Proteotranscriptomic Analysis and Toxicity Assay Suggest the Functional Distinction between Venom Gland Chambers in Twin-Spotted Assassin Bug, Platymeris biguttatus
Abstract
:Simple Summary
Abstract
1. Introduction
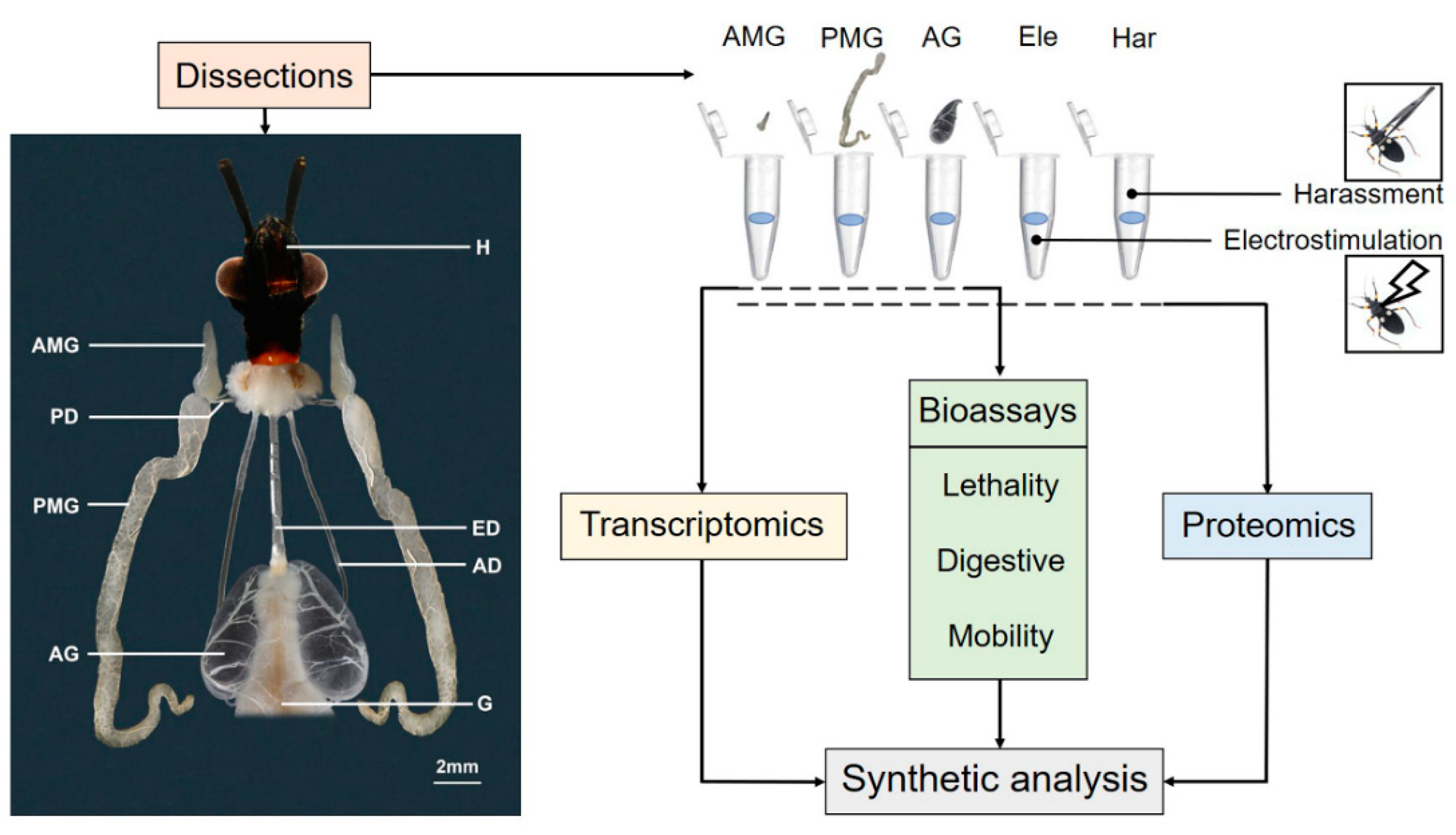
2. Materials and Methods
2.1. Salivary Venom Sampling by Stimulation Treatments
2.2. Salivary Venom Collection by Venom Gland Dissection
2.3. Gland Imaging
2.4. RNA-SEQ and De Novo Transcriptome Assembly
2.5. NanoLC-MS/MS Analysis and Sequence Annotation of Secreted Proteins
2.6. Toxicological Bioassays of Salivary Glands
3. Results
3.1. Morphology of the Salivary Gland
3.2. The Compositions of Secreted Protein in Different Glands
3.3. Effects of Different Venom Gland Chamber Extracts on Galleria mellonella Larvae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutertre, S.; Jin, A.H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, S.D.; Mueller, A.; Clayton, D.; Starobova, H.; Hamilton, B.R.; Payne, R.J.; Vetter, I.; King, G.F.; Undheim, E.A.B. A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci. Adv. 2018, 4, e4640. [Google Scholar] [CrossRef] [Green Version]
- Inceoglu, B.; Lango, J.; Jing, J.; Chen, L.; Doymaz, F.; Pessah, I.N.; Hammock, B.D. One scorpion, two venoms: Prevenom of Parabuthus transvaalicus acts as an alternative type of venom with distinct mechanism of action. Proc. Natl. Acad. Sci. USA 2003, 100, 922–927. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.A.; Rosenthal, M.; Undheim, E.E.A.; King, G.F. Harvesting venom toxins from assassin bugs and other heteropteran insects. J. Vis. Exp. 2018, 2018, e57729. [Google Scholar] [CrossRef]
- Walker, A.A.; Robinson, S.D.; Undheim, E.A.B.; Jin, J.; Han, X.; Fry, B.G.; Vetter, I.; King, G.F. Missiles of mass disruption: Composition and glandular origin of venom used as a projectile defensive weapon by the assassin bug Platymeris Rhadamanthus. Toxins 2019, 11, 673. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.A.; Weirauch, C.; Fry, B.G.; King, G.F. Venoms of heteropteran insects: A treasure trove of diverse pharmacological toolkits. Toxins 2016, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.S. The action and composition of the saliva of an assassin bug Platymeris rhadamanthus Gaerst. (Hemiptera, Reduviidae). J. Exp. Biol. 1961, 38, 61–77. [Google Scholar] [CrossRef]
- Cantón, P.E.; Bonning, B.C. Extraoral digestion: Outsourcing the role of the hemipteran midgut. Curr. Opin. Insect Sci. 2020, 41, 86–91. [Google Scholar] [CrossRef]
- Vanderplank, F.L. The assassin bug, Platymerus rhadamanthus Gerst (Hemiptera: Reduviidae), a useful predator of the rhinoceros beetles Oryctes boas (F.) and Oryctes monoceros (Oliv.). (Coleoptera: Scarabaeidae). J. Entomol. Soc. S. Afr. 1958, 21, 309–314. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Assumpção, T.C.; Francischetti, I.M.B. An insight into the sialomes of bloodsucking Heteroptera. Psyche 2012, 2012, 470436. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.A.; Mayhew, M.L.; Jin, J.; Herzig, V.; Undheim, E.A.B.; Sombke, A.; Fry, B.G.; Meritt, D.J.; King, G.F. The assassin bug Pristhesancus plagipennis produces two distinct venoms in separate gland lumens. Nat. Commun. 2018, 9, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M.L.; Wielsch, N.; Heckel, D.G.; Vilcinskas, A.; Vogel, H. Context-dependent venom deployment and protein composition in two assassin bugs. Ecol. Evol. 2020, 10, 9932–9947. [Google Scholar] [CrossRef]
- Haridass, E.; Ananthakrishnan, T. Functional morphology of the salivary system in some Reduviidae (Insecta-Heteroptera). Proc. Indian Acad. Sci. Anim. Sci. 1981, 90, 145–160. [Google Scholar] [CrossRef]
- Miles, P.W. The physiological division of labour in the salivary glands of Oncopeltus fasciatus (Dall.) (Heteroptera: Lygaeidae). Aust. J. Biol. Sci. 1967, 20, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Swart, C.C.; Deaton, L.E.; Felgenhauer, B.E. The salivary gland and salivary enzymes of the giant waterbugs (Heteroptera; Belostomatidae). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006, 145, 114–122. [Google Scholar] [CrossRef]
- Walker, A.A.; Madio, B.; Jin, J.; Undheim, E.A.B.; Fry, B.G.; King, G.F. Melt with this kiss: Paralyzing and liquefying venom of the assassin bug Pristhesancus plagipennis (Hemiptera: Reduviidae). Mol. Cell. Proteom. 2017, 16, 552–566. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Schwanhüusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Nilsson, T.; Warren, G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Curr. Opin. Cell Biol. 1994, 6, 517–521. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, 459–466. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Akçay, A. The calculation of LD50 using probit analysis. FASEB J. 2013, 27, 1217–1228. [Google Scholar] [CrossRef]
- Seifert, E. OriginPro 9.1: Scientific data analysis and graphing software-Software review. J. Chem. Inf. Model. 2014, 54, 1552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.M.; Sahayaraj, K. Gross morphology and histology of head and salivary apparatus of the predatory bug, Rhynocoris marginatus. J. Insect Sci. 2012, 12, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.A.; Robinson, S.D.; Yeates, D.K.; Jin, J.; Baumann, K.; Dobson, J.; Fry, B.G.; King, G.F. Entomo-venomics: The evolution, biology and biochemistry of insect venoms. Toxicon 2018, 154, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, R.M.; Sforça, M.L.; Amino, R.; Juliano, M.A.; Oyama, S.; Juliano, L.; Pertinhez, T.A.; Spisni, A.; Schenkman, S. Lytic activity and structural differences of amphipathic peptides derived from trialysin. Biochemistry 2006, 45, 1765–1774. [Google Scholar] [CrossRef]
- Amino, R.; Martins, R.M.; Procopio, J.; Hirata, I.Y.; Juliano, M.A.; Schenkman, S. Trialysin, a novel pore-forming protein from saliva of hematophagous insects activated by limited proteolysis. J. Biol. Chem. 2002, 277, 6207–6213. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, E.L.; Iwasaki, K.; Tsuji, Y. Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 997–1030. [Google Scholar] [CrossRef] [Green Version]
- Nicotra, S.; Sorio, D.; Filippi, G.; De Gioia, L.; Paterlini, V.; De Palo, E.F.; Grandori, R.; Tagliaro, F.; Santambrogio, C. Terbium chelation, a specific fluorescent tagging of human transferrin. Optimization of conditions in view of its application to the HPLC analysis of carbohydrate-deficient transferrin (CDT). Anal. Bioanal. Chem. 2017, 409, 6605–6612. [Google Scholar] [CrossRef]
- Ritchie, R.F.; Palomaki, G.E.; Neveux, L.M.; Navolotskaia, O.; Ledue, T.B.; Craig, W.Y. Reference distributions for the negative acute-phase serum proteins, albumin, transferrin and transthyretin: A practical, simple and clinically relevant approach in a large cohort. J. Clin. Lab. Anal. 1999, 13, 273–279. [Google Scholar] [CrossRef]
- Wang, J.; Sykes, B.D.; Ryan, R.O. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 1188–1193. [Google Scholar] [CrossRef] [Green Version]
- Whitten, M.M.A.; Tew, I.F.; Lee, B.L.; Ratcliffe, N.A. A novel role for an insect apolipoprotein (Apolipophorin III) in β-1,3-glucan pattern recognition and cellular encapsulation reactions. J. Immunol. 2004, 172, 2177–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras, E.; Rausell, C.; Real, M.D. Tribolium castaneum apolipophorin-III acts as an immune response protein against Bacillus thuringiensis Cry3Ba toxic activity. J. Invertebr. Pathol. 2013, 113, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Agwa, A.J.; Szanto, T.G.; Csóti, A.; Panyi, G.; Schroeder, C.I.; Walker, A.A.; King, G.F. Weaponisation ‘on the fly’: Convergent recruitment of knottin and defensin peptide scaffolds into the venom of predatory assassin flies. Insect Biochem. Mol. Biol. 2020, 118, 103310. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, F.; Tian, L.; Li, X.; Zhang, Y.; Wang, T.; Ma, L.; Song, F.; Cai, W.; Li, H. Proteotranscriptomic Analysis and Toxicity Assay Suggest the Functional Distinction between Venom Gland Chambers in Twin-Spotted Assassin Bug, Platymeris biguttatus. Biology 2022, 11, 464. https://doi.org/10.3390/biology11030464
Gao F, Tian L, Li X, Zhang Y, Wang T, Ma L, Song F, Cai W, Li H. Proteotranscriptomic Analysis and Toxicity Assay Suggest the Functional Distinction between Venom Gland Chambers in Twin-Spotted Assassin Bug, Platymeris biguttatus. Biology. 2022; 11(3):464. https://doi.org/10.3390/biology11030464
Chicago/Turabian StyleGao, Fanding, Li Tian, Xinyu Li, Yinqiao Zhang, Tianfang Wang, Ling Ma, Fan Song, Wanzhi Cai, and Hu Li. 2022. "Proteotranscriptomic Analysis and Toxicity Assay Suggest the Functional Distinction between Venom Gland Chambers in Twin-Spotted Assassin Bug, Platymeris biguttatus" Biology 11, no. 3: 464. https://doi.org/10.3390/biology11030464
APA StyleGao, F., Tian, L., Li, X., Zhang, Y., Wang, T., Ma, L., Song, F., Cai, W., & Li, H. (2022). Proteotranscriptomic Analysis and Toxicity Assay Suggest the Functional Distinction between Venom Gland Chambers in Twin-Spotted Assassin Bug, Platymeris biguttatus. Biology, 11(3), 464. https://doi.org/10.3390/biology11030464







