Induction of Stem-Cell-Derived Cardiomyogenesis by Fibroblast Growth Factor 10 (FGF10) and Its Interplay with Cardiotrophin-1 (CT-1)
Abstract
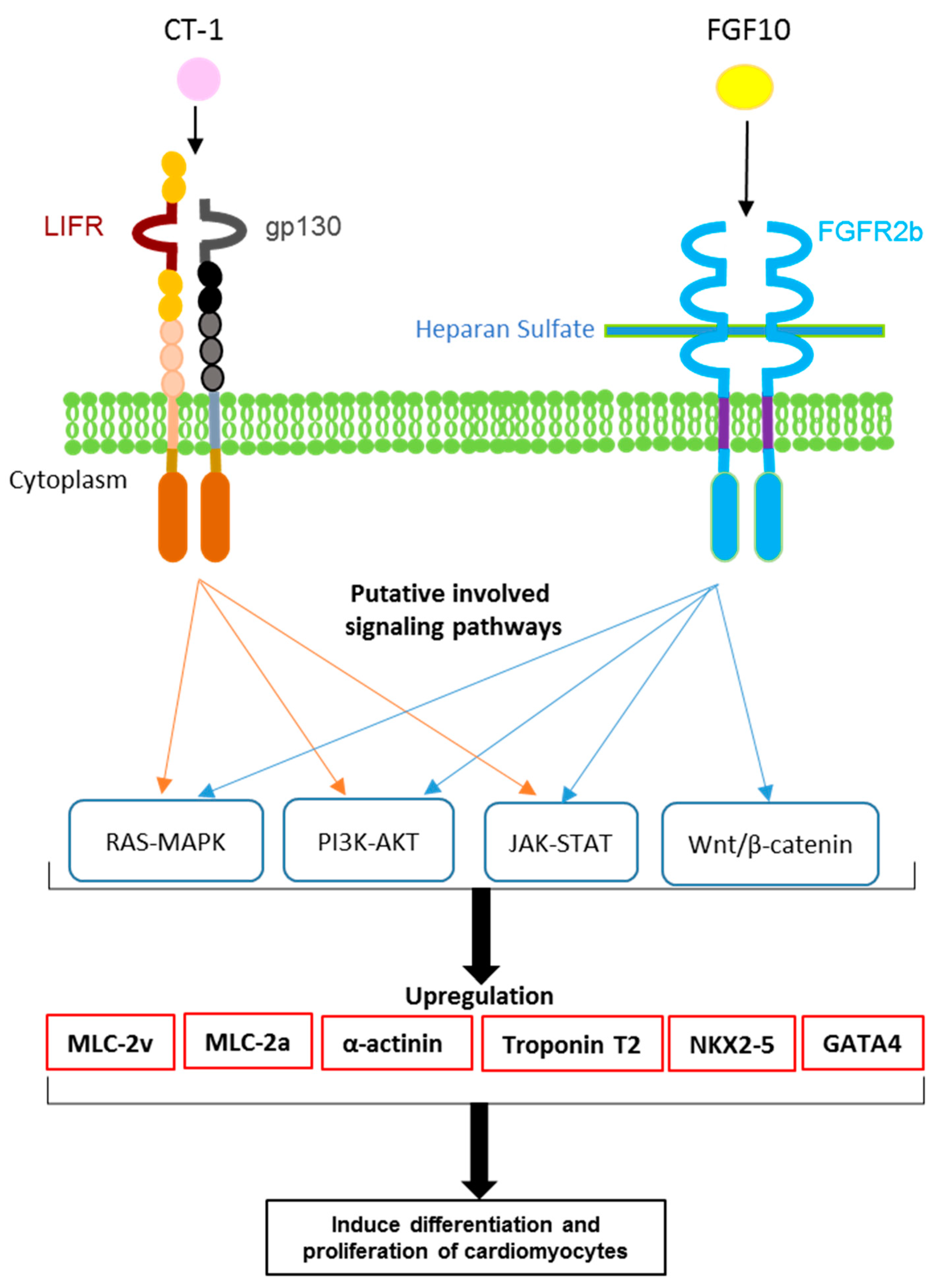
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Embryoid Body Formation and Contractile Activity Analysis during Cell Culture of ES Cells
2.3. Immunocytochemistry and Confocal Imaging
2.4. Quantitative Real-Time PCR- qPCR Analysis
2.5. Flow Cytometry
2.6. Western Blot
2.7. Statistical Analysis
3. Results
3.1. FGF 10 and CT-1 + FGF 10 Elevate Contraction Rate of Embryoid Bodies
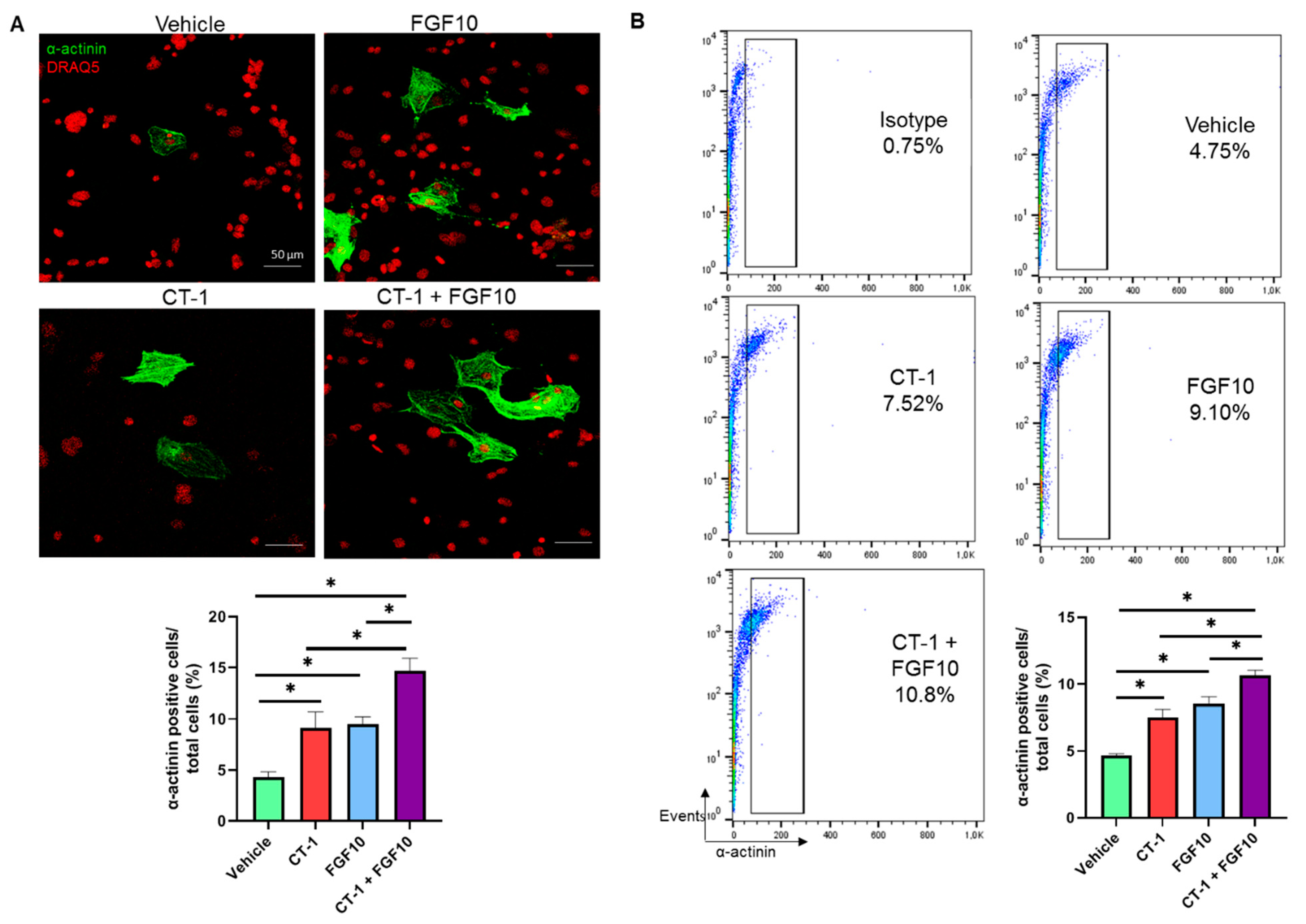
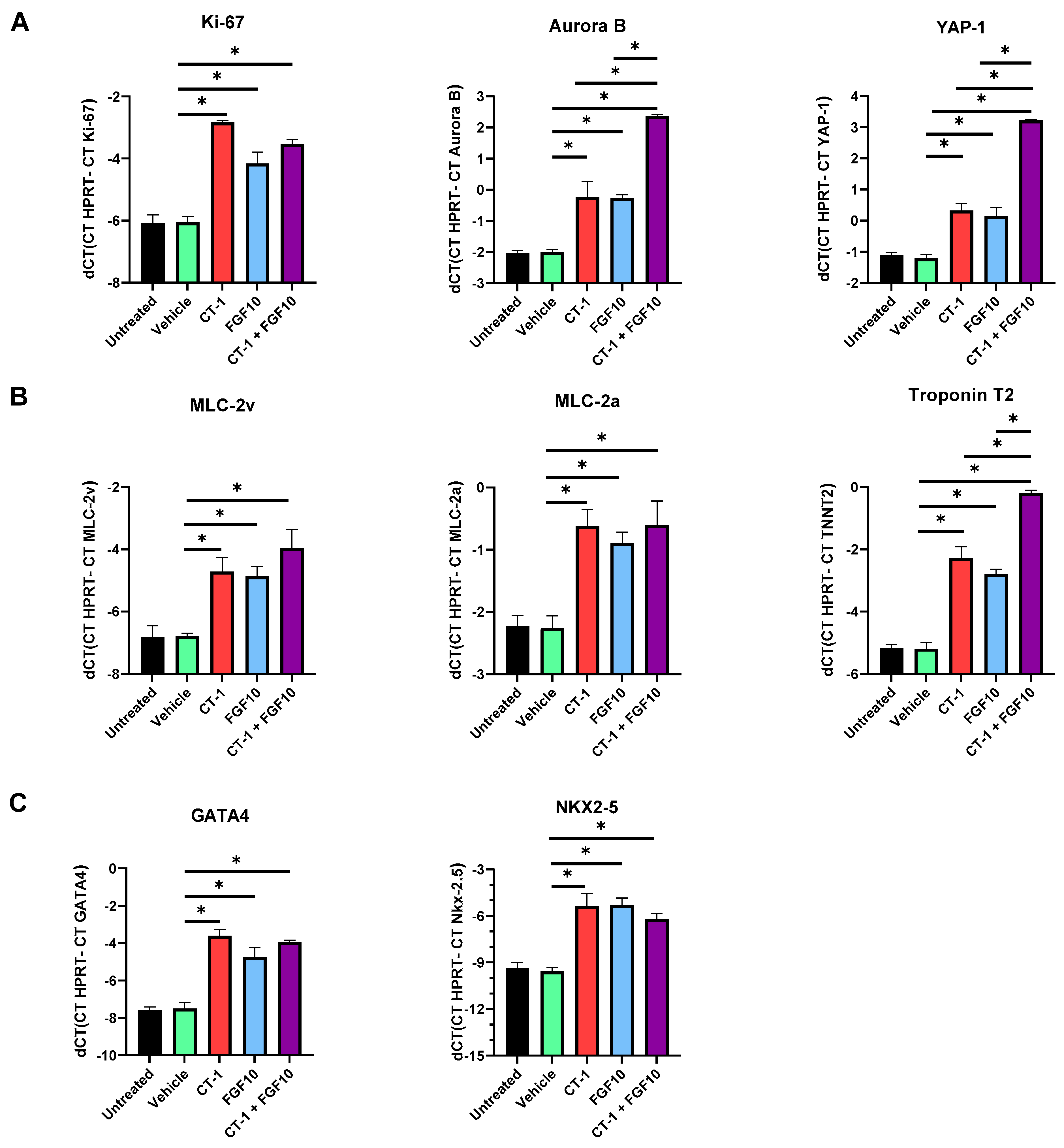
3.2. FGF10 Increases the Proliferation Rate of Cardiomyocytes
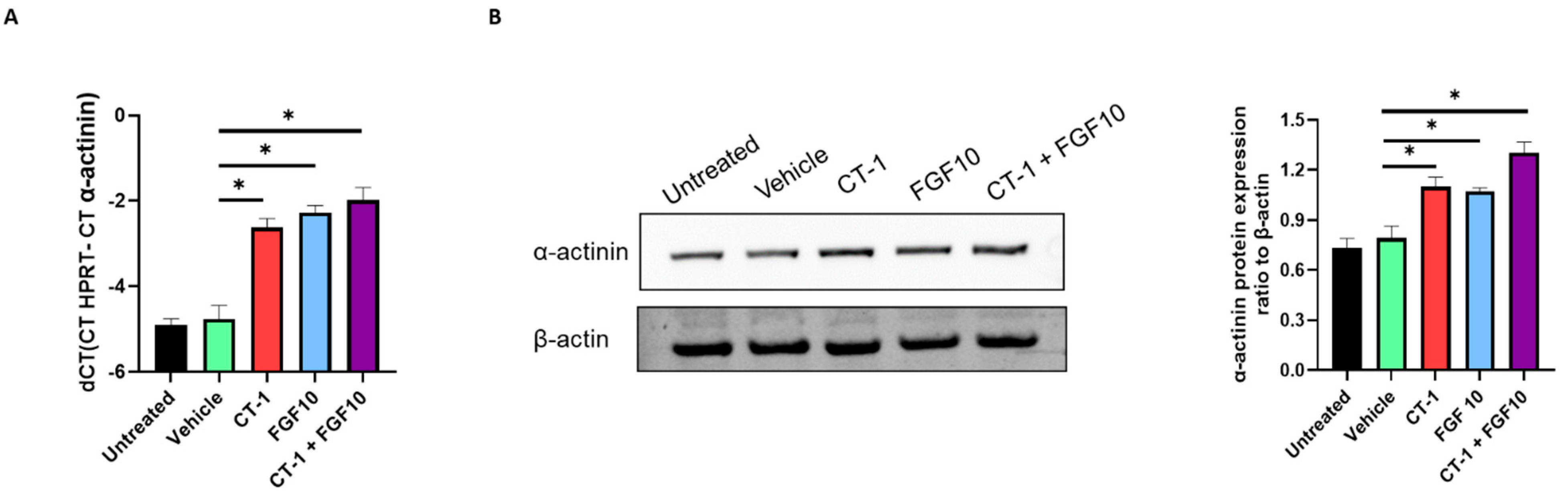
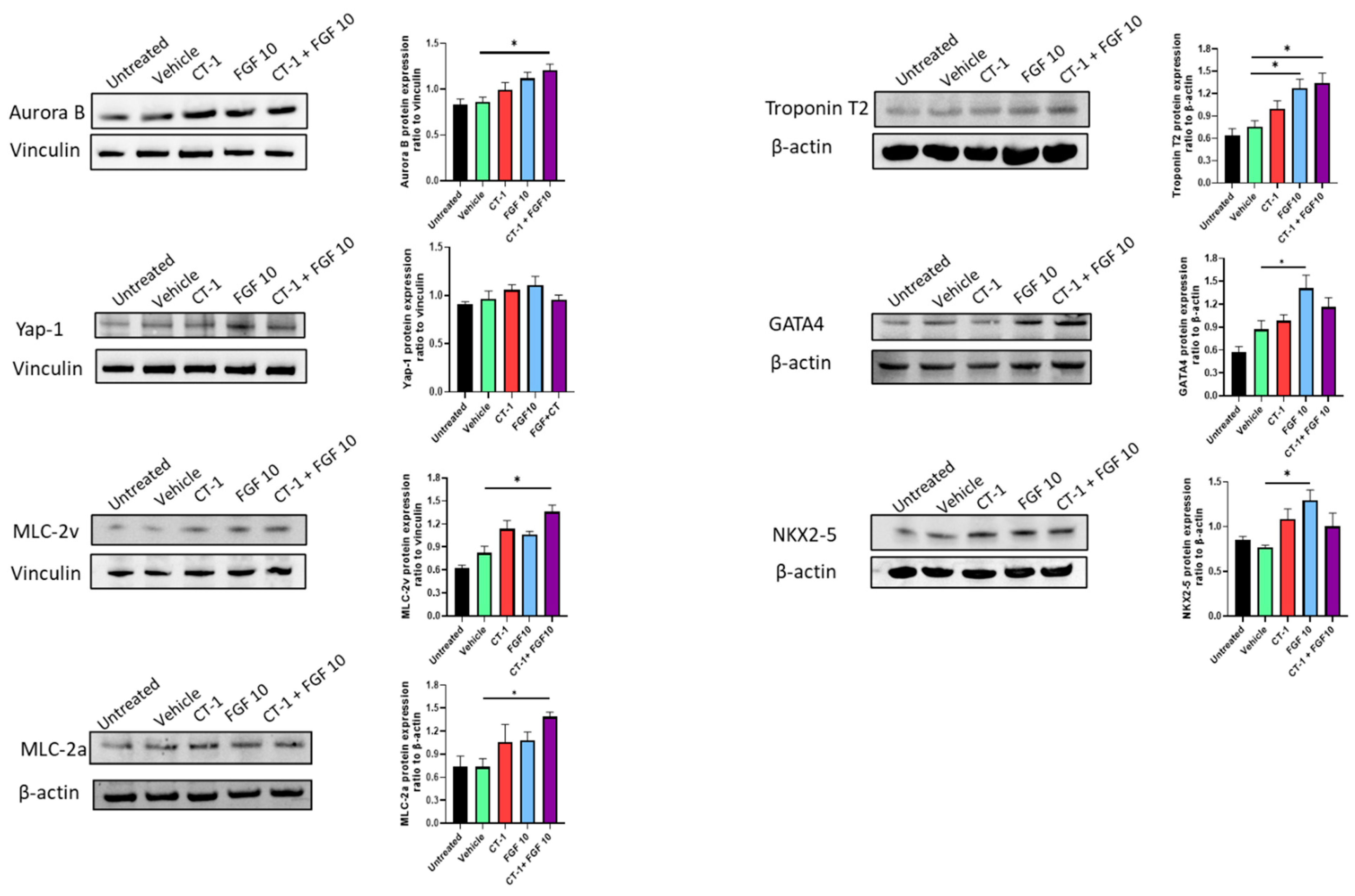
3.3. FGF10 and CT-1 Enhance Cardiomyocyte Structural, Transcriptional and Proliferation Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eschenhagen, T.; Bolli, R.; Braun, T.; Field, L.J.; Fleischmann, B.K.; Frisén, J.; Giacca, M.; Hare, J.M.; Houser, S.; Lee, R.T.; et al. Cardiomyocyte regeneration: A consensus statement. Circulation 2017, 136, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, F.; Ahmadvand, N.; Bellusci, S.; Sauer, H. The Multifunctional Contribution of FGF Signaling to Cardiac Development, Homeostasis, Disease and Repair. Front. Cell Dev. Biol. 2021, 9, 672935. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Ohta, H.; Nakayama, Y.; Konishi, M. Roles of FGF signals in heart development, health, and disease. Front. Cell Dev. Biol. 2016, 4, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marguerie, A.; Bajolle, F.; Zaffran, S.; Brown, N.A.; Dickson, C.; Buckingham, M.E.; Kelly, R.G. Congenital heart defects in Fgfr2-IIIb and Fgf10 mutant mice. Cardiovasc. Res. 2006, 71, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Rochais, F.; Sturny, R.; Chao, C.M.; Mesbah, K.; Bennett, M.; Mohun, T.J.; Bellusci, S.; Kelly, R.G. FGF10 promotes regional foetal cardiomyocyte proliferation and adult cardiomyocyte cell-cycle re-entry. Cardiovasc. Res. 2014, 104, 432–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, R.G.; Brown, N.A.; Buckingham, M.E. The Arterial Pole of the Mouse Heart Forms from Fgf10-Expressing Cells in Pharyngeal Mesoderm. Dev. Cell 2001, 1, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Miyagawa-Tomita, S.; Vincent, S.D.; Kelly, R.G.; Moon, A.M.; Buckingham, M.E. Role of mesodermal FGF8 and FGF10 overlaps in the development of the arterial pole of the heart and pharyngeal arch arteries. Circ. Res. 2010, 106, 495–503. [Google Scholar] [CrossRef] [Green Version]
- Urness, L.D.; Bleyl, S.B.; Wright, T.J.; Moon, A.M.; Mansour, S.L. Redundant and dosage sensitive requirements for Fgf3 and Fgf10 in cardiovascular development. Dev. Biol. 2011, 356, 383–397. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Zhou, S.; Wang, Q.; Meng, Z.; Peng, J.; Zhou, Y.; Song, W.; Wang, J.; Chen, S. Mutations in fibroblast growth factor (FGF8) and FGF10 identified in patients with conotruncal defects. J. Transl. Med. 2020, 18, 283. [Google Scholar] [CrossRef]
- Sheikh, F.; Fandrich, R.R.; Kardami, E.; Cattini, P.A. Overexpression of long or short FGFR-1 results in FGF-2-mediated proliferation in neonatal cardiac myocyte cultures. Cardiovasc. Res. 1999, 42, 696–705. [Google Scholar] [CrossRef] [Green Version]
- Engel, F.B.; Hsieh, P.C.H.; Lee, R.T.; Keating, M.T. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc. Natl. Acad. Sci. USA 2006, 103, 15546–15551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennbacken, K.; Wågberg, F.; Karlsson, U.; Eriksson, J.; Magnusson, L.; Chimienti, M.; Ricchiuto, P.; Bernström, J.; Ding, M.; Ross-Thriepland, D.; et al. Phenotypic screen with the human secretome identifies FGF16 as inducing proliferation of iPSC-derived cardiac progenitor cells. Int. J. Mol. Sci. 2019, 20, 6037. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, X.; Capasso, J.M.; Gerdes, A.M. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell. Cardiol. 1996, 28, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; D’Agostino, G.; Loo, S.J.; Wang, C.X.; Su, L.P.; Tan, S.H.; Tee, G.Z.; Pua, C.J.; Pena, E.M.; Cheng, R.B.; et al. Early regenerative capacity in the porcine heart. Circulation 2018, 138, 2798–2808. [Google Scholar] [CrossRef] [PubMed]
- Vega-Hernández, M.; Kovacs, A.; de Langhe, S.; Ornitz, D.M. FGF10/FGFR2b signaling is essential for cardiac fibroblast development and growth of the myocardium. Development 2011, 138, 3331–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.S.K.; Li, H.J.; Hsueh, Y.C.; Lee, D.S.; Chen, J.H.; Hwang, S.M.; Chen, C.Y.; Shih, E.; Hsieh, P.C.H. Fibroblast growth factor-10 promotes cardiomyocyte differentiation from embryonic and induced pluripotent stem cells. PLoS ONE 2010, 5, e14414. [Google Scholar] [CrossRef] [Green Version]
- Kuwahara, K.; Saito, Y.; Harada, M.; Ishikawa, M.; Ogawa, E.; Miyamoto, Y.; Hamanaka, I.; Kamitani, S.; Kajiyama, N.; Takahashi, N.; et al. Involvement of cardiotrophin-1 in cardiac myocyte-nonmyocyte interactions during hypertrophy of rat cardiac myocytes in vitro. Circulation 1999, 100, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Sauer, H.; Neukirchen, W.; Rahimi, G.; Grünheck, F.; Hescheler, J.; Wartenberg, M. Involvement of reactive oxygen species in cardiotrophin-1-induced proliferation of cardiomyocytes differentiated from murine embryonic stem cells. Exp. Cell Res. 2004, 294, 313–324. [Google Scholar] [CrossRef]
- Mascheck, L.; Sharifpanah, F.; Tsang, S.Y.; Wartenberg, M.; Sauer, H. Stimulation of cardiomyogenesis from mouse embryonic stem cells by nuclear translocation of cardiotrophin-1. Int. J. Cardiol. 2015, 193, 23–33. [Google Scholar] [CrossRef]
- Jiang, Z.S.; Jeyaraman, M.; Wen, G.B.; Fandrich, R.R.; Dixon, I.M.C.; Cattini, P.A.; Kardami, E. High- but not low-molecular weight FGF-2 causes cardiac hypertrophy in vivo; possible involvement of cardiotrophin-1. J. Mol. Cell. Cardiol. 2007, 42, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.D.; Wang, Z.; Lepore, J.J.; Min, M.L.; Taketo, M.M.; Epstein, D.J.; Morrisey, E.E. Wnt/β-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Investig. 2007, 117, 1794–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzahor, E.; Poss, K.D. Cardiac regeneration strategies: Staying young at heart. Science 2017, 356, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali Darehzereshki, N.R. FGF10 Signaling Enhances Epicardial Cell Expansion during Neonatal Mouse Heart Repair. J. Cardiovasc. Dis. Diagn. 2013, 1, 101. [Google Scholar] [CrossRef]
- Leone, M.; Magadum, A.; Engel, F.B. Cardiomyocyte proliferation in cardiac development and regeneration: A guide to methodologies and interpretations. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1237–H1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbayo, E.; Gavira, J.J.; De Yebenes, M.G.; Pelacho, B.; Abizanda, G.; Lana, H.; Blanco-Prieto, M.J.; Prosper, F. Catheter-based intramyocardial injection of FGF1 or NRG1-loaded MPs improves cardiac function in a preclinical model of ischemia-reperfusion. Sci. Rep. 2016, 6, 25932. [Google Scholar] [CrossRef]
- Pascual-Gil, S.; Simón-Yarza, T.; Garbayo, E.; Prósper, F.; Blanco-Prieto, M.J. Cytokine-loaded PLGA and PEG-PLGA microparticles showed similar heart regeneration in a rat myocardial infarction model. Int. J. Pharm. 2017, 523, 531–533. [Google Scholar] [CrossRef]
- Bortolotti, F.; Ruozi, G.; Falcione, A.; Doimo, S.; Dal Ferro, M.; Lesizza, P.; Zentilin, L.; Banks, L.; Zacchigna, S.; Giacca, M. In Vivo Functional Selection Identifies Cardiotrophin-1 as a Cardiac Engraftment Factor for Mesenchymal Stromal Cells. Circulation 2017, 136, 1509–1524. [Google Scholar] [CrossRef]
- Cai, H.; Wu, F.Y.; Wang, Q.L.; Xu, P.; Mou, F.F.; Shao, S.J.; Luo, Z.R.; Zhu, J.; Xuan, S.S.; Lu, R.; et al. Self-assembling peptide modified with QHREDGS as a novel delivery system for mesenchymal stem cell transplantation after myocardial infarction. FASEB J. 2019, 33, 8306–8320. [Google Scholar] [CrossRef]

| Gene | Forward Primer (5′->3′) | Reverse Primer (5′->3′) |
|---|---|---|
| Hprt | CCTAAGATGAGCGCAAGTTGAA | CCACAGGACTAGAACACCTGCTAA |
| Actn2 | GTCAACACTCCCAAACCCGA | CTCCAACAGCTCACTCGCTA |
| Myl7 | GGCACAACGTGGCTCTTCTA | GAACACTTACCCTCCCGA GC |
| MYL2 | CTCCAAAGAGGAGATCGACCAG | TGTTTATTTGCGCACAGCCC |
| Yap-1 | GAGCAAGCCATGACTCAGGA | CTCTGGTTCATGGCAAAACGA |
| Ki-67 | CTGCGAGCTTCACCGAGAG | CAATACTCCTTCCAAACAGGCAG |
| Tnnt2 | CCACATGCCTGCTTAAAGCTC | CTCGGCTCTCCCTCTGAAC |
| Aurkb | CGGGAGAAGAAGAGCCGTTT | AGGATGTTGGGATGTTTCAGGT |
| Gata4 | GAGCAGGGGACAAGCCG | CGAAGCGGCAGTCCTGG |
| Nkx2-5 | CCCAAGTGCTCTCCTGCTTT | AGCGCGCACAGCTCTTTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khosravi, F.; Ahmadvand, N.; Wartenberg, M.; Sauer, H. Induction of Stem-Cell-Derived Cardiomyogenesis by Fibroblast Growth Factor 10 (FGF10) and Its Interplay with Cardiotrophin-1 (CT-1). Biology 2022, 11, 534. https://doi.org/10.3390/biology11040534
Khosravi F, Ahmadvand N, Wartenberg M, Sauer H. Induction of Stem-Cell-Derived Cardiomyogenesis by Fibroblast Growth Factor 10 (FGF10) and Its Interplay with Cardiotrophin-1 (CT-1). Biology. 2022; 11(4):534. https://doi.org/10.3390/biology11040534
Chicago/Turabian StyleKhosravi, Farhad, Negah Ahmadvand, Maria Wartenberg, and Heinrich Sauer. 2022. "Induction of Stem-Cell-Derived Cardiomyogenesis by Fibroblast Growth Factor 10 (FGF10) and Its Interplay with Cardiotrophin-1 (CT-1)" Biology 11, no. 4: 534. https://doi.org/10.3390/biology11040534
APA StyleKhosravi, F., Ahmadvand, N., Wartenberg, M., & Sauer, H. (2022). Induction of Stem-Cell-Derived Cardiomyogenesis by Fibroblast Growth Factor 10 (FGF10) and Its Interplay with Cardiotrophin-1 (CT-1). Biology, 11(4), 534. https://doi.org/10.3390/biology11040534






