian, F. Comparison of microbial communities and histological changes in Phase I rice straw-based Agaricus bisporus compost prepared using two composting methods. Sci. Hortic. 2014, 174, 96–104. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Baath, E. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol. Biochem. 2010, 42, 516–520. [Google Scholar] [CrossRef]
- Ellis, S.; Ritz, K. A modified high-throughput analysis of PLFAs in soil. MethodsX 2018, 5, 1491–1497. [Google Scholar] [CrossRef]
- Cowie, B.R.; Slater, G.F.; Bernier, L.; Warren, L.A. Carbon isotope fractionation in phospholipid fatty acid biomarkers of bacteria and fungi native to an acid mine drainage lake. Org. Geochem. 2009, 40, 956–962. [Google Scholar] [CrossRef]
- Yu, H.; Zeng, G.; Huang, H.; Xi, X.; Wang, R.; Huang, D.; Huang, G.L.J. Microbial community succession and lignocellulose degradation during agricultural waste composting. Biodegradation 2007, 18, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yun, J.; Meng, L.; Li, Y. Denaturing gradient gel electrophoresis analysis on bacterial community change in the phase II composting of Volvariella volvacea substrate. Wei Sheng Wu Xue Bao 2012, 52, 977–984. [Google Scholar] [PubMed]
- Balázs, M.; Rónavári, A.; Németh, A.; Bihari, Z.; Rutkai, E.; Bartos, P.; Kiss, I.; Szvetnik, A. Effect of DNA polymerases on PCR-DGGE patterns. Int. Biodeterior. Biodegrad. 2013, 84, 244–249. [Google Scholar] [CrossRef]
- Wang, S.; He, J. Two-step denaturing gradient gel electrophoresis (2S-DGGE), a gel-based strategy to capture full-length 16S rRNA gene sequences. Appl. Microbiol. Biotechnol. 2012, 95, 1305–1312. [Google Scholar] [CrossRef]
- Sklarz, M.Y.; Angel, R.; Gillor, O.; Soares, M.I.M. Evaluating amplified rDNA restriction analysis assay for identification of bacterial communities. Antonie Leeuwenhoek 2009, 96, 659–664. [Google Scholar] [CrossRef]
- Singh, A.V.; Sharma, A.; Johri, B.N. Phylogenetic profiling of culturable bacteria associated with early phase of mushroom composting assessed by amplified rDNA restriction analysis. Ann. Microbiol. 2012, 62, 675–682. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, A.V.; Johri, B.N. Functional and genetic characterization of culturable bacteria associated with late phase of mushroom composting assessed by amplified rDNA restriction analysis. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 162–175. [Google Scholar]
- Fan, T.; Sun, Y.; Peng, J.; Wu, Q.; Ma, Y.; Zhou, X. Combination of amplified rDNA restriction analysis and high-throughput sequencing revealed the negative effect of colistin sulfate on the diversity of soil microorganisms. Microbiol. Res. 2018, 206, 9–15. [Google Scholar] [CrossRef]
- Chandna, P.; Mallik, S.; Kuhad, R.C. Assessment of bacterial diversity in agricultural by-product compost by sequencing of cultivated isolates and amplified rDNA restriction analysis. Appl. Microbiol. Biotechnol. 2013, 97, 6991–7003. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Sanni, A.I.; Jeyaram, K. Rapid differentiation among Lactobacillus, Pediococcus and Weissella species from some Nigerian indigenous fermented foods. LWT-Food Sci. Technol. 2017, 77, 39–44. [Google Scholar] [CrossRef]
- Schütte, U.M.E.; Abdo, Z.; Bent, S.J.; Shyu, C.; Williams, C.J.; Pierson, J.D.; Forney, L.J. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 2008, 80, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Vajna, B.; Szili, D.; Nagy, A.; Márialigeti, K. An improved sequence-aided T-RFLP analysis of bacterial succession during oyster mushroom substrate preparation. Microb. Ecol. 2012, 64, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mo, Y.; Yang, J.; Zhou, J.; Lin, Y.; Isabwe, A.; Zhang, J.; Gao, X.; Yu, Z. Genetic diversity pattern of microeukaryotic communities and its relationship with the environment based on PCR-DGGE and T-RFLP techniques in Dongshan Bay, southeast China. Cont. Shelf. Res. 2018, 164, 1–9. [Google Scholar] [CrossRef]
- Ovreas, L. Population and community level approaches for analyzing microbial diversity in natural environments. Eco. Lett. 2000, 3, 236–251. [Google Scholar] [CrossRef]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High throughput sequencing: An overview of sequencing chemistry. Indian J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef]
- Aka, B.E.Z.; Djeni, T.N.; Amoikon, S.L.T.; Kannengiesser, J.; Ouazzani, N.; Dje, M.K. High-throughput 16S rRNA gene sequencing of the microbial community associated with palm oil mill effluents of two oil processing systems. Sci. Rep. 2021, 11, e13232. [Google Scholar] [CrossRef]
- Vieira, F.R.; Pecchia, J.A. An exploration into the bacterial community under different pasteurization conditions during substrate preparation (composting–phase II) for Agaricus bisporus cultivation. Environ. Microbiol. 2018, 75, 318–330. [Google Scholar] [CrossRef]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic analysis of microbial consortia enriched from compost: New insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol. Biofuels 2016, 9, e22. [Google Scholar] [CrossRef]
- Karczewski, K.; Riss, W.H.; Meyer, E.I. Comparison of DNA-fingerprinting (T-RFLP) and high-throughput sequencing (HTS) to assess the diversity and composition of microbial communities in groundwater ecosystems. Limnologica 2017, 67, 45–53. [Google Scholar] [CrossRef]
- Largeteau, M.L.; Regnault-Roger, C.; Savoie, J.M. Verticillium disease of Agaricus bisporus: Variations in host contribution to total fungal DNA in relation to symptom heterogeneity. Eur. J. Plant Pathol. 2007, 118, 155–164. [Google Scholar] [CrossRef]
- Wang, H.W.; Xu, M.; Cai, X.-Y.; Tian, F. Evaluation of soil microbial communities and enzyme activities in cucumber continuous cropping soil treated with spent mushroom (Flammulina velutipes) substrate. J. Soils. Sediments 2021, 21, 2938–2951. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Hu, D.D.; Ma, X.T.; Li, S.G.; Gu, J.G.; Hu, Q.X. Adopting stick spawn reduced the spawn running time and improved mushroom yield and biological efficiency of Pleurotus eryngii. Sci. Hortic. 2014, 175, 156–159. [Google Scholar] [CrossRef]
- Suarez, C.; Ratering, S.; Weigel, V.; Sacharow, J.; Bienhaus, J.; Ebert, J.; Hirza, A.; Rüh, M.; Schnell, S. Isolation of bacteria at different points of Pleurotus ostreatus cultivation and their influence in mycelial growth. Microbiol. Res. 2020, 234, 126393. [Google Scholar] [CrossRef]
- Longley, R.; Benucci, G.N.M.; Mills, G.; Bonito, G. Fungal and bacterial community dynamics in substrates during the cultivation of morels (Morchella rufobrunnea) indoors. FEMS Microbiol. Lett. 2019, 366, fnz215. [Google Scholar] [CrossRef]
- Zhang, B.; Yan, L.; Li, Q.; Zou, J.; Tan, H.; Tan, W.; Peng, W.; Li, X.; Zhang, X. Dynamic succession of substrate-associated bacterial composition and function during Ganoderma lucidum growth. PeerJ 2018, 6, e4975. [Google Scholar] [CrossRef]
- Rossouw, W.; Korsten, L. Cultivable microbiome of fresh white button mushrooms. Lett. Appl. Microbiol. 2016, 64, 164–170. [Google Scholar] [CrossRef]
- Cerda, A.; Artola, A.; Font, X.; Barrena, R.; Gea, T.; Sánche, A. Composting of food wastes: Status and challenges. Bioresour. Technol. 2018, 248, 57–67. [Google Scholar] [CrossRef]
- Guerriero, G.; Hausman, J.F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Destructuring plant biomass: Focus on fungal and extremophilic cell wall hydrolases. Plant Sci. 2015, 234, 180–193. [Google Scholar] [CrossRef]
- Martins, L.F.; Antunes, L.P.; Pascon, R.C.; de Oliveira, J.C.F.; Digiampietri, L.A.; Barbosa, D.; Peixoto, B.M.; Vallim, M.A.; Viana-Niero, C.; Ostroski, E.H.; et al. Metagenomic analysis of a tropical composting operation at the Sao Paulo Zoo Park reveals diversity of biomass degradation functions and organisms. PLoS ONE 2013, 8, e61928. [Google Scholar] [CrossRef]
- McGee, C.F. Microbial ecology of the Agaricus bisporus mushroom cropping process. Appl. Microbiol. Biotechnol. 2018, 102, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Liu, Y.; Shen, G.R.; Guo, L.X.; Zhou, X.W. Investigation and analysis of microbiological communities in natural Ophiocordyceps sinensis. Can. J. Microbiol. 2015, 61, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Chen, X.; Guo, M.Y.; Bai, X.H.; Liu, Y.; Shen, G.R.; Li, Y.L.; Lin, J.; Zhou, X.W. High-throughput sequencing-based analysis of endogenetic fungal communities inhabiting the Chinese Cordyceps reveals unexpectedly high fungal diversity. Sci. Rep. 2016, 6, e3343. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Zhou, X.; Liu, Y.; Bai, X.; Zhou, X. Composition and predictive functional analysis of bacterial communities inhabiting Chinese Cordyceps insight into conserved core microbiome. BMC Microbiol. 2019, 19, e105. [Google Scholar] [CrossRef]
- Bai, F.; Li, Z.; Umezawa, A.; Terada, N.; Jin, S. Bacterial type III secretion system as a protein delivery tool for a broad range of biomedical applications. Biotechnol. Adv. 2018, 36, 482–493. [Google Scholar] [CrossRef]
- Pent, M.; Poldmaa, K.; Bahram, M. Bacterial communities in boreal forest mushrooms are shaped both by soil parameters and host identity. Front. Microbiol. 2017, 8, e836. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Murata, H.; Shirata, A. Identification of non-Pseudomonad bacteria from fruit bodies of wild Agaricales fungi that detoxify tolaasin produced by Pseudomonas tolaasii. Biosci. Biotechnol. Biochem. 2002, 66, 2201–2208. [Google Scholar] [CrossRef]
- Cho, Y.S.; Kim, J.S.; Crowley, D.E.; Cho, B.G. Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol. Lett. 2003, 218, 271–276. [Google Scholar] [CrossRef]
- Yang, L.; Yang, H.L.; Tu, Z.C.; Wang, X.I. High-throughput sequencing of microbial community diversity and dynamics during Douchi fermentation. PLoS ONE 2016, 11, e0168166. [Google Scholar] [CrossRef]
- Nielsen, J.L.; Nguyen, H.; Meyer, R.L.; Nielsen, P.H. Identification of glucose-fermenting bacteria in a full-scale enhanced biological phosphorus removal plant by stable isotope probing. Microbiology 2012, 158, 1818–1825. [Google Scholar] [CrossRef]
- Sangeetha, T.; Guo, Z.; Liu, W.; Gao, L.; Wang, L.; Cui, M.; Chen, C.; Wang, A. Energy recovery evaluation in an up flow microbial electrolysis coupled anaerobic digestion (ME-AD) reactor: Role of electrode positions and hydraulic retention times. Appl. Energy 2017, 206, 1214–1224. [Google Scholar] [CrossRef]
- Zhong, X.Z.; Ma, S.C.; Wang, S.P.; Wang, T.T.; Sun, Z.Y.; Tang, Y.Q.; Deng, Y.; Kida, K. A comparative study of composting the solid fraction of dairy manure with or without bulking material: Performance and microbial community dynamics. Bioresour. Technol. 2018, 247, 443–452. [Google Scholar] [CrossRef] [PubMed]
- López-González, J.A.; Suárez-Estrella, F.; Vargas-García, M.C.; López, M.J.; Jurado, M.M.; Moreno, J. Dynamics of bacterial microbiota during lignocellulosic waste composting: Studies upon its structure, functionality and biodiversity. Bioresour. Technol. 2015, 175, 406–416. [Google Scholar] [CrossRef]
- Zhou, G.; Xu, X.; Qiu, X.; Zhang, J. Biochar influences the succession of microbial communities and the metabolic functions during rice straw composting with pig manure. Bioresour. Technol. 2019, 272, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhou, G.; Zhang, J.; Wang, W. Microbial community responses to biochar addition when a green waste and manure mix are composted: A molecular ecological network analysis. Bioresour. Technol. 2019, 273, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, L.; Hassan, M.; Xie, B. Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresour. Technol. 2018, 256, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Sharmin, F.; Wakelin, S.; Huygens, F.; Hargreaves, M. Firmicutes dominate the bacterial taxa within sugar-cane processing plants. Sci. Rep. 2012, 3, e3107. [Google Scholar] [CrossRef]
- Wang, C.; Guo, X.; Deng, H.; Dong, D.; Tu, Q.; Wu, W. New insights into the structure and dynamics of actinomycetal community during manure composting. Appl. Microbiol. Biotechnol. 2014, 98, 3327–3337. [Google Scholar] [CrossRef]
- Partanen, P.; Hultman, J.; Paulin, L.; Auvinen, P.; Romantschuk, M. Bacterial diversity at different stages of the composting process. BMC Microbiol. 2010, 10, e94. [Google Scholar] [CrossRef]
- Pantazaki, A.A.; Pritsa, A.A.; Kyriakidis, D.A. Biotechnologically relevant enzymes from Thermus thermophilus. Appl. Microbiol. Biotechnol. 2002, 58, 1–12. [Google Scholar] [CrossRef]
- Insam, H.; de Bertoldi, M. Compost Science and Technology. In Microbiology of the Composting Process, Waste Manage Series; Elsevier: Amsterdam, The Netherlands, 2007; Volume 8, pp. 25–48. [Google Scholar]
- de Gannes, V.; Eudoxie, G.; Hickey, W.J. Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour. Technol. 2013, 133, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Azadnia, P.; Zamani, M.H.; Ghasemi, S.A.; Babaki, A.K.; Jashni, M.K.; Taarof, N. Isolation and Identification of Thermophilic Lactobacilli from Traditional Yogurts of Tribes of Kazerun. J. Anim. Vet. Adv. 2011, 10, 774–776. [Google Scholar]
- Barbieri, E.; Ceccaroli, P.; Saltarelli, R.; Guidi, C.; Potenza, L.; Basaglia, M.; Fontana, F.; Baldan, E.; Casella, S.; Ryahi, O.; et al. New evidence for nitrogen fixation within the Italian white truffle Tuber magnatum. Fungal Biol. 2010, 114, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Aydoğdu, M.; Sülü, S.M.; Kurbetli, İ.; Sülü, G. In vitro and in vivo biological control of the green mold using different bacteria in button mushroom cultivation. Egypt. J. Biol. Pest. Control 2021, 31, e70. [Google Scholar] [CrossRef]
- Baars, J.J.P.; Scholtmeijer, K.; Sonnenberg, A.S.M.; Van Peer, A. Critical factors involved in primordia building in Agaricus bisporus: A review. Molecules 2020, 25, 2984. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Naraian, R. Enhanced growth and yield of oyster mushroom by growth-promoting bacteria Glutamicibacter arilaitensis MRC119. J. Basic Microbiol. 2020, 61, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Benucci, G.M.N.; Bonito, G.M. The Truffle microbiome: Species and geography effects on bacteria associated with fruiting bodies of Hypogeous pezizales. Microb. Ecol. 2016, 72, 4–8. [Google Scholar] [CrossRef]
- Vieira, F.R.; Pecchia, J.A. Fungal community assembly during a high-temperature composting under different pasteurization regimes used to elaborate the Agaricus bisporus substrate. Fungal Biol. 2021, 25, 826–833. [Google Scholar] [CrossRef]
- Basotra, N.; Kaur, B.; Di Falco, M.; Tsang, A.; Chadha, B.S. Mycothermus thermophilus (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresour. Technol. 2016, 222, 413–421. [Google Scholar] [CrossRef]
- Romdhane, I.B.B.; Achouri, I.M.; Belghith, H. Improvement of highly thermostable xylanases production by Talaromyces thermophilus for the agro-industrials residue hydrolysis. Appl. Biochem. Biotechnol. 2010, 162, 1635–1646. [Google Scholar] [CrossRef]
- Singh, S.; Madlala, A.M.; Prior, B.A. Thermomyces lanuginosus: Properties of strains and their hemicellulases. FEMS Microbiol. Rev. 2003, 27, 3–16. [Google Scholar] [CrossRef]
- Eastwood, D.C.; Herman, B.; Noble, R.; Dobrovin-Pennington, A.; Sreenivasaprasad, S.; Burton, K.S. Environmental regulation of reproductive phase change in Agaricus bisporus by 1-octen-3-ol, temperature and CO2. Fung. Genet. Biol. 2013, 55, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Kalkhove, S.I.C.; Lugones, L.G.; Baars, J.J.P.; Wosten, H.A.B.; Bakker, P. Effects of the mushroom-volatile 1-octen-3-ol on dry bubble disease. Appl. Microbiol. Biotechnol. 2013, 97, 5535–5543. [Google Scholar] [CrossRef] [PubMed]
- Wever, G.; van der Burg, A.M.M.; Straatsma, G. Potential of adapted mushroom compost as a growing medium in horticulture. Acta Hortic. 2005, 697, 171–177. [Google Scholar] [CrossRef]
- Velusami, B.; Curran, T.P.; Grogan, H. Hydrogen sulfide gas emissions during disturbance and removal of stored spent mushroom compost. J. Agric. Saf. Health 2013, 19, 261–275. [Google Scholar]
- Zhang, C.; Huang, T.; Shen, C.; Wang, X.; Qi, Y.; Shen, J.; Song, A.; Qiu, L.; Ai, Y. Downregulation of ethylene production increases mycelial growth and primordia formation in the button culinary-medicinal mushroom, Agaricus bisporus (agaricomycetes). Int. J. Med. Mushrooms 2016, 18, 1131–1140. [Google Scholar] [CrossRef]
- Kurtzman, R.H. Cobalt Chloride and Ethylene Affect Fruiting of Agaricus bisporus. Mycologia 1995, 87, 366–369. [Google Scholar] [CrossRef]
- Lee, E.Y.; Lee, C.W. Isolation and nitrogen removal characteristics of heterotrophic nitrification-aerobic denitrifying bacteria, Stenotrophomonas sp. CW-4Y. Korean Soc. Biotechnol. Bioeng. 2014, 29, 72–80. [Google Scholar]
- Pauliuc, I.; Botau, D. Antibacterial activity of Pleurotus ostreatus gemmotherapic extract. J. Hortic. Biotechnol. 2013, 17, 242–245. [Google Scholar]

| Mushroom Cultivation | Agricultural Materials | Biological Efficiency (%) | References |
|---|---|---|---|
| Hericium erinaceus | Sawdust | 50.3 | [21] |
| Wheat straw | 43.5 | ||
| Wheat straw | 19.5 | [22] | |
| Oak sawdust | 37.3 | ||
| Poplar sawdust | 32.4 | ||
| Common vetch straw | 28.2 | ||
| Volvariella volvacea | Paddy straw | 10.2–14.9 | [23] |
| Banana leaves | 15.2 | [24] | |
| Oil palm empty fruit bunch | 3.6–6.5 | [25] | |
| Ganoderma lucidum | Oat straw | 2.3 | [26] |
| Sawdust (Swietenia mahagoni) | 4.3–7.6 | [27] | |
| Sawdust (Dipterocarpur turbinatus) | 3.6–6.8 | ||
| Sawdust (Tectona grandis) | 0.0 | ||
| Auricularia polytricha | Sawdust | 90.0 | [28] |
| Sawdust | 113.6 | [29] | |
| Sawdust supplement with oil palm frond | 184.8 | ||
| Sawdust supplement with empty fruit bunch | 195.6 | ||
| Lentinula edodes | Rice straw | 36.1–49.7 | [30] |
| Rice straw | 48.7 | [31] | |
| Wheat straw | 66.0 | ||
| Barley straw | 64.1–88.6 | ||
| Sugarcane bagasse | 130.2–133.4 | [32] | |
| Sugarcane leaves | 82.7–97.8 | ||
| Bracts of pineapple crown | 37.5–36.3 | ||
| Pleurotus eryngii | Ramie stalk | 51.0 | [33] |
| Kenaf stalk | 52.4 | ||
| Bulrush stalk | 36.8 | ||
| Cotton seed hull | 45.2 | ||
| Wheat straw | 48.2 | [34] | |
| Rice straw | 45.9 | ||
| Corn cobs | 51.8 | ||
| Sugarcane bagasse | 41.3 | ||
| Sawdust | 35.5 | ||
| Pleurotus ostreatus | Corn cob maize residues | 14.0 | [35] |
| Composted sawdust | 60.1 | ||
| Beech sawdust | 33.5 | [36] | |
| Non-composted sawdust | 4.3 | [37] | |
| Composted sawdust | 61.0 | ||
| Composted sawdust | 107.3 | [38] | |
| Wheat straw | 52.6 | [36] | |
| Rice straw | 50.6 | [37] | |
| Banana leaves | 37.1 | ||
| Non-composted corncob | 66.8 | [39] | |
| Sawdust | 46.4 | ||
| Sugarcane bagasse | 65.6 | ||
| Wheat straw | 105.0 | [40] | |
| Wheat straw with spent coffee grounds | 101.7 | ||
| Pleurotus cystidiosus | Non-composted corncob | 50.1 | [39] |
| Sawdust | 36.2 | ||
| Sugarcane bagasse | 49.5 | ||
| Agrocybe cylindracea | Beech sawdust | 38.3 | [36] |
| Wheat straw | 61.4 | ||
| Wheat straw | 23.0–36.0 | [41] | |
| Flammulina velutipes | Rubber tree sawdust and rice straw (1:1) | 123.9 | [42] |
| Lentinus sajor-caju | Wheat straw | 74.9 | [43] |
| Rice straw | 78.3 | ||
| Soya stalk | 83.0 | ||
| Sunflower stalk | 63.1 | ||
| Agaricus bisporus | Composted wheat straw | 47.2–100.3 | [44,45] |
| Composted oat straw | 47.2–52.9 | [46] | |
| Agaricus subrufescens | Composted wheat straw | 6.6–53.7 | [44,47] |
| Agaricus blazei (A. subrufescens) | One year-fermented horse manure bedding compost | 62.1 | [48,49] |
| One year-fermented horse manure bedding compost with sawdust | 24.9–27.7 | ||
| One year-fermented horse manure bedding compost with corncob | 20.0–25.3 | ||
| One year-fermented horse manure bedding compost with woodchips | 22.6–53.1 |
| Substrate Types | Dominant Fungi | Properties Related to the Cultivation | Method of Use | Mushrooms | References | ||
|---|---|---|---|---|---|---|---|
| Phylum | Class/Genus | Species | |||||
| Soil | Ascomycota | - | - | - | High-throughput sequencing of ITS gene | C. militaris | [16] |
| Composting (phase II) | Ascomycota | Scytalidium Thermomyces | S. thermophilum T. lanuginosus T. badanensis | - | PCR and DGGE | A. subrufescens | [56] |
| Peach sawdust-b ased composting | Ascomycota | Eurotiomycetes | - | Lignocellulosic degradation | Metagenomic ITS sequencing | - | [80] |
| Sordariomycetes | - | Lignocellulosic degradation | |||||
| Compost (wheat straw) | Ascomycota | Chaetomium Malbranchea Thermomyces Torula | C. thermophile M. sulfurea T. lanuginosus T. thermophila | - - - - | Culture-dependent method | A. bisporus | [93] |
| Compost | Ascomycota | Mycothermus | Thermophilic fungi My. thermophilus | Produce lignocellulolytic enzymes | DNA recovery (Amplicon sequencing) | A. bisporus | [164,165] |
| Thermomyces | T. thermophilus | Produce hemicellulase | [166] | ||||
| Thermomyces | T. lanuginosus | Produce xylanase | [167] | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suwannarach, N.; Kumla, J.; Zhao, Y.; Kakumyan, P. Impact of Cultivation Substrate and Microbial Community on Improving Mushroom Productivity: A Review. Biology 2022, 11, 569. https://doi.org/10.3390/biology11040569
Suwannarach N, Kumla J, Zhao Y, Kakumyan P. Impact of Cultivation Substrate and Microbial Community on Improving Mushroom Productivity: A Review. Biology. 2022; 11(4):569. https://doi.org/10.3390/biology11040569
Chicago/Turabian StyleSuwannarach, Nakarin, Jaturong Kumla, Yan Zhao, and Pattana Kakumyan. 2022. "Impact of Cultivation Substrate and Microbial Community on Improving Mushroom Productivity: A Review" Biology 11, no. 4: 569. https://doi.org/10.3390/biology11040569
APA StyleSuwannarach, N., Kumla, J., Zhao, Y., & Kakumyan, P. (2022). Impact of Cultivation Substrate and Microbial Community on Improving Mushroom Productivity: A Review. Biology, 11(4), 569. https://doi.org/10.3390/biology11040569








