Association Analysis between Genetic Variants of elovl5a and elovl5b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Measuring PUFA Contents
2.2. Sequencing and Genotyping
2.3. Genetic Diversity of Common Carp elovl5a and elovl5b
2.4. Associations of cSNPs in elovl5a and elovl5b with the Contents of 12 PUFAs
2.5. Joint Effects of Significant SNPs in elovl5b and fads2b on the PUFA Contents
2.6. Estimating the Breeding Values with the cSNPs in elovl5b and fads2b on the PUFA Contents
3. Results
3.1. Genetic Diversities of Common Carp elovl5a and elovl5b
3.2. cSNPs in elovl5a and elovl5b Associated with the PUFA Contents
3.3. Joint Effect of elovl5b and fads2b on the PUFA Contents
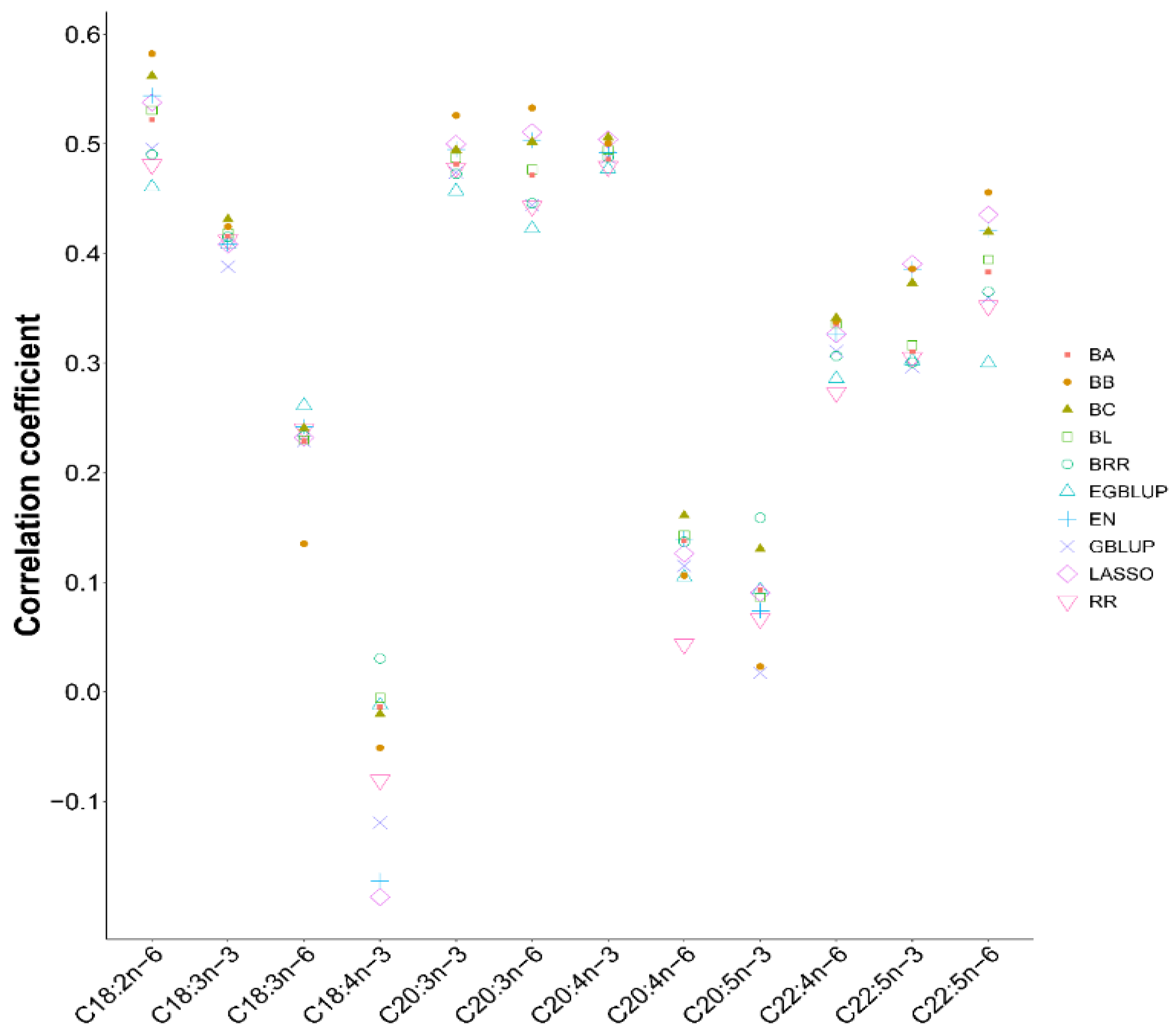
3.4. The cSNPs in fads2b and elovl5b to Predict the Breeding Values
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bentsen, H. Dietary polyunsaturated fatty acids, brain function and mental health. Microb. Ecol. Health Dis. 2017, 28, 1281916. [Google Scholar] [CrossRef]
- Hoppenbrouwers, T.; Cvejić Hogervorst, J.H.; Garssen, J.; Wichers, H.J.; Willemsen, L.E.M. Long Chain Polyunsaturated Fatty Acids (LCPUFAs) in the Prevention of Food Allergy. Front. Immunol. 2019, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Soo, H.J.; Sam, K.K.; Chong, J.; Lau, N.S.; Ting, S.Y.; Kuah, M.K.; Kwang, S.Y.; Ranjani, M.; Shu-Chien, A.C. Functional characterisation of fatty acyl desaturase, Fads2, and elongase, Elovl5, in the Boddart’s goggle-eyed goby Boleophthalmus boddarti (Gobiidae) suggests an incapacity for long-chain polyunsaturated fatty acid biosynthesis. J. Fish Biol. 2020, 97, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Chen, C.; Dong, Y.; You, C.; Wang, S.; Monroig, O.; Tocher, D.R.; Li, Y. Regulation of long-chain polyunsaturated fatty acid biosynthesis in teleost fish. Prog. Lipid Res. 2021, 82, 101095. [Google Scholar] [CrossRef] [PubMed]
- Wijendran, V.; Brenna, J.T.; Wang, D.H.; Zhu, W.; Meng, D.; Ganguli, K.; Kothapalli, K.S.D.; Requena, P.; Innis, S.; Walker, W.A. Long-chain polyunsaturated fatty acids attenuate the IL-1β-induced proinflammatory response in human fetal intestinal epithelial cells. Pediatr. Res. 2015, 78, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Bunn, S.E.; Brett, M.T.; Kainz, M.J. Polyunsaturated fatty acids in stream food webs—High dissimilarity among producers and consumers. Freshw. Biol. 2017, 62, 1325–1334. [Google Scholar] [CrossRef]
- Castro, L.F.C.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Monroig, O.; Tocher, D.R.; Navarro, J.C. Biosynthesis of polyunsaturated fatty acids in marine invertebrates: Recent advances in molecular mechanisms. Mar. Drugs 2013, 11, 3998–4018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, X.Q.; Ye, Y.Q.; Wang, Q.; Li, Q.S.; Zhao, R.; Wang, H.W.; Li, J.T. Association between the Polymorphisms of fads2a and fads2b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio). Animals 2021, 11, 1780. [Google Scholar] [CrossRef] [PubMed]
- Renaville, B.; Tulli, F.; Bruno, M.; Tibaldi, E.; Messina, M. Fatty acid desaturase 2 (FADS2) insertion/deletion polymorphism impact on muscle fatty acid profile in European grayling (Thymallus thymallus). Br. J. Nutr. 2013, 110, 1559–1564. [Google Scholar] [CrossRef]
- Proskura, W.S.; Liput, M.; Zaborski, D.; Sobek, Z.; Yu, Y.H.; Cheng, Y.H.; Dybus, A. The effect of polymorphism in the FADS2 gene on the fatty acid composition of bovine milk. Arch. Anim. Breed. 2019, 62, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gan, Z.W.; Ding, Z.; Wu, Y.X.; Chen, X.Y.; Tian, H.M.; Liu, G.L.; Yang, Y.T.; Xie, L. Genetic Variants in the ELOVL5 but not ELOVL2 Gene Associated with Polyunsaturated Fatty Acids in Han Chinese Breast Milk. Biomed. Environ. Sci. 2017, 30, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Shimizu, Y.; Tanaka, A.; Nogi, T.; Tabuchi, I.; Oyama, K.; Taniguchi, M.; Mannen, H.; Sasazaki, S. The SNP in the promoter region of the bovine ELOVL5 gene influences economic traits including subcutaneous fat thickness. Mol. Biol. Rep. 2013, 40, 3231–3237. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, F.; Liu, T.; Yuan, L.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; et al. Ovine ELOVL5 and FASN genes polymorphisms and their correlations with sheep tail fat deposition. Gene 2022, 807, 145954. [Google Scholar] [CrossRef]
- Monroig, Ó.; Tocher, D.R.; Hontoria, F.; Navarro, J.C. Functional characterisation of a Fads2 fatty acyl desaturase with Δ6/Δ8 activity and an Elovl5 with C16, C18 and C20 elongase activity in the anadromous teleost meagre (Argyrosomus regius). Aquaculture 2013, 412–413, 14–22. [Google Scholar] [CrossRef]
- Ren, H.T.; Huang, Y.; Tang, Y.K.; Yu, J.H.; Xu, P. Two Elovl5-like elongase genes in Cyprinus carpio var. Jian: Gene characterization, mRNA expression, and nutritional regulation. Mol. Biol. 2015, 49, 592–600. [Google Scholar] [CrossRef]
- Li, J.T.; Wang, Q.; Huang Yang, M.D.; Li, Q.S.; Cui, M.S.; Dong, Z.J.; Wang, H.W.; Yu, J.H.; Zhao, Y.J.; Yang, C.R.; et al. Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat. Genet. 2021, 53, 1493–1503. [Google Scholar] [CrossRef]
- Li, Y.; Monroig, O.; Zhang, L.; Wang, S.; Zheng, X.; Dick, J.R.; You, C.; Tocher, D.R. Vertebrate fatty acyl desaturase with Delta4 activity. Proc. Natl. Acad. Sci. USA 2010, 107, 16840–16845. [Google Scholar] [CrossRef]
- Weckx, S.; Del-Favero, J.; Rademakers, R.; Claes, L.; Cruts, M.; De Jonghe, P.; Van Broeckhoven, C.; De Rijk, P. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005, 15, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.; Zhang, Z.; Kroon, D.; Casstevens, T.; Ramdoss, Y.; Buckler, E. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Frichot, E.; François, O.; O’Meara, B. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Francis, R.M. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Dereeper, A.; Nicolas, S.; Le Cunff, L.; Bacilieri, R.; Doligez, A.; Peros, J.P.; Ruiz, M.; This, P. SNiPlay: A web-based tool for detection, management and analysis of SNPs. Application to grapevine diversity projects. BMC Bioinform. 2011, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Blay, S.; Graham, J.; McNeney, B. LDheatmap: AnRFunction for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide Polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]
- Porth, I.; Klapšte, J.; Skyba, O.; Hannemann, J.; McKown, A.D.; Guy, R.D.; DiFazio, S.P.; Muchero, W.; Ranjan, P.; Tuskan, G.A.; et al. Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 2013, 200, 710–726. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jin, L.; Zhang, G.; Lu, Y.; Shen, Y.; Bao, J. Association mapping of grain color, phenolic content, flavonoid content and antioxidant capacity in dehulled rice. Theor. Appl. Genet. 2011, 122, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.N.; Rogers, C.E. Symbolic Description of Factorial Models for Analysis of Variance. Appl. Stat. 1973, 22, 392–399. [Google Scholar] [CrossRef]
- Charmet, G.; Tran, L.G.; Auzanneau, J.; Rincent, R.; Bouchet, S. BWGS: A R package for genomic selection and its application to a wheat breeding programme. PLoS ONE 2020, 15, e0222733. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.A.; van der Werf, J. Genomic best linear unbiased prediction (gBLUP) for the estimation of genomic breeding values. Methods Mol. Biol. 2013, 1019, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Reif, J.C. Modeling Epistasis in Genomic Selection. Genetics 2015, 201, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Li, Z.; Sillanpää, M.J. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor. Appl. Genet. 2012, 125, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Addendum: Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 2005, 67, 768. [Google Scholar] [CrossRef]
- De Los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The Bayesian Lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Iheshiulor, O.O.M.; Woolliams, J.A.; Svendsen, M.; Solberg, T.; Meuwissen, T.H.E. Simultaneous fitting of genomic-BLUP and Bayes-C components in a genomic prediction model. Genet. Sel. Evol. 2017, 49, 63. [Google Scholar] [CrossRef] [PubMed]
- Psofakis, P.; Karapanagiotidis, I.T.; Malandrakis, E.E.; Golomazou, E.; Exadactylos, A.; Mente, E. Effect of fishmeal replacement by hydrolyzed feather meal on growth performance, proximate composition, digestive enzyme activity, haematological parameters and growth-related gene expression of gilthead seabream (Sparus aurata). Aquaculture 2020, 521, 735006. [Google Scholar] [CrossRef]
- He, Z.; Zhang, R.; Jiang, F.; Zhang, H.; Zhao, A.; Xu, B.; Jin, L.; Wang, T.; Jia, W.; Jia, W.; et al. FADS1-FADS2 genetic polymorphisms are associated with fatty acid metabolism through changes in DNA methylation and gene expression. Clin. Epigenet. 2018, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Walle, P.; Männistö, V.; de Mello, V.D.; Vaittinen, M.; Perfilyev, A.; Hanhineva, K.; Ling, C.; Pihlajamäki, J. Liver DNA methylation of FADS2 associates with FADS2 genotype. Clin. Epigenet. 2019, 11, 10. [Google Scholar] [CrossRef]
- Walkiewicz, K.; Benitez Cardenas, A.S.; Sun, C.; Bacorn, C.; Saxer, G.; Shamoo, Y. Small changes in enzyme function can lead to surprisingly large fitness effects during adaptive evolution of antibiotic resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 21408–21413. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, X.; Peng, C.; Shi, Y.; Li, H.; Xu, Z.; Zhu, W. D3DistalMutation: A Database to Explore the Effect of Distal Mutations on Enzyme Activity. J. Chem. Inf. Model. 2021, 61, 2499–2508. [Google Scholar] [CrossRef]
- Baby, S.; Hyeong, K.E.; Lee, Y.M.; Jung, J.H.; Oh, D.Y.; Nam, K.C.; Kim, T.H.; Lee, H.K.; Kim, J.J. Evaluation of genome based estimated breeding values for meat quality in a berkshire population using high density single nucleotide polymorphism chips. Asian-Australas. J. Anim. Sci. 2014, 27, 1540–1547. [Google Scholar] [CrossRef][Green Version]
- Brito, L.F.; Clarke, S.M.; McEwan, J.C.; Miller, S.P.; Pickering, N.K.; Bain, W.E.; Dodds, K.G.; Sargolzaei, M.; Schenkel, F.S. Prediction of genomic breeding values for growth, carcass and meat quality traits in a multi-breed sheep population using a HD SNP chip. BMC Genet. 2017, 18, 7. [Google Scholar] [CrossRef]
- Liu, J.J.; Liang, A.X.; Campanile, G.; Plastow, G.; Zhang, C.; Wang, Z.; Salzano, A.; Gasparrini, B.; Cassandro, M.; Yang, L.G. Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J. Dairy Sci. 2018, 101, 433–444. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Lee, H.J.; Go, M.J.; Jang, H.B.; Choi, N.H.; Bae, J.B.; Castillo-Fernandez, J.E.; Bell, J.T.; Spector, T.D.; Lee, H.J.; et al. Genome-wide methylation analysis identifies ELOVL5 as an epigenetic biomarker for the risk of type 2 diabetes mellitus. Sci. Rep. 2018, 8, 14862. [Google Scholar] [CrossRef]
- Jin, M.; Monroig, Ó.; Navarro, J.C.; Tocher, D.R.; Zhou, Q.C. Molecular and functional characterisation of two elovl4 elongases involved in the biosynthesis of very long-chain (>C24) polyunsaturated fatty acids in black seabream Acanthopagrus schlegelii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 212, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.T.; Hamill, R.M.; O’Halloran, A.M.; Davey, G.C.; McBryan, J.; Mullen, A.M.; McGee, C.; Gispert, M.; Southwood, O.I.; Sweeney, T. SNP variation in the promoter of the PRKAG3 gene and association with meat quality traits in pig. BMC Genet. 2012, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Tian, H.; Lu, T.; Yu, M.; Xu, W.; Liu, G.; Xie, L. DHA intake interacts with ELOVL2 and ELOVL5 genetic variants to influence polyunsaturated fatty acids in human milk. J. Lipid Res. 2019, 60, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.C.; Song, L.; Guo, H.Y.; Guo, L.; Zhang, N.; Liu, B.S.; Jiang, S.G.; Zhang, D.C. Elovl4a participates in LC-PUFA biosynthesis and is regulated by PPARαβ in golden pompano Trachinotus ovatus (Linnaeus 1758). Sci. Rep. 2019, 9, 4684. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Marchesi, J.A.; Puig-Oliveras, A.; Revilla, M.; Estelle, J.; Alves, E.; Folch, J.M.; Ballester, M. Epigenetic regulation of the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs. Genet. Sel. Evol. 2015, 47, 20. [Google Scholar] [CrossRef] [PubMed]
| Locus | Gene | Position */Exon | Ho | He | PIC | MAF | Genotype | Amino Acid Change |
|---|---|---|---|---|---|---|---|---|
| E5a.29 | elovl5a | 29/1 | 0.1225 | 0.1649 | 0.1513 | 0.0907 | CC | T-I |
| CT | ||||||||
| TT | ||||||||
| E5a.87 | elovl5a | 87/2 | 0.0493 | 0.0480 | 0.0469 | 0.0246 | AG | L-L |
| GG | ||||||||
| E5a.180 | elovl5a | 180/2 | 0.1029 | 0.0976 | 0.0929 | 0.0515 | AC | S-S |
| CC | ||||||||
| E5a.351 | elovl5a | 351/4 | 0.0735 | 0.0708 | 0.0683 | 0.0368 | CC | Y-Y |
| CT | ||||||||
| E5a.429 | elovl5a | 429/4 | 0.0588 | 0.0843 | 0.0808 | 0.0441 | CC | H-H |
| CT | ||||||||
| TT | ||||||||
| E5a.552 | elovl5a | 552/5 | 0.4608 | 0.4538 | 0.3508 | 0.348 | CC | Y-Y |
| CT | ||||||||
| TT | ||||||||
| E5a.582 | elovl5a | 582/5 | 0.1569 | 0.2001 | 0.1801 | 0.1127 | AA | P-P |
| AG | ||||||||
| GG | ||||||||
| E5a.651 | elovl5a | 651/6 | 0.0294 | 0.029 | 0.0286 | 0.0147 | AA | T-T |
| GA | ||||||||
| E5a.798 | elovl5a | 798/7 | 0.0837 | 0.0802 | 0.077 | 0.0419 | AG | S-S |
| GG | ||||||||
| E5a.810 | elovl5a | 810/7 | 0.4724 | 0.4697 | 0.3594 | 0.3769 | AA | I-I |
| AT | ||||||||
| TT | ||||||||
| E5b.172 | elovl5b | 172/2 | 0.1822 | 0.1656 | 0.1519 | 0.0911 | CC | P-S |
| CT | ||||||||
| E5b.174 | elovl5b | 174/2 | 0.1413 | 0.1313 | 0.1227 | 0.0706 | CA | P-S |
| CC | ||||||||
| E5b.195 | elovl5b | 195/2 | 0.2156 | 0.1924 | 0.1739 | 0.1078 | AA | L-L |
| AC | ||||||||
| E5b.333 | elovl5b | 333/4 | 0.9071 | 0.4988 | 0.3744 | 0.4758 | CC | N-N |
| CT | ||||||||
| TT | ||||||||
| E5b.424 | elovl5b | 424/4 | 0.4387 | 0.3587 | 0.2944 | 0.2342 | CC | L-L |
| CT | ||||||||
| TT | ||||||||
| E5b.711 | elovl5b | 711/6 | 0.5576 | 0.435 | 0.3404 | 0.3197 | CC | T-T |
| TC | ||||||||
| TT | ||||||||
| E5b.782 | elovl5b | 782/7 | 0.145 | 0.1345 | 0.1254 | 0.0725 | GA | R-Q |
| GG | ||||||||
| E5b.813 | elovl5b | 813/7 | 0.2193 | 0.2181 | 0.1943 | 0.1245 | CC | N-N |
| CT | ||||||||
| TT |
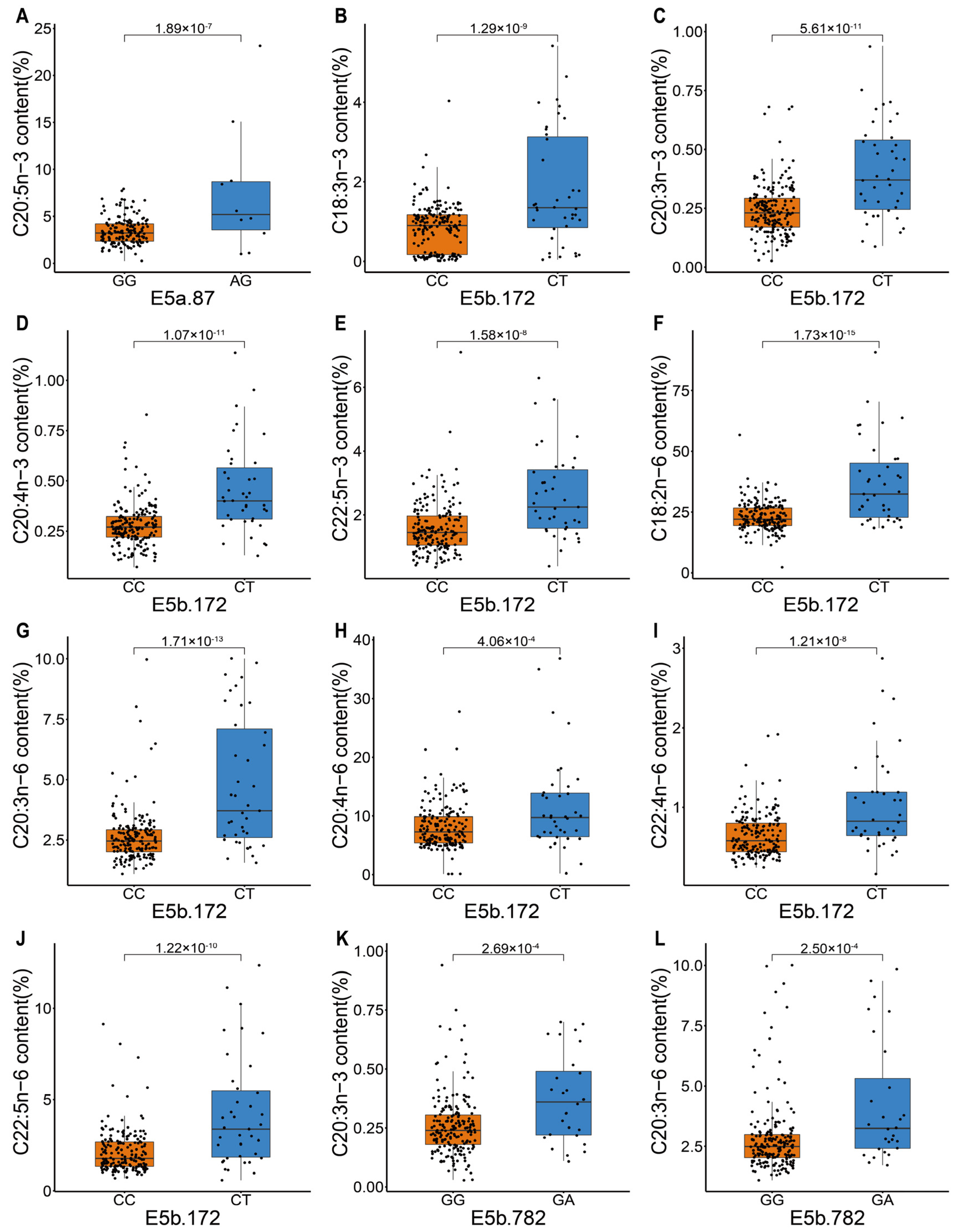
| Trait | Marker | Perm_p Value of GLM | MarkerR2 | FDR of ANOVA | MM | Mm |
|---|---|---|---|---|---|---|
| C18:3n-3 | E5b.172 | 4.44 × 10−3 | 6.90 | 1.29 × 10−9 | 0.84 ± 0.59 * | 1.79 ± 1.46 |
| C20:3n-3 | E5b.172 | 2.01 × 10−3 | 3.96 | 5.61 × 10−11 | 0.24 ± 0.11 | 0.41 ± 0.2 |
| E5b.782 | 3.54 × 10−2 | 2.15 | 2.69 × 10−4 | 0.26 ± 0.13 | 0.38 ± 0.19 | |
| C20:4n-3 | E5b.172 | 1.54 × 10−3 | 7.12 | 1.07 × 10−11 | 0.28 ± 0.11 | 0.46 ± 0.22 |
| C20:5n-3 | E5a.87 | 3.12 × 10−3 | 4.13 | 1.89 × 10−7 | 3.45 ± 1.46 | 7.57 ± 6.88 |
| C22:5n-3 | E5b.172 | 7.71 × 10−3 | 3.70 | 1.58 × 10−8 | 1.58 ± 0.82 | 2.61 ± 1.36 |
| C18:2n-6 | E5b.172 | 3.60 × 10−4 | 2.86 | 1.73 × 10−15 | 23.21 ± 5.75 | 37.02 ± 17 |
| C20:3n-6 | E5b.172 | 6.70 × 10−4 | 3.37 | 1.71 × 10−13 | 2.63 ± 1.12 | 4.82 ± 2.71 |
| E5b.782 | 3.51 × 10−2 | 4.48 | 2.50 × 10−4 | 2.83 ± 1.51 | 4.25 ± 2.53 | |
| C20:4n-6 | E5b.172 | 3.67 × 10−2 | 2.10 | 4.06 × 10−4 | 8.11 ± 3.79 | 11.61 ± 8.04 |
| C22:4n-6 | E5b.172 | 7.23 × 10−3 | 3.44 | 1.21 × 10−8 | 0.65 ± 0.28 | 1.05 ± 0.62 |
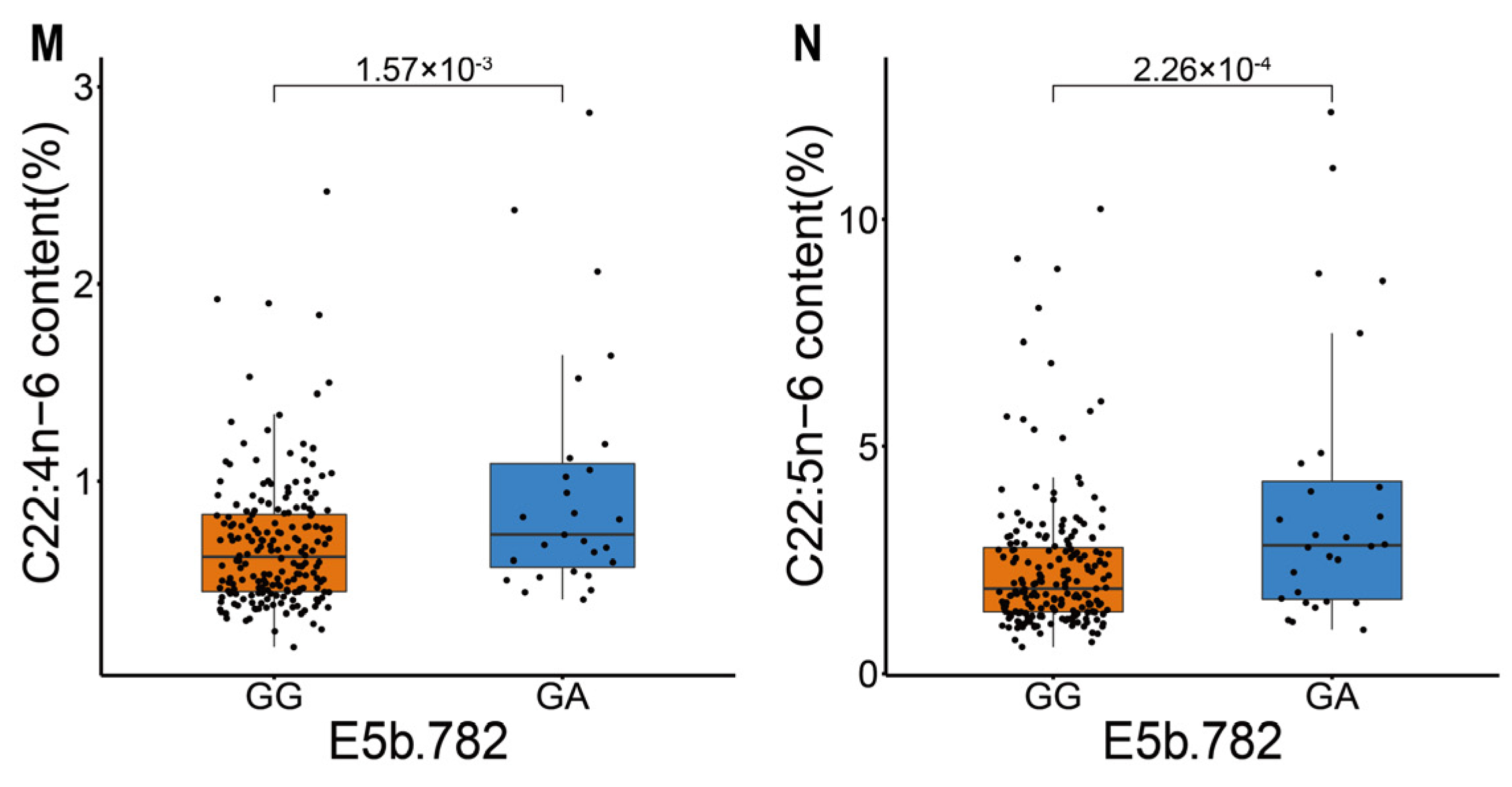
| E5b.782 | 4.51 × 10−2 | 1.83 | 1.57 × 10−3 | 0.68 ± 0.33 | 0.97 ± 0.62 | |
| C22:5n-6 | E5b.172 | 2.30 × 10−3 | 4.07 | 1.22 × 10−10 | 2.15 ± 1.2 | 4.23 ± 3.02 |
| E5b.782 | 3.46 × 10−2 | 1.83 | 2.26 × 10−4 | 2.32 ± 1.51 | 3.84 ± 3.06 |
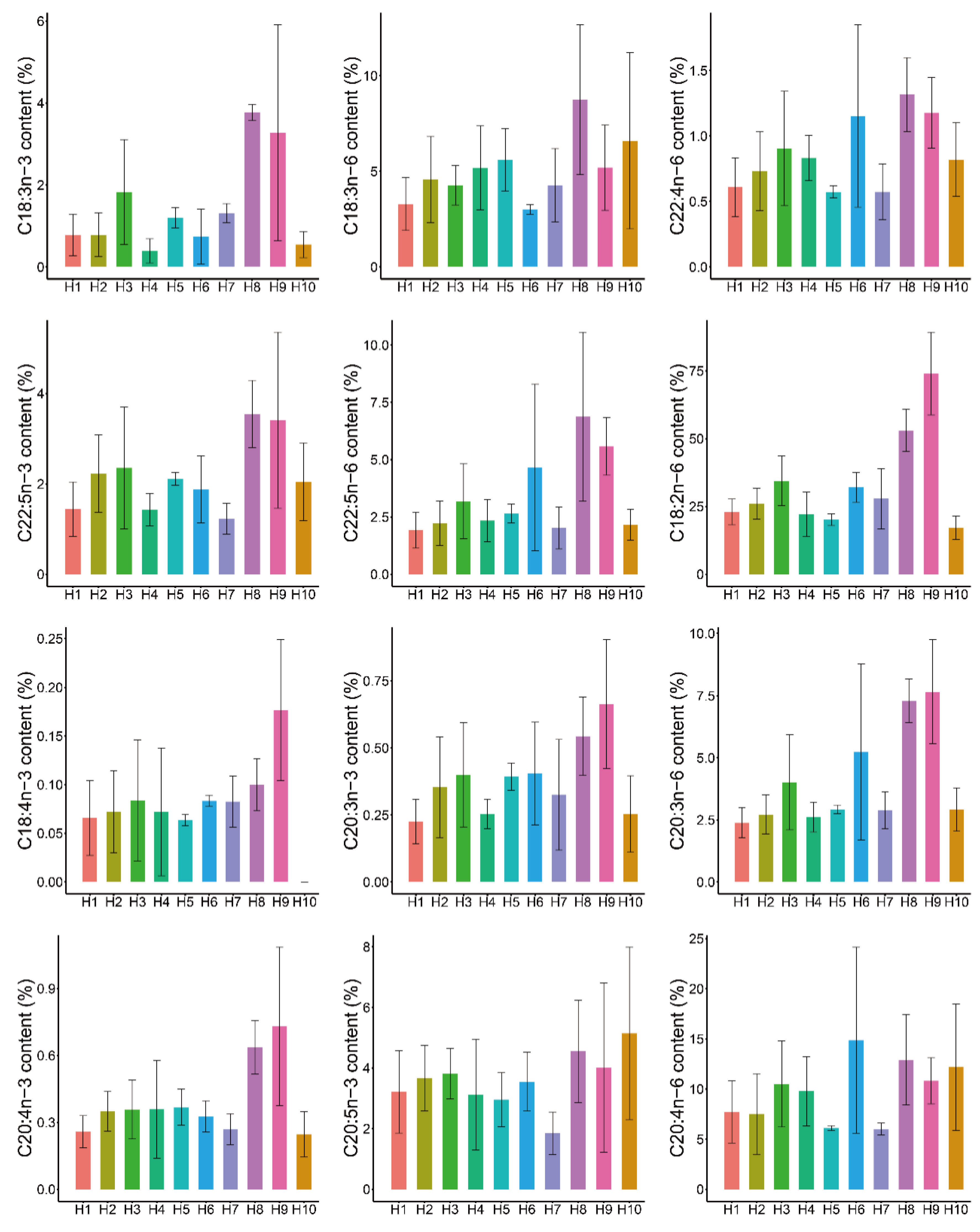
| PUFA | fads2b.751 | fads2b.1197 | E5b.172 | E5b.782 | Genotype Combination |
|---|---|---|---|---|---|
| C18:3n-3 | 5 | NA | 6.9 | NA | 33.6 |
| C20:3n-3 | 13 | 4.3 | 3.96 | 2.15 | 32.73 |
| C20:4n-3 | 11 | 5 | 7.12 | NA | 33.22 |
| C22:5n-3 | 11 | 5 | 3.7 | NA | 22.63 |
| C18:2n-6 | NA | 4 | 2.86 | NA | 54.95 |
| C20:3n-6 | 11 | 4 | 3.37 | 4.48 | 37.59 |
| C22:4n-6 | 7 | NA | 3.44 | 1.83 | 21.26 |
| C22:5n-6 | 9 | 3 | 4.07 | 1.83 | 28.19 |
| C18:4n-3 | NA | NA | NA | NA | 4.24 |
| C18:3n-6 | NA | NA | NA | NA | 5.02 |
| C20:5n-3 | NA | NA | NA | NA | 8.02 |
| C20:4n-6 | NA | NA | 2.1 | NA | 13.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Q.-S.; Ye, Y.-Q.; Wang, Q.; Sun, X.-Q.; Zhao, R.; Li, J.-T. Association Analysis between Genetic Variants of elovl5a and elovl5b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio). Biology 2022, 11, 466. https://doi.org/10.3390/biology11030466
Zhang Y, Li Q-S, Ye Y-Q, Wang Q, Sun X-Q, Zhao R, Li J-T. Association Analysis between Genetic Variants of elovl5a and elovl5b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio). Biology. 2022; 11(3):466. https://doi.org/10.3390/biology11030466
Chicago/Turabian StyleZhang, Yan, Qing-Song Li, Yu-Qing Ye, Qi Wang, Xiao-Qing Sun, Ran Zhao, and Jiong-Tang Li. 2022. "Association Analysis between Genetic Variants of elovl5a and elovl5b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio)" Biology 11, no. 3: 466. https://doi.org/10.3390/biology11030466
APA StyleZhang, Y., Li, Q.-S., Ye, Y.-Q., Wang, Q., Sun, X.-Q., Zhao, R., & Li, J.-T. (2022). Association Analysis between Genetic Variants of elovl5a and elovl5b and Poly-Unsaturated Fatty Acids in Common Carp (Cyprinus carpio). Biology, 11(3), 466. https://doi.org/10.3390/biology11030466






