Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review
Abstract
Simple Summary
Abstract
Highlights
- Dissolution of As-bearing minerals by dissimilatory arsenic-reducing bacteria (DARB) releases As into the environment.
- Arsenic toxicity has a wide range of effects on plants and other environmental components.
- Shewanella spec.-mediated As detoxification could be a valuable tool for As remediation.
- Shewanella spec.-mediated methylation could limit the availability of toxic arsenicals in the environment.
- Flavins secreted by Shewanella oneidensis MR-1 facilitate As sequestration and detoxification.
1. Introduction
2. Arsenic Species and Their Behaviour in the Environment
Influence of Environmental Factors on the Mobilisation of As
3. Arsenic Toxicity and Its Consequences
4. Mitigation and Remediation Strategies for Arsenic
5. Iron and Its Impact on the Fate of Arsenic
Effect of Dissimilatory Iron-Reducing Bacteria’s Transformation of Iron on the Fate of Arsenic Soils
6. The Impact of Bacteria on Arsenic Transformation and Cycling
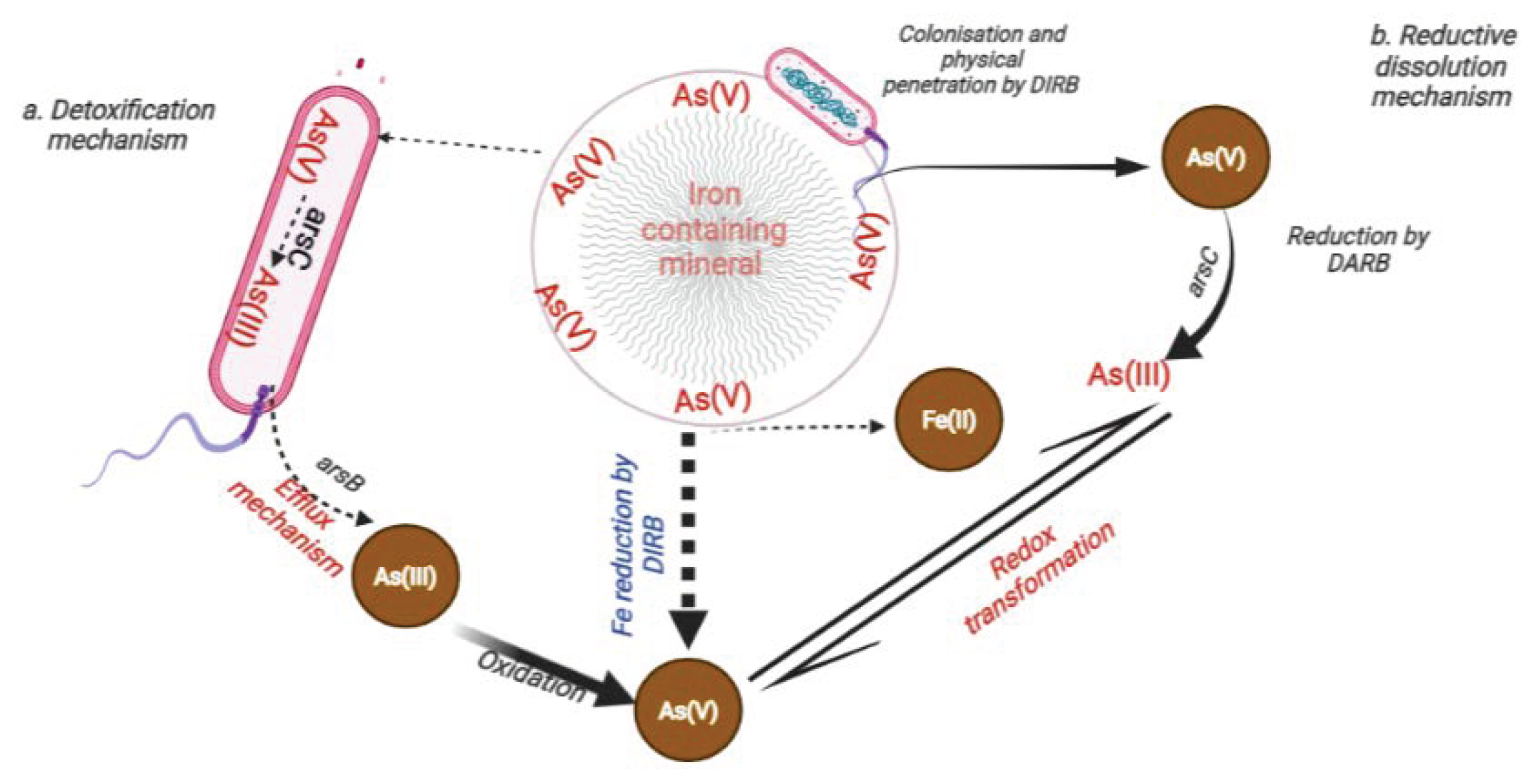
6.1. Influence of DARB on the Transformation and Mobilisation of Arsenic in the Environment
6.1.1. Uptake of Nutrients from As-Bearing Minerals by DARB
6.1.2. Dissolution of As-Bearing Minerals by DARB
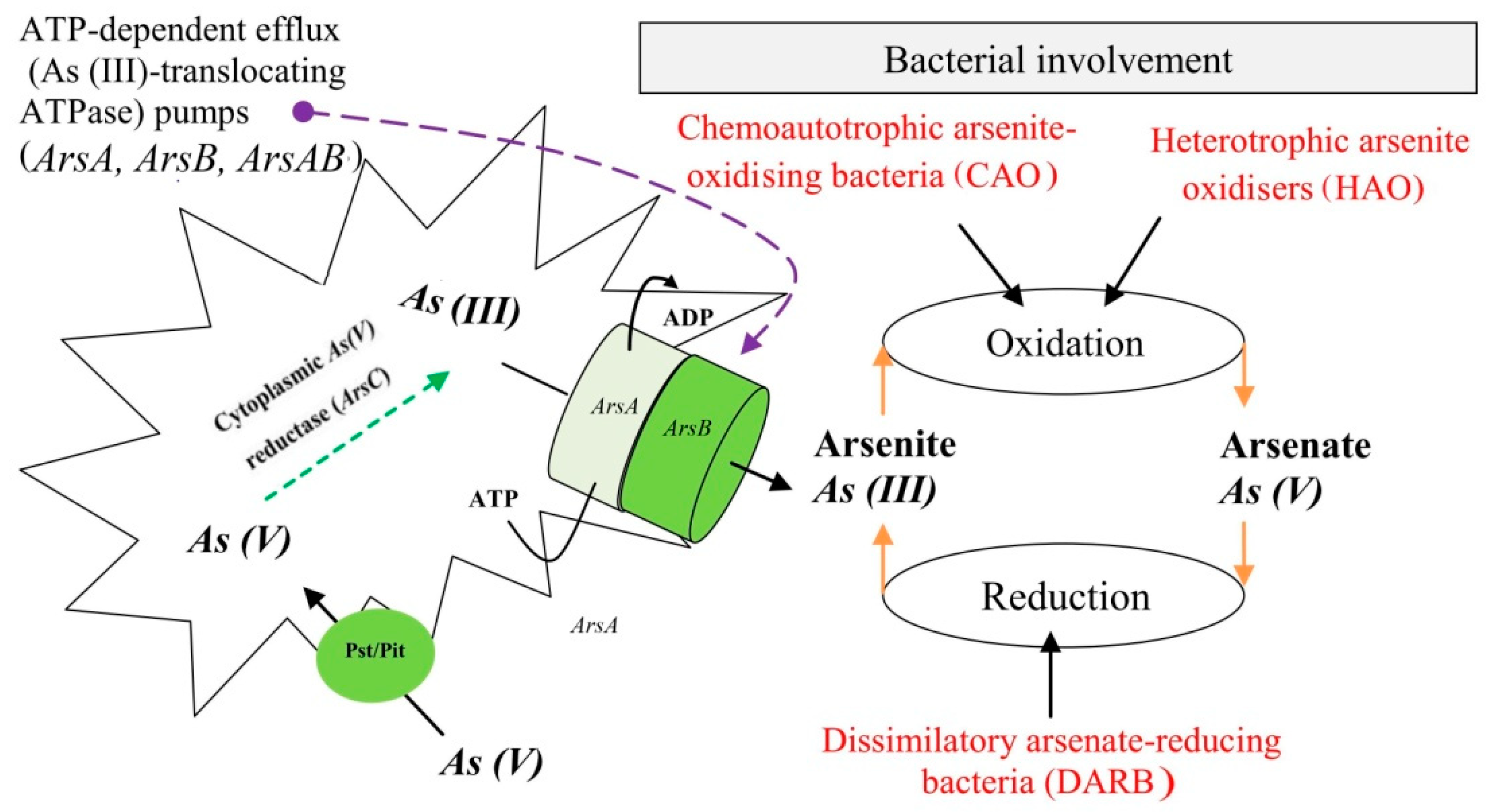
6.2. Heterotrophic Arsenite-Oxidising Bacteria (HAO)
6.3. Chemoautotrophic Arsenite-Oxidising Bacteria (CAO)
7. Removal and Detoxification of Transformed Arsenic
7.1. Mechanism of Arsenic Detoxification from DARB Cells
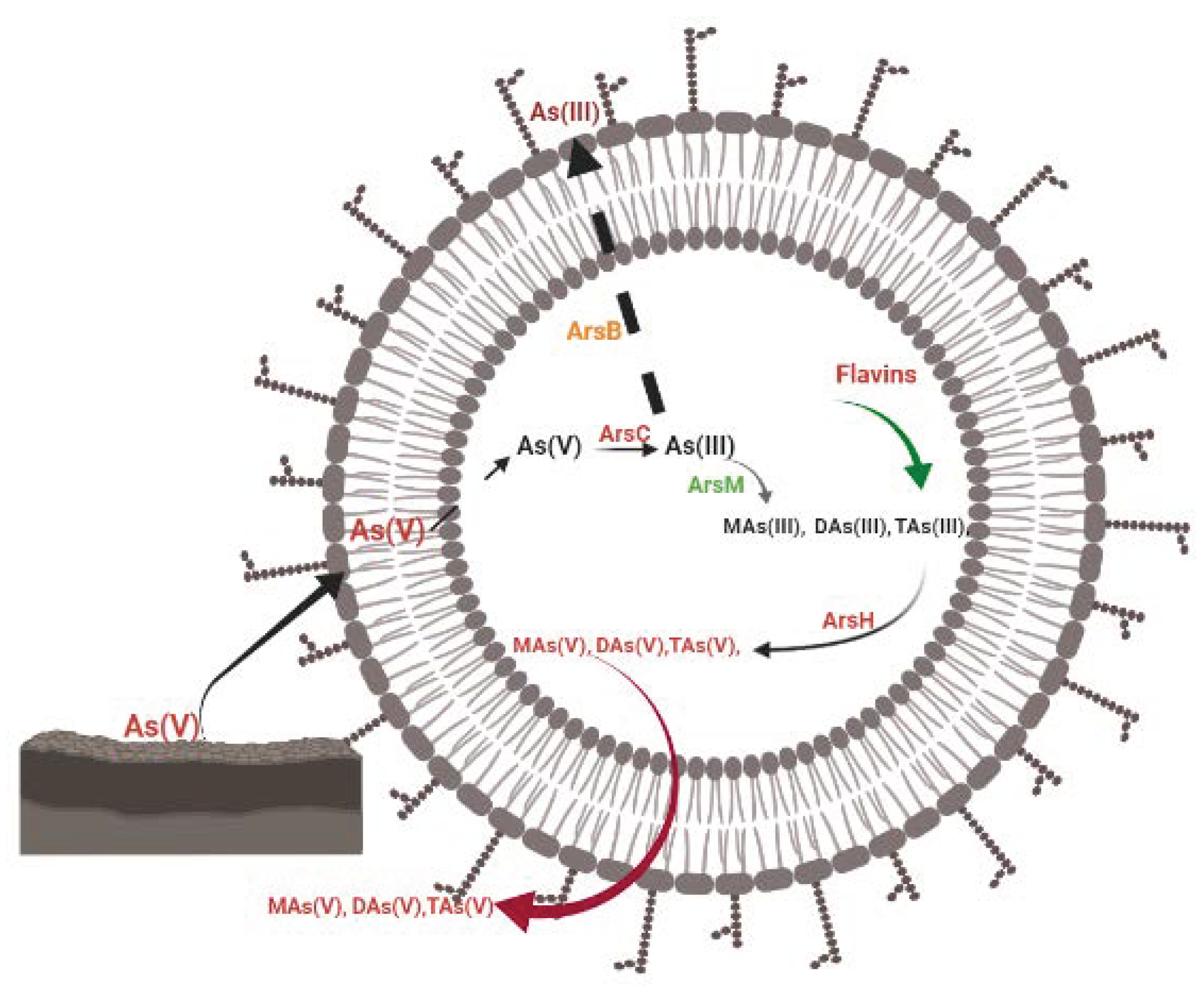
7.1.1. Ejection or Expulsion of As(III) from the Cell
7.1.2. As(III) Methylation to Trivalent Organoarsenicals
8. Contribution of Shewanella oneidensis MR-1 to As Redox Cycling and Remediation
8.1. Biology and Distribution of Shewanella Genus
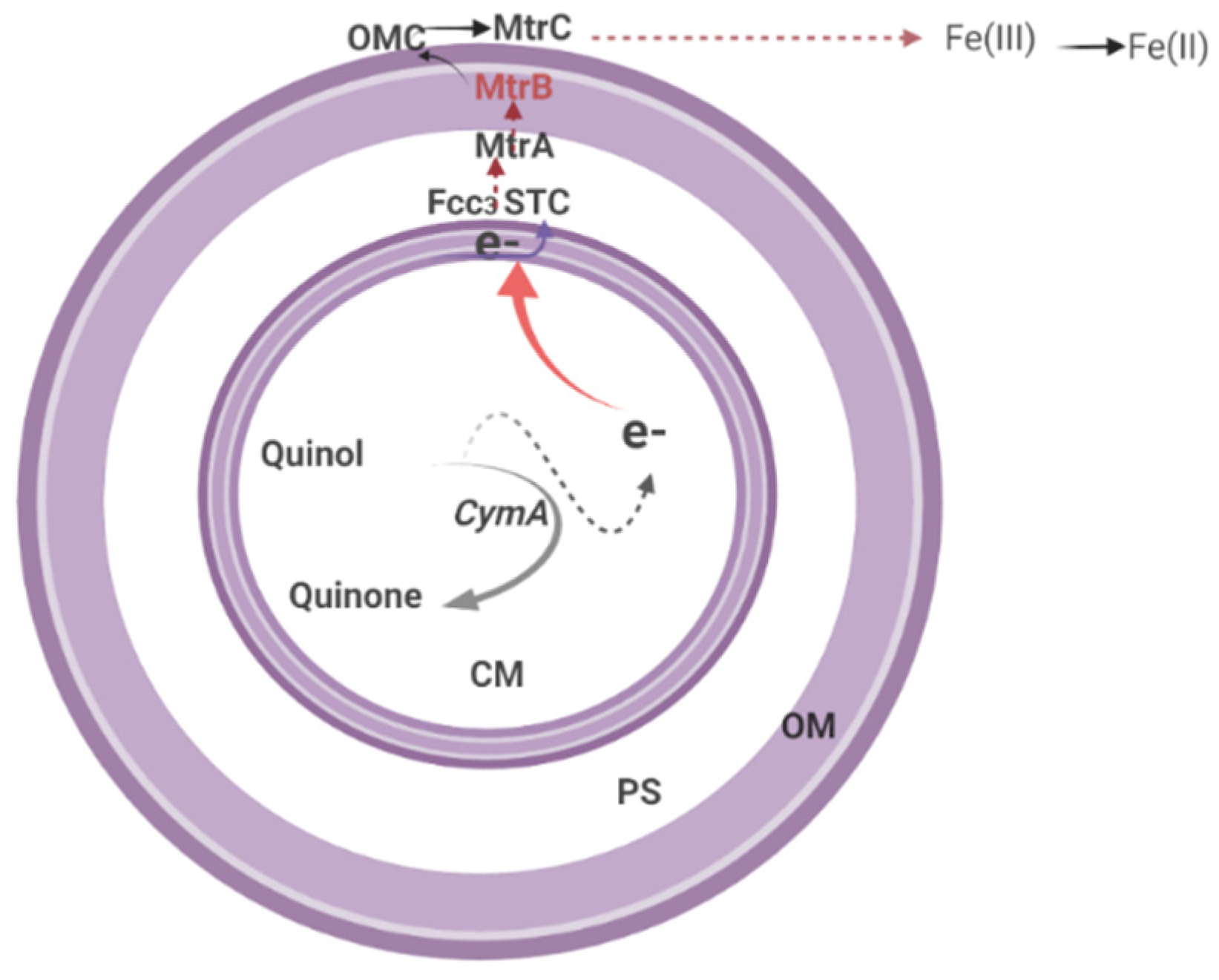
Extracellular Electron Transfer (EET)
8.2. EET Process in Shewanella oneidensis MR-1
9. Flavin’s Contributions to the Detoxification and Sequestration of Toxic Arsenite
10. Concluding Remarks
11. Future Direction
12. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garelick, H.; Jones, H.; Dybowska, A.; Valsami-Jones, E. Arsenic pollution sources. Rev. Environ. Contam. Toxicol. 2009, 197, 17–60. [Google Scholar] [CrossRef]
- Schnug, E.; Windmann, H.; Lottermoser, B.G.; Ulrich, A.E.; Bol, R.; Maekawa, M.; Haneklaus, S.H. Elemental loads with phosphate fertilizers—A constraint for soil productivity? In Soil Constraints and Productivity; Bolan, N.S., Kirkham, M.B., Eds.; CRC-Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Kratz, S.; Godlinski, F.; Schnug, E. Heavy metal loads to agricultural soils in Germany from the application of commercial phosphorus fertilizers and their contribution to background concentration in soils. In The New Uranium Mining Boom; Merkel, B., Schipek, M., Eds.; Springer Geology, Springer: Berlin/Heidelberg, Germany, 2011; pp. 755–762. [Google Scholar] [CrossRef]
- Vieira, C.; Morais, S.; Ramos, S.; Delerue-Matos, C.; Oliveira, M.B.P.P. Mercury, cadmium, lead and arsenic levels in three pelagic fish species from the Atlantic Ocean: Intra-and inter-specific variability and human health risks for consumption. Food Chem. Toxicol. 2011, 49, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Cobbina, S.J.; Chen, Y.; Zhou, Z.; Wu, X.; Zhao, T.; Zhang, Z.; Feng, W.; Wang, W.; Li, Q.; Wu, X.; et al. Toxicity assessment due to sub-chronic exposure to individual and mixtures of four toxic heavy metals. J. Hazard. Mater. 2015, 294, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sar, P. Microcosm based analysis of arsenic release potential of Bacillus sp. strain IIIJ3-1 under varying redox conditions. World J. Microbiol. Biotechnol. 2020, 36, 87. [Google Scholar] [CrossRef] [PubMed]
- Brammer, H.; Ravenscroft, P. Arsenic in groundwater: A threat to sustainable agriculture in South and South-east Asia. Environ. Int. 2009, 35, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Lukasz, D.; Liwia, R.; Aleksandra, M.; Aleksandra, S. Dissolution of arsenic minerals mediated by dissimilatory arsenate reducing bacteria: Estimation of the physiological potential for arsenic mobilization. BioMed Res. Int. 2014, 2014, 841892. [Google Scholar] [CrossRef] [PubMed]
- Drewniak, L.; Sklodowska, A. Arsenic-transforming microbes and their role in biomining processes. Environ. Sci. Pollut. Res. Int. 2013, 20, 7728–7739. [Google Scholar] [CrossRef]
- Wu, T.L.; Cui, X.D.; Cui, P.X.; Ata-Ul-Karim, S.T.; Sun, Q.; Liu, C.; Fan, T.T.; Gong, H.; Zhou, D.M.; Wang, Y.L. Speciation and location of arsenic and antimony in rice samples around antimony mining area. Environ. Pollut. 2019, 252, 1439–1447. [Google Scholar] [CrossRef]
- Yamamura, S.; Kurasawa, H.; Kashiwabara, Y.; Hori, T.; Aoyagi, T.; Nakajima, N.; Amachi, S. Soil microbial communities involved in reductive dissolution of arsenic from arsenate-laden minerals with different carbon sources. Environ. Sci. Technol. 2019, 53, 12398–12406. [Google Scholar] [CrossRef]
- Cai, X.; Wang, P.; Li, Z.; Li, Y.; Yin, N.; Du, H.; Cui, Y. Mobilization and transformation of arsenic from ternary complex OM-Fe (III)-As (V) in the presence of As (V)-reducing bacteria. J. Hazard. Mater. 2020, 381, 120975. [Google Scholar] [CrossRef]
- Schwertmann, U.; Taylor, R.M. Iron oxides. In Minerals in Soil Environments; SSSA Book Series 1; Dixon, J.B., Weed, S.B., Eds.; SSSA: Madison, WI, USA, 1989; pp. 379–438. [Google Scholar]
- Johnson, K.S.; Gordon, R.M.; Coale, K.H. What controls dissolved iron concentrations in the world ocean. Mar. Chem. 1997, 57, 137–161. [Google Scholar] [CrossRef]
- Huang, J.; Jones, A.; Waite, T.D.; Chen, Y.; Huang, X.; Rosso, K.M.; Kappler, A.; Mansor, M.; Tratnyek, P.G.; Zhang, H. Fe (II) redox chemistry in the environment. Chem. Rev. 2021, 121, 8161–8233. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.G.; Konhauser, K.O. Iron in Earth surface systems: A major player in chemical and biological processes. Elements 2011, 7, 83–88. [Google Scholar] [CrossRef]
- Chuan, M.C.; Shu, G.Y.; Liu, J.C. Solubility of heavy metals in a contaminated soil: Effects of redox potential and pH. Water Air Soil Pollut. 1996, 90, 543–556. [Google Scholar] [CrossRef]
- Burns, R.G. Rates and mechanisms of chemical weathering of ferromagnesian silicate minerals on Mars. Geochimi. Cosmochim. Acta 1993, 57, 4555–4574. [Google Scholar] [CrossRef]
- Salmon, T.P.; Rose, A.L.; Neilan, B.A.; Waite, T.D. The FeL model of iron acquisition: Nondissociative reduction of ferric complexes in the marine environment. Limnol. Oceanogr. 2006, 51, 1744–1754. [Google Scholar] [CrossRef]
- Caccavo Jr, F.; Blakemore, R.P.; Lovley, D.R. A hydrogen-oxidizing, Fe (III)-reducing microorganism from the Great Bay Estuary, New Hampshire. Appl. Environ. Microbiol. 1992, 58, 3211–3216. [Google Scholar] [CrossRef] [PubMed]
- Turpeinen, R.; Pantsar-Kallio, M.; Häggblom, M.; Kairesalo, T. Influence of microbes on the mobilization, toxicity and biomethylation of arsenic in soil. Sci. Total Environ. 1999, 236, 173–180. [Google Scholar] [CrossRef]
- Oremland, R.S.; Hoeft, S.E.; Santini, J.M.; Bano, N.; Hollibaugh, R.A.; Hollibaugh, J.T. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 2002, 68, 4795–4802. [Google Scholar] [CrossRef]
- Smedley, P.L.; Kinniburgh, D.G. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 2002, 17, 517–568. [Google Scholar] [CrossRef]
- Sharma, V.K.; Sohn, M. Aquatic arsenic: Toxicity, speciation, transformations, and remediation. Environ. Int. 2009, 35, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Dhar, R.K.; Zheng, Y.; Saltikov, C.W.; Radloff, K.A.; Mailloux, B.J.; Ahmed, K.M.; van Geen, A. Microbes enhance mobility of arsenic in Pleistocene aquifer sand from Bangladesh. Environ. Sci. Technol. 2011, 45, 2648–2654. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, T.; Yamaguchi, N.; Makino, T.; Sakurai, K.; Kimura, K.; Kudo, K.; Amachi, S. Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ. Sci. Technol. 2013, 47, 6263–6271. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.P.; Jaiswal, D.K. Book review: Advances in biodegradation and bioremediation of industrial waste. Front. Microbiol. 2016, 6, 1555. [Google Scholar] [CrossRef]
- Zhao, M.; Cui, Z.; Fu, L.; Ndayisenga, F.; Zhou, D. Shewanella drive Fe (III) reduction to promote electro-Fenton reactions and enhance Fe inner-cycle. ACS EST Water 2020, 1, 613–620. [Google Scholar] [CrossRef]
- Bousserrhine, N.; Gasser, U.G.; Jeanroy, E.; Berthelin, J. Bacterial and chemical reductive dissolution of Mn-, Co-, Cr-, and Al-substituted goethites. Geomicrobiol. J. 1999, 16, 245–258. [Google Scholar] [CrossRef]
- Wielinga, B.; Mizuba, M.M.; Hansel, C.M.; Fendorf, S. Iron promoted reduction of chromate by dissimilatory iron-reducing bacteria. Environ. Sci. Technol. 2001, 35, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Kocar, B.D.; Herbel, M.J.; Tufano, K.J.; Fendorf, S. Contrasting effects of dissimilatory iron (III) and arsenic (V) reduction on arsenic retention and transport. Environ. Sci. Technol. 2006, 40, 6715–6721. [Google Scholar] [CrossRef]
- Luo, Q.; Tsukamoto, T.K.; Zamzow, K.L.; Miller, G.C. Arsenic, selenium, and sulfate removal using an ethanol-enhanced sulfate-reducing bioreactor. Mine Water Environ. 2008, 27, 100–108. [Google Scholar] [CrossRef]
- Huang, J.H.; Voegelin, A.; Pombo, S.A.; Lazzaro, A.; Zeyer, J.; Kretzschmar, R. Influence of arsenate adsorption to ferrihydrite, goethite, and boehmite on the kinetics of arsenate reduction by Shewanella putrefaciens strain CN-32. Environ. Sci. Technol. 2011, 45, 7701–7709. [Google Scholar] [CrossRef]
- Farooq, M.A.; Islam, F.; Ali, B.; Najeeb, U.; Mao, B.; Gill, R.A.; Yan, G.; Siddiqu, K.H.M.; Zhou, W. Arsenic toxicity in plants: Cellular and molecular mechanisms of its transport and metabolism. Environ. Exp. Bot. 2016, 132, 42–52. [Google Scholar] [CrossRef]
- Kawa, Y.K.; Wang, J.; Chen, X.; Zhu, X.; Zeng, X.C.; Wang, Y. Reductive dissolution and release of arsenic from arsenopyrite by a novel arsenate-respiring bacterium from the arsenic-contaminated soils. Int. Biodeter. Biodegr. 2019, 143, 104712. [Google Scholar] [CrossRef]
- Qu, C.; Chen, W.; Hu, X.; Cai, P.; Chen, C.; Yu, X.Y.; Huang, Q. Heavy metal behaviour at mineral-organo interfaces: Mechanisms, modelling and influence factors. Environ. Int. 2019, 131, 104995. [Google Scholar] [CrossRef]
- Xue, Q.; Ran, Y.; Tan, Y.; Peacock, C.L.; Du, H. Arsenite and arsenate binding to ferrihydrite organo-mineral coprecipitate: Implications for arsenic mobility and fate in natural environments. Chemosphere 2019, 224, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Shi, M.; Shi, L. Degradation of organic contaminants and steel corrosion by the dissimilatory metal-reducing microorganisms Shewanella and Geobacter spp. Int. Biodeter. Biodegr. 2020, 147, 104842. [Google Scholar] [CrossRef]
- Oremland, R.S.; Stolz, J.F. The ecology of arsenic. Science 2003, 300, 939–944. [Google Scholar] [CrossRef]
- Tian, H.; Shi, Q.; Jing, C. Arsenic biotransformation in solid waste residue: Comparison of contributions from bacteria with arsenate and iron reducing pathways. Environ. Sci. Technol. 2015, 49, 2140–2146. [Google Scholar] [CrossRef]
- Wang, S.; Mulligan, C.N. Effect of natural organic matter on arsenic release from soils and sediments into groundwater. Environ. Geochem. Health 2006, 28, 197–214. [Google Scholar] [CrossRef]
- Hirano, S. Biotransformation of arsenic and toxicological implication of arsenic metabolites. Arch. Toxicol. 2020, 94, 2587–2601. [Google Scholar] [CrossRef]
- Krafft, T.; Macy, J.M. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 1998, 255, 647–653. [Google Scholar] [CrossRef]
- Zhao, J.S.; Manno, D.; Thiboutot, S.; Ampleman, G.; Hawari, J. Shewanella canadensis sp. nov. and Shewanella atlantica sp. nov., manganese dioxide-and hexahydro-1,3,5-trinitro-1,3,5-triazine-reducing, psychrophilic marine bacteria. Int. J. Syst. Evol. Microbiol. 2007, 57, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Hu, S.; Lin, J.; Liu, T.; Li, X.; Li, F. Quantifying microbially mediated kinetics of ferrihydrite transformation and arsenic reduction: Role of the arsenate-reducing gene expression pattern. Environ. Sci. Technol. 2020, 54, 6621–6631. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, O.N.; Méjean, V.; Iobbi-Nivol, C. The Shewanella genus: Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 2020, 44, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, M.; Lu, G.; Si, Y. Biotransformation and biomethylation of arsenic by Shewanella oneidensis MR-1. Chemosphere 2016, 145, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Rosen, B.P.; Zhang, Y.; Wang, G.; Franke, S.; Rensing, C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Tufano, K.J.; Reyes, C.; Saltikov, C.W.; Fendorf, S. Reductive processes controlling arsenic retention: Revealing the relative importance of iron and arsenic reduction. Environ. Sci. Technol. 2008, 42, 8283–8289. [Google Scholar] [CrossRef]
- Von Canstein, H.; Ogawa, J.; Shimizu, S.; Lloyd, J.R. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 2008, 74, 615–623. [Google Scholar] [CrossRef]
- Pi, K.; Markelova, E.; Zhang, P.; Van Cappellen, P. Arsenic oxidation by flavin-derived reactive species under oxic and anoxic conditions: Oxidant formation and pH dependence. Environ. Sci. Technol. 2019, 53, 10897–10905. [Google Scholar] [CrossRef]
- Zou, L.; Huang, Y.H.; Long, Z.E.; Qiao, Y. On-going applications of Shewanella species in microbial electrochemical system for bioenergy, bioremediation and biosensing. World J. Microbiol. Biotechnol. 2019, 35, 9. [Google Scholar] [CrossRef]
- Andres, J.; Bertin, P.N. The microbial genomics of arsenic. FEMS Microbiol. Rev. 2016, 40, 299–322. [Google Scholar] [CrossRef]
- Hussain, M.M.; Wang, J.; Bibi, I.; Shahid, M.; Niazi, N.K.; Iqbal, J.; Mian, I.A.; Shaheen, S.M.; Bashir, S.; Shah, N.S. Arsenic speciation and biotransformation pathways in the aquatic ecosystem: The significance of algae. J. Hazard. Mater. 2021, 403, 124027. [Google Scholar] [CrossRef] [PubMed]
- Adeloju, S.B.; Khan, S.; Patti, A.F. Arsenic contamination of groundwater and its implications for drinking water quality and human health in under-developed countries and remote communities—A review. Appl. Sci. 2021, 11, 1926. [Google Scholar] [CrossRef]
- Sundman, A.; Karlsson, T.; Sjöberg, S.; Persson, P. Complexation and precipitation reactions in the ternary As (V)–Fe (III)–OM (organic matter) system. Geochim. Cosmochim. Acta 2014, 145, 297–314. [Google Scholar] [CrossRef]
- Al-Abed, S.R.; Jegadeesan, G.; Purandare, J.; Allen, D. Arsenic release from iron rich mineral processing waste: Influence of pH and redox potential. Chemosphere 2007, 66, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Mikutta, C.; Kretzschmar, R. Spectroscopic evidence for ternary complex formation between arsenate and ferric iron complexes of humic substances. Environ. Sci. Technol. 2011, 45, 9550–9557. [Google Scholar] [CrossRef]
- Liu, G.; Cai, Y. Studying arsenite–humic acid complexation using size exclusion chromatography–inductively coupled plasma mass spectrometry. J. Hazard. Mater. 2013, 262, 1223–1229. [Google Scholar] [CrossRef]
- Liu, G.; Fernandez, A.; Cai, Y. Complexation of arsenite with humic acid in the presence of ferric iron. Environ. Sci. Technol. 2011, 45, 3210–3216. [Google Scholar] [CrossRef]
- Cassone, G.; Chillé, D.; Foti, C.; Giuffré, O.; Ponterio, R.C.; Sponer, J.; Saija, F. Stability of hydrolytic arsenic species in aqueous solutions: As3+ vs. As5+. Phys. Chem. Chem. Phys. 2018, 20, 23272–23280. [Google Scholar] [CrossRef]
- Quinodóz, F.B.; Maldonado, L.; Blarasin, M.; Matteoda, E.; Lutri, V.; Cabrera, A.; Giacobone, D. The development of a conceptual model for arsenic mobilization in a fluvio-eolian aquifer using geochemical and statistical methods. Environ. Earth Sci. 2019, 78, 206. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Darma, A.; Yang, J.; Bloem, E.; Możdżen, K.; Zandi, P. Arsenic biotransformation and mobilization: The role of bacterial strains and other environmental variables. Environ. Sci. Pollut. Res. Int. 2021, 29, 1763–1787. [Google Scholar] [CrossRef] [PubMed]
- O’Day, P.A.; Vlassopoulos, D.; Root, R.; Rivera, N. The influence of sulfur and iron on dissolved arsenic concentrations in the shallow subsurface under changing redox conditions. Proc. Natl. Acad. Sci. USA 2004, 101, 13703–13708. [Google Scholar] [CrossRef] [PubMed]
- Hanh, H.T.; Kim, J.Y.; Bang, S.; Kim, K.W. Sources and fate of As in the environment. Geosyst. Eng. 2010, 13, 35–42. [Google Scholar] [CrossRef]
- Smith, J.V. Geology, mineralogy, and human welfare. Proc. Natl. Acad. Sci. USA 1999, 96, 3348–3349. [Google Scholar] [CrossRef]
- Meharg, A.A. Variation in arsenic accumulation–hyperaccumulation in ferns and their allies: Rapid report. New Phytol. 2003, 157, 25–31. [Google Scholar] [CrossRef]
- Majumder, A.; Bhattacharyya, K.; Kole, S.C.; Ghosh, S. Efficacy of indigenous soil microbes in arsenic mitigation from contaminated alluvial soil of India. Environ. Sci. Pollut. Res. Int. 2013, 20, 5645–5653. [Google Scholar] [CrossRef]
- Ferreira, P.M.; Majuste, D.; Freitas, E.T.F.; Caldeira, C.L.; Dantas, M.S.S.; Ciminelli, V.S.T. Galvanic effect of pyrite on arsenic release from arsenopyrite dissolution in oxygen-depleted and oxygen-saturated circumneutral solutions. J. Hazard. Mater. 2021, 412, 125236. [Google Scholar] [CrossRef]
- Rasheed, H.; Slack, R.; Kay, P. Human health risk assessment for arsenic: A critical review. Crit. Rev. Environ. Sci. Technol. 2016, 46, 1529–1583. [Google Scholar] [CrossRef]
- Guo, H.; Liu, Z.; Ding, S.; Hao, C.; Xiu, W.; Hou, W. Arsenate reduction and mobilization in the presence of indigenous aerobic bacteria obtained from high arsenic aquifers of the Hetao basin, Inner Mongolia. Environ. Pollut. 2015, 203, 50–59. [Google Scholar] [CrossRef]
- Beiyuan, J.; Awad, Y.M.; Beckers, F.; Tsang, D.C.; Ok, Y.S.; Rinklebe, J. Mobility and phytoavailability of As and Pb in a contaminated soil using pine sawdust biochar under systematic change of redox conditions. Chemosphere 2017, 178, 110–118. [Google Scholar] [CrossRef]
- Marshall, G.; Ferreccio, C.; Yuan, Y.; Bates, M.N.; Steinmaus, C.; Selvin, S.; Liaw, J.; Smith, A.H. Fifty-year study of lung and bladder cancer mortality in Chile related to arsenic in drinking water. J. Natl. Cancer Inst. 2007, 99, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Zhang, H.; Junaid, M.; Mao, K.; Xu, N.; Chang, C.; Rasool, A.; Aslam, M.W.; Ali, J.; Yang, Z. Insights into the mechanisms of arsenic-selenium interactions and the associated toxicity in plants, animals, and humans: A critical review. Crit. Rev. Environ. Sci. Technol. 2021, 51, 704–750. [Google Scholar] [CrossRef]
- Garg, N.; Singla, P. Arsenic toxicity in crop plants: Physiological effects and tolerance mechanisms. Environ. Chem. Lett. 2011, 9, 303–321. [Google Scholar] [CrossRef]
- Patel, M.; Kumari, A.; Parida, A.K. Arsenic tolerance mechanisms in plants and potential role of arsenic hyperaccumulating plants for phytoremediation of arsenic-contaminated soil. In Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives II; Hasanuzzaman, M., Ed.; Springer: Singapore, 2020; pp. 137–162. [Google Scholar]
- Senila, M.; Levei, E.; Cadar, O.; Senila, L.R.; Roman, M.; Puskas, F.; Sima, M. Assessment of availability and human health risk posed by arsenic contaminated well waters from Timis-Bega area, Romania. J. Anal. Methods Chem. 2017, 2017, 3037651. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Khan, W.U.; Shah, A.A.; Yasin, N.A.; Naz, S.; Ali, A.; Tahir, A.; Batool, A.I. Synergistic effects of nitric oxide and silicon on promoting plant growth, oxidative stress tolerance and reduction of arsenic uptake in Brassica juncea. Chemosphere 2021, 262, 128384. [Google Scholar] [CrossRef]
- González, A.Z.I.; Krachler, M.; Cheburkin, A.K.; Shotyk, W. Spatial distribution of natural enrichments of arsenic, selenium, and uranium in a minerotrophic peatland, Gola di Lago, Canton Ticino, Switzerland. Environ. Sci. Technol. 2006, 40, 6568–6574. [Google Scholar] [CrossRef]
- Kwon, J.C.; Lee, J.S.; Jung, M.C. Arsenic contamination in agricultural soils surrounding mining sites in relation to geology and mineralization types. Appl. Geochem. 2012, 27, 1020–1026. [Google Scholar] [CrossRef]
- Li, Y.; Guo, H.; Hao, C. Arsenic release from shallow aquifers of the Hetao basin, Inner Mongolia: Evidence from bacterial community in aquifer sediments and groundwater. Ecotoxicology 2014, 23, 1900–1914. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Air Quality Guidelines for Europe, 2nd ed.; European Series; WHO: Copenhagen, Denmark, 2000. Available online: https://www.euro.who.int/__data/assets/pdf_file/0014/123071/AQG2ndEd_6_1_Arsenic.PDF (accessed on 10 February 2022).
- Weerasundara, L.; Ok, Y.S.; Bundschuh, J. Selective removal of arsenic in water: A critical review. Environ. Pollut. 2020, 268, 115668. [Google Scholar] [CrossRef]
- Alka, S.; Shahir, S.; Ibrahim, N.; Ndejiko, M.J.; Vo, D.V.N.; Manan, F.A. Arsenic removal technologies and future trends: A mini review. J. Clean. Product. 2021, 278, 123805. [Google Scholar] [CrossRef]
- Bissen, M.; Frimmel, F.H. Arsenic—A review. Part II: Oxidation of arsenic and its removal in water treatment. Acta Hydrochim. Hydrobiol. 2003, 31, 97–107. [Google Scholar] [CrossRef]
- Ojuederie, O.B.; Babalola, O.O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef]
- Yu, G.; Liu, J.; Long, Y.; Chen, Z.; Sunahara, G.I.; Jiang, P.; You, S.; Lin, H.; Xiao, H. Phytoextraction of cadmium-contaminated soils: Comparison of plant species and low molecular weight organic acids. Int. J. Phytoremediation 2020, 22, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Haneklaus, S.; Schnug, E. A critical evaluation of phytoextraction on uranium-contaminated agricultural soils. In Loads and Fate of Fertilizer Derived Uranium; Schnug, E., de Kok, L.J., Eds.; Margraf Publishers Scientific Books: Weikersheim, Germany, 2008; pp. 111–125. [Google Scholar]
- Yildirim, D.; Sasmaz, A. Phytoremediation of As, Ag, and Pb in contaminated soils using terrestrial plants grown on Gumuskoy mining area (Kutahya Turkey). J. Geochem. Explor. 2017, 182, 228–234. [Google Scholar] [CrossRef]
- Baker, A.J.; McGrath, S.P.; Reeves, R.D.; Smith, J.A.C. Metal hyperaccumulator plants: A review of the ecology and physiology of a biological resource for phytoremediation of metal-polluted soils. In Phytoremediation of Contaminated Soils; Terry, N., Vangronsveld, J., Banuelos, G., Eds.; CRC Press: Boca Raton, FL, USA, 2020; pp. 85–107. [Google Scholar]
- Malayeri, B.E.; Chehregani, A.; Mohsenzadeh, F.; Kazemeini, F.; Asgari, M. Plants growing in a mining area: Screening for metal accumulator plants possibly useful for bioremediation. Toxicol. Environ. Chem. 2013, 95, 434–444. [Google Scholar] [CrossRef]
- Koptsik, G.N. Problems and prospects concerning the phytoremediation of heavy metal polluted soils: A review. Eurasian Soil Sci. 2014, 47, 923–939. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Zeng, G.; Zhou, L.; Wang, X.; Wang, Y.; Wang, C.; Hu, X.; Xu, W. Enhanced efficiency of cadmium removal by Boehmeria nivea (L.) Gaud. in the presence of exogenous citric and oxalic acids. J. Environ. Sci. 2014, 26, 2508–2516. [Google Scholar] [CrossRef]
- Liu, D.; Islam, E.; Li, T.; Yang, X.; Jin, X.; Mahmood, Q. Comparison of synthetic chelators and low molecular weight organic acids in enhancing phytoextraction of heavy metals by two ecotypes of Sedum alfredii Hance. J. Hazard. Mater. 2008, 153, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H.; et al. Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Basu, S.; Rabara, R.C.; Negi, S.; Shukla, P. Engineering PGPMOs through gene editing and systems biology: A solution for phytoremediation? Trends Biotechnol. 2018, 36, 499–510. [Google Scholar] [CrossRef]
- Lovley, D.R. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat. Rev. Microbiol. 2003, 1, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Hau, H.H.; Gralnick, J.A. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 2007, 61, 237–258. [Google Scholar] [CrossRef]
- Nealson, K.H.; Scott, J. Ecophysiology of the genus Shewanella. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 1133–1151. [Google Scholar] [CrossRef]
- Bencheikh-Latmani, R.; Williams, S.M.; Haucke, L.; Criddle, C.S.; Wu, L.; Zhou, J.; Tebo, B.M. Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr (VI) and U (VI) reduction. Appl. Environ. Microbiol. 2005, 71, 7453–7460. [Google Scholar] [CrossRef] [PubMed]
- Wiatrowski, H.A.; Ward, P.M.; Barkay, T. Novel reduction of mercury (II) by mercury-sensitive dissimilatory metal-reducing bacteria. Environ. Sci. Technol. 2006, 40, 6690–6696. [Google Scholar] [CrossRef] [PubMed]
- Saltikov, C.W.; Wildman Jr, R.A.; Newman, D.K. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J. Bacteriol. 2005, 187, 7390–7396. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Yang, Y.; Shi, W.; Peng, Z.; Chen, X.; Zhu, X.; Wang, Y. Microbially mediated methylation of arsenic in the arsenic-rich soils and sediments of Jianghan plain. Front. Microbiol. 2018, 9, 1389. [Google Scholar] [CrossRef] [PubMed]
- Saltikov, C.W.; Cifuentes, A.; Venkateswaran, K.; Newman, D.K. The ars detoxification system is advantageous but not required for As (V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl. Environ. Microbiol. 2003, 69, 2800–2809. [Google Scholar] [CrossRef]
- Yan, G.; Chen, X.; Du, S.; Deng, Z.; Wang, L.; Chen, S. Genetic mechanisms of arsenic detoxification and metabolism in bacteria. Curr. Genet. 2019, 65, 329–338. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Shi, L. Molecular underpinnings for microbial extracellular electron transfer during biogeochemical cycling of earth elements. Sci. China Life Sci. 2019, 62, 1275–1286. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Pu, M.; Xu, P.; Liang, G.; Yu, D. Oryza sativa positive regulator of iron deficiency response 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant Cell Environ. 2020, 43, 261–274. [Google Scholar] [CrossRef]
- Kang, C.H.; Kwon, Y.J.; So, J.S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Zandi, P.; Yang, J.; Xia, X.; Yu, T.; Li, Q.; Możdżeń, K.; Barabasz-Krasny, B.; Yaosheng, W. Do sulfur addition and rhizoplane iron plaque affect chromium uptake by rice (Oryza sativa L.) seedlings in culture solution? J. Hazard. Mater. 2020, 388, 121803. [Google Scholar] [CrossRef] [PubMed]
- Zandi, P.; Yang, J.; Xia, X.; Barabasz-Krasny, B.; Możdżeń, K.; Puła, J.; Bloem, E.; Wang, Y.; Hussain, S.; Hashemi, S.M.; et al. Sulphur nutrition and iron plaque formation on roots of rice seedlings and their consequences for immobilisation and uptake of chromium in solution culture. Plant Soil 2021, 462, 365–388. [Google Scholar] [CrossRef]
- Bigham, J.M.; Fitzpatrick, R.W.; Schulze, D.G. Iron oxides. In Soil Mineralogy with Environmental Applications, SSSA Book Ser. 7; Dixon, J.B., Schulze, D.G., Eds.; SSSA: Madison, WI, USA, 2002; pp. 323–366. [Google Scholar]
- Shi, L.; Squier, T.C.; Zachara, J.M.; Fredrickson, J.K. Respiration of metal (hydr) oxides by Shewanella and Geobacter: A key role for multihaem c-type cytochromes. Mol. Microbiol. 2007, 65, 12–20. [Google Scholar] [CrossRef]
- Weber, F.A.; Hofacker, A.F.; Voegelin, A.; Kretzschmar, R. Temperature dependence and coupling of iron and arsenic reduction and release during flooding of a contaminated soil. Environ. Sci. Technol. 2010, 44, 116–122. [Google Scholar] [CrossRef]
- Morin, G.; Ostergren, J.D.; Juillot, F.; Ildefonse, P.; Calas, G.; Brown, G.E. XAFS determination of the chemical form of lead in smelter-contaminated soils and mine tailings: Importance of adsorption processes. Am. Mineral. 1999, 84, 420–434. [Google Scholar] [CrossRef]
- Ona-Nguema, G.; Morin, G.; Wang, Y.; Menguy, N.; Juillot, F.; Olivi, L.; Aquilanti, G.; Abdelmoula, M.; Ruby, C.; Bargar, J.R.; et al. Arsenite sequestration at the surface of nano-Fe(OH)2, ferrous-carbonate hydroxide, and green-rust after bioreduction of arsenic-sorbed lepidocrocite by Shewanella putrefaciens. Geochim. Cosmochim. Acta 2009, 73, 1359–1381. [Google Scholar] [CrossRef]
- Hohmann, C.; Morin, G.; Ona-Nguema, G.; Guigner, J.M.; Brown Jr, G.E.; Kappler, A. Molecular-level modes of As binding to Fe (III) (oxyhydr) oxides precipitated by the anaerobic nitrate-reducing Fe (II)-oxidizing Acidovorax sp. strain BoFeN1. Geochim. Cosmochim. Acta 2011, 75, 4699–4712. [Google Scholar] [CrossRef]
- Dia, A.; Lauga, B.; Davranche, M.; Fahy, A.; Duran, R.; Nowack, B.; Petitjean, P.; Henin, O.; Martin, S.; Marsac, R.; et al. Bacteria-mediated reduction of As (V)-doped lepidocrocite in a flooded soil sample. Chem. Geol. 2015, 406, 34–44. [Google Scholar] [CrossRef]
- Anderson, C.R.; Cook, G.M. Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr. Microbiol. 2004, 48, 341–347. [Google Scholar] [CrossRef]
- Manning, B.A.; Fendorf, S.E.; Goldberg, S. Surface structures and stability of arsenic (III) on goethite: Spectroscopic evidence for inner-sphere complexes. Environ. Sci. Technol. 1998, 32, 2383–2388. [Google Scholar] [CrossRef]
- Raven, K.P.; Jain, A.; Loeppert, R.H. Arsenite and arsenate adsorption on ferrihydrite: Kinetics, equilibrium, and adsorption envelopes. Environ. Sci. Technol. 1998, 32, 344–349. [Google Scholar] [CrossRef]
- Adra, A.; Morin, G.; Ona-Nguema, G.; Brest, J. Arsenate and arsenite adsorption onto Al-containing ferrihydrites. Implications for arsenic immobilization after neutralization of acid mine drainage. Appl. Geochem. 2016, 64, 2–9. [Google Scholar] [CrossRef]
- Reardon, C.L.; Dohnalkova, A.C.; Nachimuthu, P.; Kennedy, D.W.; Saffarini, D.A.; Arey, B.W.; Shi, L.; Wang, Z.; Moore, D.; McLean, J.S.; et al. Role of outer-membrane cytochromes MtrC and OmcA in the biomineralization of ferrihydrite by Shewanella oneidensis MR-1. Geobiology 2010, 8, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Adams, R.; Newsome, L.; Moore, K.L.; Lyon, I.C.; Lloyd, J.R. Dissimilatory Fe (III) reduction controls on arsenic mobilization: A combined biogeochemical and NanoSIMS imaging approach. Front. Microbiol. 2021, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Rosso, K.M.; Smith, D.M.; Dupuis, M. An ab initio model of electron transport in hematite (α-Fe2O3) basal planes. J. Chem. Phys. 2003, 118, 6455–6466. [Google Scholar] [CrossRef]
- Shi, L.; Rosso, K.M.; Zachara, J.M.; Fredrickson, J.K. Mtr extracellular electron-transfer pathways in Fe (III)-reducing or Fe (II)-oxidizing bacteria: A genomic perspective. Biochem. Soc. Trans. 2012, 40, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Zachara, J.M.; Fredrickson, J.K.; Li, S.M.; Kennedy, D.W.; Smith, S.C.; Gassman, P.L. Bacterial reduction of crystalline Fe (super 3+) oxides in single phase suspensions and subsurface materials. Am. Mineral. 1998, 83, 1426–1443. [Google Scholar] [CrossRef]
- Brown, G.E., Jr.; Henrich, V.; Casey, W.; Clark, D.; Eggleston, C.; Andrew Felmy, A.F.; Goodman, D.W.; Gratzel, M.; Maciel, G.; McCarthy, M.I.; et al. Metal oxide surfaces and their interactions with aqueous solutions and microbial organisms and microbial organisms are part of the bioresource and agricultural engineering commons. Chem. Rev. 1999, 99, 77–174. [Google Scholar] [CrossRef]
- Jönsson, J.; Sherman, D.M. Sorption of As (III) and As (V) to siderite, green rust (fougerite) and magnetite: Implications for arsenic release in anoxic groundwaters. Chem. Geol. 2008, 255, 173–181. [Google Scholar] [CrossRef]
- Lovley, D.R.; Lonergan, D.J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl. Environ. Microbiol. 1990, 56, 1858–1864. [Google Scholar] [CrossRef] [PubMed]
- Dinh, H.T.; Kuever, J.; Mußmann, M.; Hassel, A.W.; Stratmann, M.; Widdel, F. Iron corrosion by novel anaerobic microorganisms. Nature 2004, 427, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chang, W.; Cui, T.; Xu, D.; Zhang, D.; Lou, Y.; Qian, H.; Song, H.; Mol, A.; Cao, F.; et al. Adaptive bidirectional extracellular electron transfer during accelerated microbiologically influenced corrosion of stainless steel. Commun. Mater. 2021, 2, 67. [Google Scholar] [CrossRef]
- Lovley, D.R. Dissimilatory metal reduction. Annu. Rev. Microbiol. 1993, 47, 263–290. [Google Scholar] [CrossRef] [PubMed]
- Hull, E.A.; Barajas, M.; Burkart, K.A.; Fung, S.R.; Jackson, B.P.; Barrett, P.M.; Neumann, R.B.; Olden, J.D.; Gawel, J.E. Human health risk from consumption of aquatic species in arsenic-contaminated shallow urban lakes. Sci. Total Environ. 2021, 770, 145318. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Lee, J.H.; Kim, M.G.; Myung, N.V.; Fredrickson, J.K.; Sadowsky, M.J.; Hur, H.G. Biogenic formation of As-S nanotubes by diverse Shewanella strains. Appl. Environ. Microbiol. 2009, 75, 6896–6899. [Google Scholar] [CrossRef]
- Jiang, S.; Lee, J.H.; Kim, D.; Kanaly, R.A.; Kim, M.G.; Hur, H.G. Differential arsenic mobilization from As-bearing ferrihydrite by iron-respiring Shewanella strains with different arsenic-reducing activities. Environ. Sci. Technol. 2013, 47, 8616–8623. [Google Scholar] [CrossRef]
- Myers, C.R.; Nealson, K.H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese (IV) and iron (III) in Shewanella putrefaciens MR-1. J. Bacteriol. 1990, 172, 6232–6238. [Google Scholar] [CrossRef]
- Venkateswaran, K.; Moser, D.P.; Dollhopf, M.E.; Lies, D.P.; Saffarini, D.A.; MacGregor, B.J.; Ringelberg, D.B.; White, D.C.; Nishijima, M.; Sano, H.; et al. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 705–724. [Google Scholar] [CrossRef]
- Cullen, W.; Reimer, K.J. Arsenic speciation in the environment. Chem. Rev. 1989, 89, 713–764. [Google Scholar] [CrossRef]
- Sekar, R.; DiChristina, T.J. Microbially driven Fenton reaction for degradation of the widespread environmental contaminant 1, 4-dioxane. Environ. Sci. Technol. 2014, 48, 12858–12867. [Google Scholar] [CrossRef]
- White, G.F.; Edwards, M.J.; Gomez-Perez, L.; Richardson, D.J.; Butt, J.N.; Clarke, T.A. Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 2016, 68, 87–138. [Google Scholar] [CrossRef]
- Liu, H.; Li, P.; Wang, H.; Qing, C.; Tan, T.; Shi, B.; Zhang, G.; Jiang, Z.; Wang, Y.; Hasan, S.Z. Arsenic mobilization affected by extracellular polymeric substances (EPS) of the dissimilatory iron reducing bacteria isolated from high arsenic groundwater. Sci. Total Environ. 2020, 735, 139501. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Guo, W.; Wang, L.; Jing, C. Extracellular polymeric substances from Shewanella oneidensis MR-1 biofilms mediate the transformation of ferrihydrite. Sci. Total Environ. 2021, 784, 147245. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.C.; Blum, J.D.; Klaue, B.; Karagas, M.R. Arsenic occurrence in New Hampshire drinking water. Environ. Sci. Technol. 1999, 33, 1328–1333. [Google Scholar] [CrossRef]
- Cummings, D.E.; Caccavo, F.; Fendorf, S.; Rosenzweig, R.F. Arsenic mobilization by the dissimilatory Fe (III)-reducing bacterium Shewanella alga BrY. Environ. Sci. Technol. 1999, 33, 723–729. [Google Scholar] [CrossRef]
- O’Loughlin, E.J. Effects of electron transfer mediators on the bioreduction of lepidocrocite (γ-FeOOH) by Shewanella putrefaciens CN32. Environ. Sci. Technol. 2008, 42, 6876–6882. [Google Scholar] [CrossRef]
- Muehe, E.M.; Morin, G.; Scheer, L.; Pape, P.L.; Esteve, I.; Daus, B.; Kappler, A. Arsenic (V) incorporation in vivianite during microbial reduction of arsenic (V)-bearing biogenic Fe (III)(oxyhydr) oxides. Environ. Sci. Technol. 2016, 50, 2281–2291. [Google Scholar] [CrossRef]
- Wang, J.; Xie, Z.; Wang, Y.; Yang, Y.; Chen, M. Synergy between indigenous bacteria and extracellular electron shuttles enhances transformation and mobilization of Fe (III)/As (V). Sci. Total Environ. 2021, 783, 147002. [Google Scholar] [CrossRef]
- Bentley, R.; Chasteen, T.G. Microbial methylation of metalloids: Arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 2002, 66, 250–271. [Google Scholar] [CrossRef]
- Kulp, T.R. Arsenic and primordial life. Nat. Geosci. 2014, 7, 785–786. [Google Scholar] [CrossRef]
- Cervantes, C.; Ji, G.; Ramírez, J.L.; Silver, S. Resistance to arsenic compounds in microorganisms. FEMS Microbiol. Rev. 1994, 15, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, H.; Shafique, M.; Rehman, Y. Genes and biochemical pathways involved in microbial transformation of arsenic. In Arsenic Toxicity: Challenges and Solutions; Kumar, N., Ed.; Springer: Singapore, 2021; pp. 391–413. [Google Scholar]
- Kruger, M.C.; Bertin, P.N.; Heipieper, H.J.; Arsène-Ploetze, F. Bacterial metabolism of environmental arsenic--mechanisms and biotechnological applications. Appl. Microbiol. Biotechnol. 2013, 97, 3827–3841. [Google Scholar] [CrossRef]
- Sathendra, E.R.; Kumar, R.P.; Baskar, G. Microbial transformation of heavy metals. In Waste Bioremediation: Energy, Environment, and Sustainability; Varjani, S., Gnansounou, E., Gurunathan, B., Pant, D., Zakaria, Z., Eds.; Springer: Singapore, 2018; pp. 249–263. [Google Scholar] [CrossRef]
- Pepi, M.; Volterrani, M.; Renzi, M.; Marvasi, M.; Gasperini, S.; Franchi, E.; Focardi, S.E. Arsenic-resistant bacteria isolated from contaminated sediments of the Orbetello Lagoon, Italy, and their characterization. J. Appl. Microbiol. 2007, 103, 2299–2308. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Oremland, R.S. Microbial transformations of arsenic in the environment: From soda lakes to aquifers. Elements 2006, 2, 85–90. [Google Scholar] [CrossRef]
- Cutting, R.S.; Coker, V.S.; Telling, N.D.; Kimber, R.L.; van der Laan, G.; Pattrick, R.A.D.; Vaughan, D.J.; Arenholz, E.; Lloyd, J.R. Microbial reduction of arsenic-doped schwertmannite by Geobacter sulfurreducens. Environ. Sci. Technol. 2012, 46, 12591–12599. [Google Scholar] [CrossRef]
- Banfield, J.F.; Barker, W.W.; Welch, S.A.; Taunton, A. Biological impact on mineral dissolution: Application of the lichen model to understanding mineral weathering in the rhizosphere. Proc. Natl. Acad. Sci. USA 1999, 96, 3404–3411. [Google Scholar] [CrossRef]
- Welch, S.A.; Barker, W.W.; Banfield, J.F. Microbial extracellular polysaccharides and plagioclase dissolution. Geochim. Cosmochim. Acta 1999, 63, 1405–1419. [Google Scholar] [CrossRef]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. Int. 2016, 23, 3984–3999. [Google Scholar] [CrossRef]
- Raymond, K.N.; Müller, G.; Matzanke, B.F. Complexation of iron by siderophores a review of their solution and structural chemistry and biological function. In Structural Chemistry. Topics in Current Chemistry; Bayley, H., Houk, K.N., Hughes, G., Hunter, C.A., Ishihara, K., Krische, M.J., Lehn, J.M., Luque, R., Olivucci, M., Siegel, J.S., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; Volume 123, pp. 49–102. [Google Scholar]
- Nevin, K.P.; Lovley, D.R. Mechanisms for Fe (III) oxide reduction in sedimentary environments. Geomicrobiol. J. 2002, 19, 141–159. [Google Scholar] [CrossRef]
- Aiken, A.M.; Peyton, B.M.; Apel, W.A.; Petersen, J.N. Heavy metal-induced inhibition of Aspergillus niger nitrate reductase: Applications for rapid contaminant detection in aqueous samples. Anal. Chim. Acta 2003, 480, 131–142. [Google Scholar] [CrossRef]
- Yarnell, A. Nature’s tiniest geoengineers. Chem. Eng. News 2003, 81, 24–25. [Google Scholar] [CrossRef]
- Nair, A.; Juwarkar, A.A.; Singh, S.K. Production and characterization of siderophores and its application in arsenic removal from contaminated soil. Water Air Soil Pollut. 2007, 180, 199–212. [Google Scholar] [CrossRef]
- Chibuike, G.U.; Obiora, S.C. Heavy metal polluted soils: Effect on plants and bioremediation methods. Appl. Environ. Soil Sci. 2014, 2014, 752708. [Google Scholar] [CrossRef]
- Retamal-Morales, G.; Mehnert, M.; Schwabe, R.; Tischler, D.; Zapata, C.; Chávez, R.; Schlömann, M.; Levicán, G. Detection of arsenic-binding siderophores in arsenic-tolerating actinobacteria by a modified CAS assay. Ecotoxicol. Environ. Saf. 2018, 157, 176–181. [Google Scholar] [CrossRef]
- Laverman, A.M.; Blum, J.S.; Schaefer, J.K.; Phillips, E.; Lovley, D.R.; Oremland, R.S. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl. Environ. Microbiol. 1995, 61, 3556–3561. [Google Scholar] [CrossRef]
- Zobrist, J.; Dowdle, P.R.; Davis, J.A.; Oremland, R.S. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ. Sci. Technol. 2000, 34, 4747–4753. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Sukhacheva, M.V.; Muyzer, G. Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes. Extremophiles 2012, 16, 597–605. [Google Scholar] [CrossRef]
- Bini, E.; Rauschenbach, I.; Narasingarao, P.; Starovoytov, V.; Hauser, L.; Jeffries, C.D.; Land, M.; Bruce, D.; Detter, C.; Goodwin, L.; et al. Complete genome sequence of Desulfurispirillum indicum strain S5(T). Stand. Genom. Sci. 2011, 5, 371–378. [Google Scholar] [CrossRef]
- Muehe, E.M.; Scheer, L.; Daus, B.; Kappler, A. Fate of arsenic during microbial reduction of biogenic versus abiogenic As–Fe (III)–mineral coprecipitates. Environ. Sci. Technol. 2013, 47, 8297–8307. [Google Scholar] [CrossRef]
- Chen, G.; Ke, Z.; Liang, T.; Liu, L.; Wang, G. Shewanella oneidensis MR-1-induced Fe (III) reduction facilitates roxarsone transformation. PLoS ONE 2016, 11, e0154017. [Google Scholar] [CrossRef]
- Conley, B.E.; Intile, P.J.; Bond, D.R.; Gralnick, J.A. Divergent Nrf family proteins and MtrCAB homologs facilitate extracellular electron transfer in Aeromonas hydrophila. Appl. Environ. Microbiol. 2018, 84, e02134-18. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, P.; Pirbadian, S.; El-Naggar, M.Y.; Jensen, G.J. Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc. Natl. Acad. Sci. USA 2018, 115, 3246–3255. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, H.P.; Zhu, L. Structures of nitroaromatic compounds induce Shewanella oneidensis MR-1 to adopt different electron transport pathways to reduce the contaminants. J. Hazard. Mater. 2020, 384, 121495. [Google Scholar] [CrossRef]
- Silver, S.; Phung, L.T.; Rosen, B.P. Arsenic metabolism: Resistance, reduction, and oxidation. In Environmental Chemistry of Arsenic; Frankenberger, W.T., Jr., Ed.; Marcel Dekker: New York, NY, USA, 2002; pp. 247–272. [Google Scholar]
- Ehrlich, H.L. Inorganic energy sources for chemolithotrophic and mixotrophic bacteria. Geomicrobiol. J. 1978, 1, 65–83. [Google Scholar] [CrossRef]
- Oremland, R.S.; Stolz, J.F. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005, 13, 45–49. [Google Scholar] [CrossRef]
- Biswas, R.; Vivekanand, V.; Saha, A.; Ghosh, A.; Sarkar, A. Arsenite oxidation by a facultative chemolithotrophic Delftia spp. BAs29 for its potential application in groundwater arsenic bioremediation. Int. Biodeter. Biodegrad. 2019, 136, 55–62. [Google Scholar] [CrossRef]
- Debiec-Andrzejewska, K.; Krucon, T.; Piatkowska, K.; Drewniak, L. Enhancing the plant’s growth and arsenic uptake from soil using arsenite-oxidising bacteria. Environ. Pollut. 2020, 264, 114692. [Google Scholar] [CrossRef]
- Butcher, B.G.; Deane, S.M.; Rawlings, D.E. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 1826–1833. [Google Scholar] [CrossRef]
- Schäfer, G. Membrane-associated energy transduction in bacteria and archaea, 2nd ed. In Encyclopedia of Biological Chemistry; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: New York, NY, USA, 2013; pp. 28–35. [Google Scholar]
- Gihring, T.M.; Banfield, J.F. Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett. 2001, 204, 335–340. [Google Scholar] [CrossRef]
- Santini, J.M.; Sly, L.I.; Schnagl, R.D.; Macy, J.M. A new chemolithoautotrophic arsenite-oxidising bacterium isolated from a gold mine: Phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol. 2000, 66, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Franchi, E.; Cosmina, P.; Pedron, F.; Rosellini, I.; Barbafieri, M.; Petruzzelli, G.; Vocciante, M. Improved arsenic phytoextraction by combined use of mobilizing chemicals and autochthonous soil bacteria. Sci. Total Environ. 2019, 655, 328–336. [Google Scholar] [CrossRef]
- Páez-Espino, D.; Tamames, J.; de Lorenzo, V.; Cánovas, D. Microbial responses to environmental arsenic. Biometals 2009, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.; Phung, L.T. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 2005, 71, 599–608. [Google Scholar] [CrossRef]
- Rosen, B.P. Biochemistry of arsenic detoxification. FEBS Lett. 2002, 529, 86–92. [Google Scholar] [CrossRef]
- Jackson, C.R.; Dugas, S.L. Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 2003, 3, 18. [Google Scholar] [CrossRef]
- Krumova, K.; Nikolovska, M.; Groudeva, V. Characterization of arsenic-transforming bacteria from arsenic contaminated sites in Bulgaria. Biotechnol. Biotechnol. Equip. 2008, 22, 729–735. [Google Scholar] [CrossRef][Green Version]
- Dey, S.; Rosen, B.P. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J. Bacteriol. 1995, 177, 385–389. [Google Scholar] [CrossRef]
- Tisa, L.S.; Rosen, B.P. Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J. Biol. Chem. 1990, 265, 190–194. [Google Scholar] [CrossRef]
- Kuroda, M.; Dey, S.; Sanders, O.I.; Rosen, B.P. Alternate energy coupling of ArsB, the membrane subunit of the Ars anion-translocating ATPase. J. Biol. Chem. 1997, 272, 326–331. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the alpha-and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Rosen, B.P. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 1991, 5, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Walmsley, A.R.; Rosen, B.P. An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. USA 2006, 103, 15617–15622. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Rawat, S.; Stemmler, T.L.; Rosen, B.P. Arsenic binding and transfer by the ArsD As (III) metallochaperone. Biochemistry 2010, 49, 3658–3666. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Yang, J.; Rosen, B.P. ArsD: An As (III) metallochaperone for the ArsAB As (III)-translocating ATPase. J. Bioenerg. Biomembr. 2007, 39, 453–458. [Google Scholar] [CrossRef]
- Hayakawa, T.; Kobayashi, Y.; Cui, X.; Hirano, S. A new metabolic pathway of arsenite: Arsenic–glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch. Toxicol. 2005, 79, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Stolz, J.F.; Basu, P.; Santini, J.M.; Oremland, R.S. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 2006, 60, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Costa, M. Arsenic: A global environmental challenge. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 47–63. [Google Scholar] [CrossRef]
- Hamon, R.E.; Lombi, E.; Fortunati, P.; Nolan, A.L.; McLaughlin, M.J. Coupling speciation and isotope dilution techniques to study arsenic mobilization in the environment. Environ. Sci. Technol. 2004, 38, 1794–1798. [Google Scholar] [CrossRef]
- MacDonell, M.T.; Colwell, R.R. Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. Syst. Appl. Microbiol. 1985, 6, 171–182. [Google Scholar] [CrossRef]
- Ikeda, S.; Takamatsu, Y.; Tsuchiya, M.; Suga, K.; Tanaka, Y.; Kouzuma, A.; Watanabe, K. Shewanella oneidensis MR-1 as a bacterial platform for electro-biotechnology. Essays Biochem. 2021, 65, 355–364. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Romine, M.F.; Beliaev, A.S.; Auchtung, J.M.; Driscoll, M.E.; Gardner, T.S.; Nealson, K.H.; Osterman, A.L.; Pinchuk, G.; Reed, J.L.; et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008, 6, 592–603. [Google Scholar] [CrossRef]
- Lemaire, O.N.; Honoré, F.A.; Tempel, S.; Fortier, E.M.; Leimkühler, S.; Méjean, V.; Méjean, V.; Iobbi-Nivol, C. Insights into chromate resistance of Shewanella decolorationis LDS1. Appl. Environ. Microbiol. 2019, 85, e00777-19. [Google Scholar] [CrossRef]
- Brutinel, E.D.; Gralnick, J.A. Shuttling happens: Soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 2012, 93, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Euzéby, J.P. List of bacterial names with standing in nomenclature: A folder available on the Internet. Int. J. Syst. Bacteriol. 1997, 47, 590–592. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature. Nucleic Acid Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature (bacterio. net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Beblawy, S.; Bursac, T.; Paquete, C.; Louro, R.; Clarke, T.A.; Gescher, J. Extracellular reduction of solid electron acceptors by Shewanella oneidensis. Mol. Microbiol. 2018, 109, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Besold, J.; Biswas, A.; Suess, E.; Scheinost, A.C.; Rossberg, A.; Mikutta, C.; Kretzschmar, R.; Gustafsson, J.P.; Planer-Friedrich, B. Monothioarsenate transformation kinetics determining arsenic sequestration by sulfhydryl groups of peat. Environ. Sci. Technol. 2018, 52, 7317–7326. [Google Scholar] [CrossRef] [PubMed]
- Yam, H.M.; Leong, S.K.W.; Qiu, X.; Zaiden, N. Bioremediation of Arsenic-contaminated water through application of bioengineered Shewanella oneidensis. In IRC-SET: Proceedings of the 6th IRC Conference on Science, Engineering and Technology; Guo, H., Ren, H., Kim, N., Eds.; Springer Nature: Singapore, 2021; pp. 559–574. [Google Scholar] [CrossRef]
- Laso-Pérez, R.; Hahn, C.; van Vliet, D.M.; Tegetmeyer, H.E.; Schubotz, F.; Smit, N.T.; Pape, T.; Sahling, H.; Bohrmann, G.; Boetius, A.; et al. Anaerobic degradation of non-methane alkanes by “Candidatus Methanoliparia” in hydrocarbon seeps of the Gulf of Mexico. mBio 2019, 10, e01814-19. [Google Scholar] [CrossRef]
- Coursolle, D.; Gralnick, J.A. Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 2010, 77, 995–1008. [Google Scholar] [CrossRef]
- Marritt, S.J.; Lowe, T.G.; Bye, J.; McMillan, D.G.; Shi, L.; Fredrickson, J.; Zachara, J.; Richardson, D.J.; Cheesman, M.R.; Jeuken, L.J.C.; et al. A functional description of CymA, an electron-transfer hub supporting anaerobic respiratory flexibility in Shewanella. Biochem. J. 2012, 444, 465–474. [Google Scholar] [CrossRef]
- Rahman, M.A.; Hasegawa, H.; Lim, R.P. Bioaccumulation, biotransformation and trophic transfer of arsenic in the aquatic food chain. Environ. Res. 2012, 116, 118–135. [Google Scholar] [CrossRef]
- Tanaka, K.; Yokoe, S.; Igarashi, K.; Takashino, M.; Ishikawa, M.; Hori, K.; Kato, S. Extracellular electron transfer via outer membrane cytochromes in a methanotrophic bacterium Methylococcus capsulatus (Bath). Front. Microbiol. 2018, 9, 2905. [Google Scholar] [CrossRef]
- White, G.F.; Shi, Z.; Shi, L.; Wang, Z.; Dohnalkova, A.C.; Marshall, M.J.; Fredrickson, J.K.; Zachara, J.M.; Butt, J.N.; Richardson, D.J.; et al. Rapid electron exchange between surface-exposed bacterial cytochromes and Fe (III) minerals. Proc. Natl. Acad. Sci. USA 2013, 110, 6346–6351. [Google Scholar] [CrossRef]
- Schicklberger, M.; Sturm, G.; Gescher, J. Genomic plasticity enables a secondary electron transport pathway in Shewanella oneidensis. Appl. Environ. Microbiol. 2013, 79, 1150–1159. [Google Scholar] [CrossRef]
- Chong, G.W.; Pirbadian, S.; El-Naggar, M.Y. Surface-induced formation and redox-dependent staining of outer membrane extensions in Shewanella oneidensis MR-1. Front. Energy Res. 2019, 7, 87. [Google Scholar] [CrossRef]
- Creasey, R.C.; Mostert, A.B.; Nguyen, T.A.; Virdis, B.; Freguia, S.; Laycock, B. Microbial nanowires–electron transport and the role of synthetic analogues. Acta Biomater. 2018, 69, 1–30. [Google Scholar] [CrossRef]
- Edwards, M.J.; White, G.F.; Norman, M.; Tome-Fernandez, A.; Ainsworth, E.; Shi, L.; Fredrickson, J.K.; Zachara, J.M.; Butt, J.N.; Richardson, D.J.; et al. Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci. Rep. 2015, 5, 11677. [Google Scholar] [CrossRef]
- Reguera, G.; Kashefi, K. The electrifying physiology of Geobacter bacteria, 30 years on. Adv. Microb. Physiol. 2019, 74, 1–96. [Google Scholar] [CrossRef]
- Field, S.J.; Dobbin, P.S.; Cheesman, M.R.; Watmough, N.J.; Thomson, A.J.; Richardson, D.J. Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane multiheme c-type cytochromes from Shewanella frigidimarina NCIMB400. J. Biol. Chem. 2000, 275, 8515–8522. [Google Scholar] [CrossRef]
- Pitts, K.E.; Dobbin, P.S.; Reyes-Ramirez, F.; Thomson, A.J.; Richardson, D.J.; Seward, H.E. Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA: Expression in Escherichia coli confers the ability to reduce soluble Fe (III) chelates. J. Biol. Chem. 2003, 278, 27758–27765. [Google Scholar] [CrossRef]
- Hartshorne, R.S.; Reardon, C.L.; Ross, D.; Nuester, J.; Clarke, T.A.; Gates, A.J.; Mills, P.C.; Fredrickson, J.K.; Zachara, J.M.; Shi, L.; et al. Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. USA 2009, 106, 22169–22174. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Paulsen, I.T.; Nelson, K.E.; Gaidos, E.J.; Nelson, W.C.; Read, T.D.; Eisen, J.A.; Seshadri, R.; Ward, N.; Methe, B.; et al. Genome sequence of the dissimilatory metal ion–reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002, 20, 1118–1123. [Google Scholar] [CrossRef]
- Albers, S.V.; Meyer, B.H. The archaeal cell envelope. Nat. Rev. Microbiol. 2011, 9, 414–426. [Google Scholar] [CrossRef]
- Shi, M.; Xia, K.; Peng, Z.; Jiang, Y.; Dong, Y.; Shi, L. Differential degradation of BDE-3 and BDE-209 by the Shewanella oneidensis MR-1-mediated Fenton reaction. Int. Biodeter. Biodegrad. 2021, 158, 105165. [Google Scholar] [CrossRef]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef]
- Gorby, Y.; McLean, J.; Korenevsky, A.; Rosso, K.; EL-Naggar, M.Y.; Beveridge, T.J. Redox-reactive membrane vesicles produced by Shewanella. Geobiology 2008, 6, 232–241. [Google Scholar] [CrossRef]
- Wang, F.; Wang, J.; Jian, H.; Zhang, B.; Li, S.; Wang, F.; Zeng, X.; Gao, L.; Bartlett, D.H.; Yu, J.; et al. Environmental adaptation: Genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS ONE 2008, 3, e1937. [Google Scholar] [CrossRef]
- Gralnick, J.A.; Vali, H.; Lies, D.P.; Newman, D.K. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. USA 2006, 103, 4669–4674. [Google Scholar] [CrossRef]
- Jiao, Y.; Newman, D.K. The pio operon is essential for phototrophic Fe (II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 2007, 189, 1765–1773. [Google Scholar] [CrossRef]
- Neyens, E.; Baeyens, J. A review of classic Fenton’s peroxidation as an advanced oxidation technique. J. Hazard. Mater. 2003, 98, 33–50. [Google Scholar] [CrossRef]
- El-Bahr, S.M. Biochemistry of free radicals and oxidative stress. Sci. Int. 2013, 1, 111–117. [Google Scholar] [CrossRef]
- Liu, W.J.; Zhu, Y.G.; Smith, F.A.; Smith, S.E. Do iron plaque and genotypes affect arsenate uptake and translocation by rice seedlings (Oryza sativa L.) grown in solution culture? J. Exp. Bot. 2004, 55, 1707–1713. [Google Scholar] [CrossRef]
- Liu, W.J.; Zhu, Y.G.; Hu, Y.; Williams, P.N.; Gault, A.G.; Meharg, A.A.; Charnock, J.M.; Smith, F.A. Arsenic sequestration in iron plaque, its accumulation and speciation in mature rice plants (Oryza sativa L.). Environ. Sci. Technol. 2006, 40, 5730–5736. [Google Scholar] [CrossRef]
- Wang, S.; Li, Q.; Stubbings, W.A.; Li, L.; Qin, J.; Li, H. The effect of Fenton reaction using H2O2 and water control on the distribution and accumulation of As speciation within the soil-rice system. Chemosphere 2021, 274, 129633. [Google Scholar] [CrossRef]
- Huijbers, M.M.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef]
- Chen, J.; Bhattacharjee, H.; Rosen, B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 2015, 96, 1042–1052. [Google Scholar] [CrossRef]
- Hervás, M.; Lόpez-Maury, L.; Leon, P.; Sanchez-Riego, A.M.; Florencio, F.J.; Navarro, J.A. ArsH from the cyanobacterium Synechocystis sp. PCC 6803 is an efficient NADPH-dependent quinone reductase. Biochemistry 2012, 51, 1178–1187. [Google Scholar] [CrossRef]
- Xue, X.M.; Yan, Y.; Xu, H.J.; Wang, N.; Zhang, X.; Ye, J. ArsH from Synechocystis sp. PCC 6803 reduces chromate and ferric iron. FEMS Microbiol. Lett. 2014, 356, 105–112. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darma, A.; Yang, J.; Zandi, P.; Liu, J.; Możdżeń, K.; Xia, X.; Sani, A.; Wang, Y.; Schnug, E. Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review. Biology 2022, 11, 472. https://doi.org/10.3390/biology11030472
Darma A, Yang J, Zandi P, Liu J, Możdżeń K, Xia X, Sani A, Wang Y, Schnug E. Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review. Biology. 2022; 11(3):472. https://doi.org/10.3390/biology11030472
Chicago/Turabian StyleDarma, Aminu, Jianjun Yang, Peiman Zandi, Jin Liu, Katarzyna Możdżeń, Xing Xia, Ali Sani, Yihao Wang, and Ewald Schnug. 2022. "Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review" Biology 11, no. 3: 472. https://doi.org/10.3390/biology11030472
APA StyleDarma, A., Yang, J., Zandi, P., Liu, J., Możdżeń, K., Xia, X., Sani, A., Wang, Y., & Schnug, E. (2022). Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review. Biology, 11(3), 472. https://doi.org/10.3390/biology11030472








