Intracellular Localization and Gene Expression Analysis Provides New Insights on LEA Proteins’ Diversity in Anhydrobiotic Cell Line
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Expression Vectors
2.3. Transfection and Protein Expression
2.4. Visualization of PvLEA Proteins Localization
2.5. RNA-Seq
2.6. Bioinformatic Analysis
3. Results
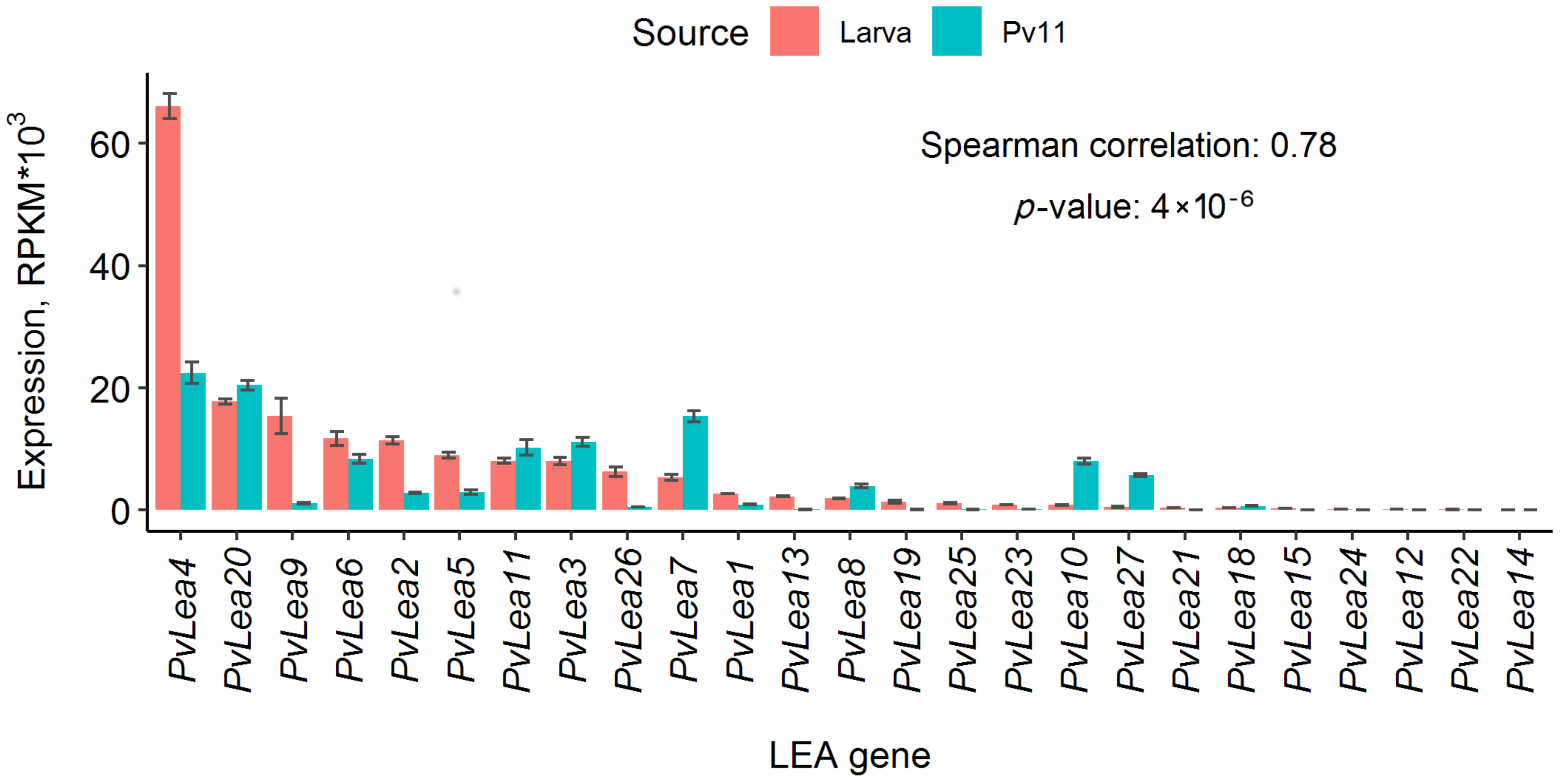
3.1. All Known PvLea Genes except PvLea16 and PvLea17 Are Expressed in Pv11 Cells
3.2. PvLea Genes Become Induced in Pv11 Cells on a Course of the Anhydrobiosis Cycle
3.3. Subcellular Localization of Some PvLEA Proteins Differs between Pv11 and Other Cell Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hand, S.C.; Menze, M.A. Molecular Approaches for Improving Desiccation Tolerance: Insights from the Brine Shrimp Artemia franciscana. Planta 2015, 242, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Cornette, R.; Yamamoto, N.; Yamamoto, M.; Kobayashi, T.; Petrova, N.A.; Gusev, O.; Shimura, S.; Kikawada, T.; Pemba, D.; Okuda, T. A New Anhydrobiotic Midge from Malawi, Polypedilum pembai Sp.n. (Diptera: Chironomidae), Closely Related to the Desiccation Tolerant Midge, Polypedilum vanderplanki Hinton. Syst. Entomol. 2017, 42, 814–825. [Google Scholar] [CrossRef]
- Cornette, R.; Kikawada, T. The Induction of Anhydrobiosis in the Sleeping Chironomid: Current Status of Our Knowledge. IUBMB Life 2011, 63, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Czernik, M.; Fidanza, A.; Luongo, F.P.; Valbonetti, L.; Scapolo, P.A. Late Embryogenesis Abundant (LEA) Proteins Confer Water Stress Tolerance to Mammalian Somatic Cells. Cryobiology 2020, 92, 189–196. [Google Scholar] [CrossRef]
- Hand, S.C.; Moore, D.S.; Patil, Y. Challenges during Diapause and Anhydrobiosis: Mitochondrial Bioenergetics and Desiccation Tolerance. IUBMB Life 2018, 70, 1251–1259. [Google Scholar] [CrossRef]
- Rashed, M.Z.; Belott, C.J.; Janis, B.R.; Menze, M.A.; Williams, S.J. New Insights into Anhydrobiosis Using Cellular Dielectrophoresis-Based Characterization. Biomicrofluidics 2019, 13, 064113. [Google Scholar] [CrossRef]
- Wolkers, W.F.; McCready, S.; Brandt, W.F.; Lindsey, G.G.; Hoekstra, F.A. Isolation and Characterization of a D-7 LEA Protein from Pollen That Stabilizes Glasses In Vitro. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1544, 196–206. [Google Scholar] [CrossRef]
- Hand, S.C.; Menze, M.A.; Toner, M.; Boswell, L.; Moore, D. LEA Proteins during Water Stress: Not Just for Plants Anymore. Annu. Rev. Physiol. 2011, 73, 115–134. [Google Scholar] [CrossRef]
- Yuen, F.; Watson, M.; Barker, R.; Grillo, I.; Heenan, R.K.; Tunnacliffe, A.; Routh, A.F. Preferential Adsorption to Air–Water Interfaces: A Novel Cryoprotective Mechanism for LEA Proteins. Biochem. J. 2019, 476, 1121–1135. [Google Scholar] [CrossRef]
- Gusev, O.A.; Suetsugu, Y.; Cornette, R.; Kawashima, T.; Logacheva, M.D.; Kondrashov, A.S.; Penin, A.A.; Hatanaka, R.; Kikuta, S.; Shimura, S.; et al. Comparative Genome Sequencing Reveals Genomic Signature of Extreme Desiccation Tolerance in the Anhydrobiotic Midge. Nat. Commun. 2014, 5, 4784. [Google Scholar] [CrossRef]
- Watanabe, K.; Imanishi, S.; Akiduki, G.; Cornette, R.; Okuda, T. Air-Dried Cells from the Anhydrobiotic Insect, Polypedilum vanderplanki, Can Survive Long Term Preservation at Room Temperature and Retain Proliferation Potential after Rehydration. Cryobiology 2016, 73, 93–98. [Google Scholar] [CrossRef]
- Nakahara, Y.; Imanishi, S.; Mitsumasu, K.; Kanamori, Y.; Iwata, K.; Watanabe, M.; Kikawada, T.; Okuda, T. Cells from an Anhydrobiotic Chironomid Survive Almost Complete Desiccation. Cryobiology 2010, 60, 138–146. [Google Scholar] [CrossRef]
- Hatanaka, R.; Gusev, O.A.; Cornette, R.; Shimura, S.; Kikuta, S.; Okada, J.; Okuda, T.; Kikawada, T. Diversity of the Expression Profiles of Late Embryogenesis Abundant (LEA) Protein Encoding Genes in the Anhydrobiotic Midge Polypedilum vanderplanki. Planta 2015, 242, 451–459. [Google Scholar] [CrossRef]
- Sogame, Y.; Kikawada, T. Current Findings on the Molecular Mechanisms Underlying Anhydrobiosis in Polypedilum vanderplanki. Curr. Opin. Insect Sci. 2017, 19, 16–21. [Google Scholar] [CrossRef]
- Miyata, Y.; Tokumoto, S.; Sogame, Y.; Deviatiiarov, R.; Okada, J.; Cornette, R.; Gusev, O.; Shagimardanova, E.; Sakurai, M.; Kikawada, T. Identification of a Novel Strong Promoter from the Anhydrobiotic Midge, Polypedilum vanderplanki, with Conserved Function in Various Insect Cell Lines. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdös, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Sogame, Y.; Okada, J.; Kikuta, S.; Miyata, Y.; Cornette, R.; Gusev, O.; Kikawada, T. Establishment of Gene Transfer and Gene Silencing Methods in a Desiccation-Tolerant Cell Line, Pv11. Extremophiles 2017, 21, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Voronina, T.A.; Nesmelov, A.A.; Kondratyeva, S.A.; Deviatiiarov, R.M.; Miyata, Y.; Tokumoto, S.; Cornette, R.; Gusev, O.A.; Kikawada, T.; Shagimardanova, E.I. New Group of Transmembrane Proteins Associated with Desiccation Tolerance in the Anhydrobiotic Midge Polypedilum vanderplanki. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Nakahara, Y.; Watanabe, M.; Fujita, A.; Kanamori, Y.; Tanaka, D.; Iwata, K.I.; Furuki, T.; Sakurai, M.; Kikawada, T.; Okuda, T. Effects of Dehydration Rate on Physiological Responses and Survival after Rehydration in Larvae of the Anhydrobiotic Chironomid. J. Insect Physiol. 2008, 54, 1220–1225. [Google Scholar] [CrossRef]
- Hatanaka, R.; Hagiwara-Komoda, Y.; Furuki, T.; Kanamori, Y.; Fujita, M.; Cornette, R.; Sakurai, M.; Okuda, T.; Kikawada, T. An Abundant LEA Protein in the Anhydrobiotic Midge, PvLEA4, Acts as a Molecular Shield by Limiting Growth of Aggregating Protein Particles. Insect Biochem. Mol. Biol. 2013, 43, 1055–1067. [Google Scholar] [CrossRef]
- Goyal, K.; Walton, L.J.; Browne, J.A.; Burnell, A.M.; Tunnacliffe, A. Molecular Anhydrobiology: Identifying Molecules Implicated in Invertebrate Anhydrobiosis. Integr. Comp. Biol. 2005, 45, 702–709. [Google Scholar] [CrossRef]
- Furuki, T.; Shimizu, T.; Chakrabortee, S.; Yamakawa, K.; Hatanaka, R.; Takahashi, T.; Kikawada, T.; Okuda, T.; Mihara, H.; Tunnacliffe, A.; et al. Effects of Group 3 LEA Protein Model Peptides on Desiccation-Induced Protein Aggregation. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 891–897. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) Proteins and Their Encoding Genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 6–24. [Google Scholar] [CrossRef]
- Boswell, L.C.; Hand, S.C. Intracellular Localization of Group 3 LEA Proteins in Embryos of Artemia franciscana. Tissue Cell 2014, 46, 514–519. [Google Scholar] [CrossRef]
- Ma, J.; Goryaynov, A.; Sarma, A.; Yang, W. Self-Regulated Viscous Channel in the Nuclear Pore Complex. Proc. Natl. Acad. Sci. USA 2012, 109, 7326–7331. [Google Scholar] [CrossRef]
- Junod, S.L.; Kelich, J.M.; Ma, J.; Yang, W. Nucleocytoplasmic Transport of Intrinsically Disordered Proteins Studied by High-Speed Super-Resolution Microscopy. Protein Sci. 2020, 29, 1459–1472. [Google Scholar] [CrossRef]
- Seibel, N.M.; Eljouni, J.; Nalaskowski, M.M.; Hampe, W. Nuclear Localization of Enhanced Green Fluorescent Protein Homomultimers. Anal. Biochem. 2007, 368, 95–99. [Google Scholar] [CrossRef]
- Ribbeck, K.; Görlich, D. Kinetic Analysis of Translocation through Nuclear Pore Complexes. EMBO J. 2001, 20, 1320. [Google Scholar] [CrossRef]
- Pancsa, R.; Fuxreiter, M. Interactions via Intrinsically Disordered Regions: What Kind of Motifs? IUBMB Life 2012, 64, 513–520. [Google Scholar] [CrossRef]
- Bauer, N.C.; Doetsch, P.W.; Corbett, A.H. Mechanisms Regulating Protein Localization. Traffic 2015, 16, 1039–1061. [Google Scholar] [CrossRef]
- França, M.B.; Panek, A.D.; Eleutherio, E.C.A. Oxidative Stress and Its Effects during Dehydration. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 146, 621–631. [Google Scholar] [CrossRef]
- Menze, M.A.; Hand, S.C. How Do Animal Mitochondria Tolerate Water Stress? Commun. Integr. Biol. 2009, 2, 428–430. [Google Scholar] [CrossRef][Green Version]
- Gusev, O.A.; Nakahara, Y.; Vanyagina, V.; Malutina, L.; Cornette, R.; Sakashita, T.; Hamada, N.; Kikawada, T.; Kobayashi, Y.; Okuda, T. Anhydrobiosis-Associated Nuclear DNA Damage and Repair in the Sleeping Chironomid: Linkage with Radioresistance. PLoS ONE 2010, 5, e14008. [Google Scholar] [CrossRef]



| Protein | WoLF PSORT * | CHO ** | Pv11 | Sf9 | ||
|---|---|---|---|---|---|---|
| C–Terminal AcGFP1 | N–Terminal AcGFP1 | C–Terminal AcGFP1 | N–Terminal AcGFP1 | C–Terminal AcGFP1 | ||
| PvLEA1 | ER | Cell membrane | ||||
| PvLEA2 | Nucleus | Cytosol and nucleus | ||||
| PvLEA3 | ER | Possibly ER | ER/ membrane | ER | ER/ membrane | Cytosol |
| PvLEA4 | Nucleus | Cytosol and nucleus | ||||
| PvLEA5 | Cytosol | |||||
| PvLEA6 | Cytosol | Cytosol | Cytosol and nucleus | Cytosol | Cytosol and nucleus | Cytosol |
| PvLEA7 | Nucleus | Cytosol and nucleus | ||||
| PvLEA8 | Cytosol | Cytosol and nucleus | ||||
| PvLEA9 | Cytosol and nucleus | |||||
| PvLEA10 | Nucleus | |||||
| PvLEA11 | Cytosol | |||||
| PvLEA12 | ||||||
| PvLEA13 | ||||||
| PvLEA14 | ||||||
| PvLEA15 | ||||||
| PvLEA16 | ||||||
| PvLEA17 | Mitochondria | |||||
| PvLEA18 | Cytosol | |||||
| PvLEA19 | Cytosol | Cytosol and nucleus | ||||
| PvLEA20 | Nucleus | |||||
| PvLEA21 | Cytosol | Cytosol | ||||
| PvLEA22 | ER | |||||
| PvLEA23 | Nucleus | Cytosol | ||||
| PvLEA24 | Cytosol | N/A | ||||
| PvLEA25 | Cytosol and nucleus | Cytosol and nucleus | ||||
| PvLEA26 | ||||||
| PvLEA27 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondratyeva, S.A.; Voronina, T.A.; Nesmelov, A.A.; Miyata, Y.; Tokumoto, S.; Cornette, R.; Vorontsova, M.V.; Kikawada, T.; Gusev, O.A.; Shagimardanova, E.I. Intracellular Localization and Gene Expression Analysis Provides New Insights on LEA Proteins’ Diversity in Anhydrobiotic Cell Line. Biology 2022, 11, 487. https://doi.org/10.3390/biology11040487
Kondratyeva SA, Voronina TA, Nesmelov AA, Miyata Y, Tokumoto S, Cornette R, Vorontsova MV, Kikawada T, Gusev OA, Shagimardanova EI. Intracellular Localization and Gene Expression Analysis Provides New Insights on LEA Proteins’ Diversity in Anhydrobiotic Cell Line. Biology. 2022; 11(4):487. https://doi.org/10.3390/biology11040487
Chicago/Turabian StyleKondratyeva, Sabina A., Taisiya A. Voronina, Alexander A. Nesmelov, Yugo Miyata, Shoko Tokumoto, Richard Cornette, Maria V. Vorontsova, Takahiro Kikawada, Oleg A. Gusev, and Elena I. Shagimardanova. 2022. "Intracellular Localization and Gene Expression Analysis Provides New Insights on LEA Proteins’ Diversity in Anhydrobiotic Cell Line" Biology 11, no. 4: 487. https://doi.org/10.3390/biology11040487
APA StyleKondratyeva, S. A., Voronina, T. A., Nesmelov, A. A., Miyata, Y., Tokumoto, S., Cornette, R., Vorontsova, M. V., Kikawada, T., Gusev, O. A., & Shagimardanova, E. I. (2022). Intracellular Localization and Gene Expression Analysis Provides New Insights on LEA Proteins’ Diversity in Anhydrobiotic Cell Line. Biology, 11(4), 487. https://doi.org/10.3390/biology11040487







