Evaluation of the Saline–Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
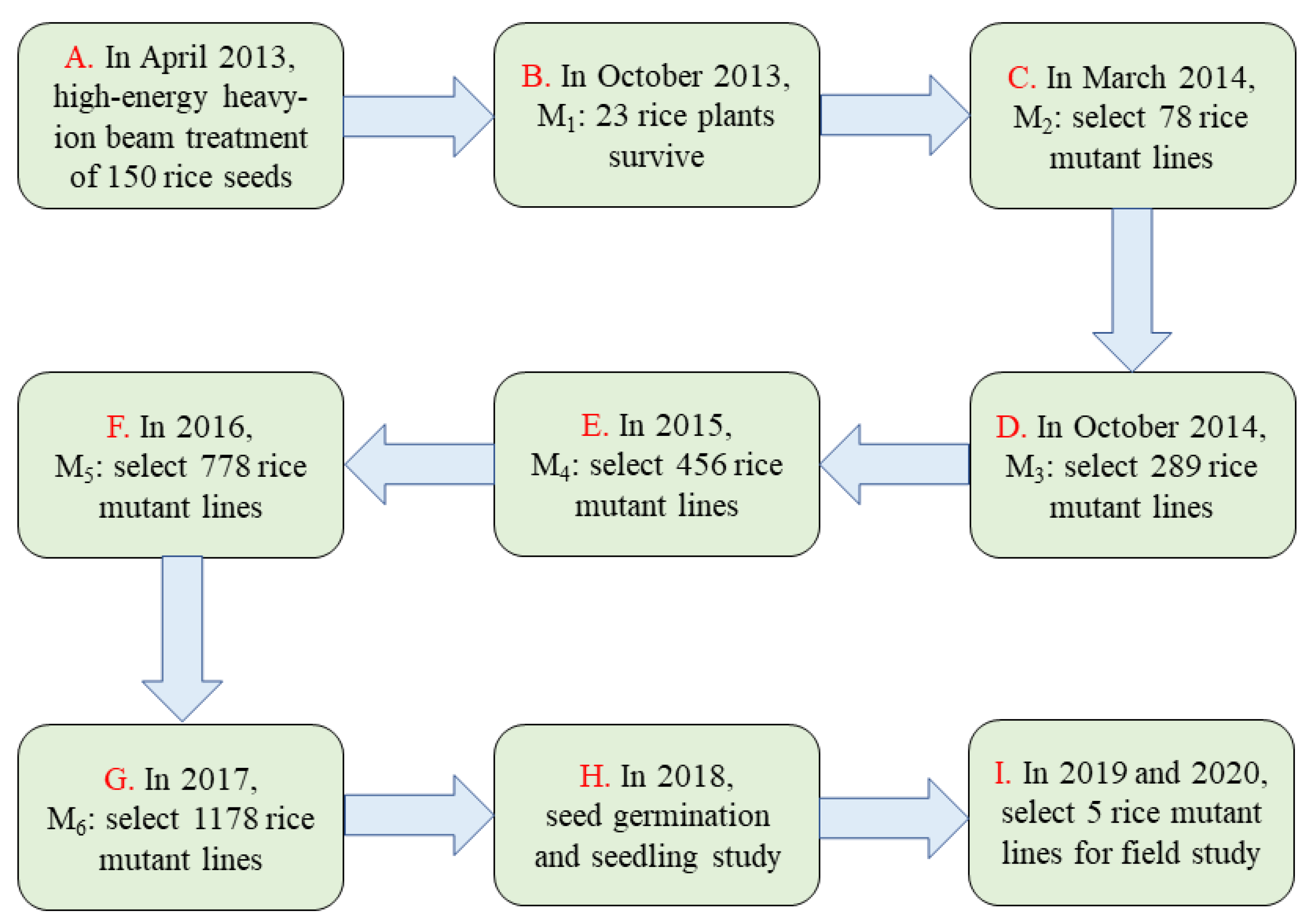
2.1. Plant Materials and Irradiation Treatments
2.2. Breeding of the Mutant Population
2.3. Seed Germination of Mutant Lines under Saline–Alkaline Stress
2.4. Seedling Growth of Mutant Lines under Saline–Alkaline Stress
2.5. Field Growth of Mutant Lines under Saline–Alkaline Stress
2.6. Field Performance of M89
2.7. Statistical Analysis
3. Results
3.1. Seed Germination of Mutant Lines under Saline–Alkaline Stress
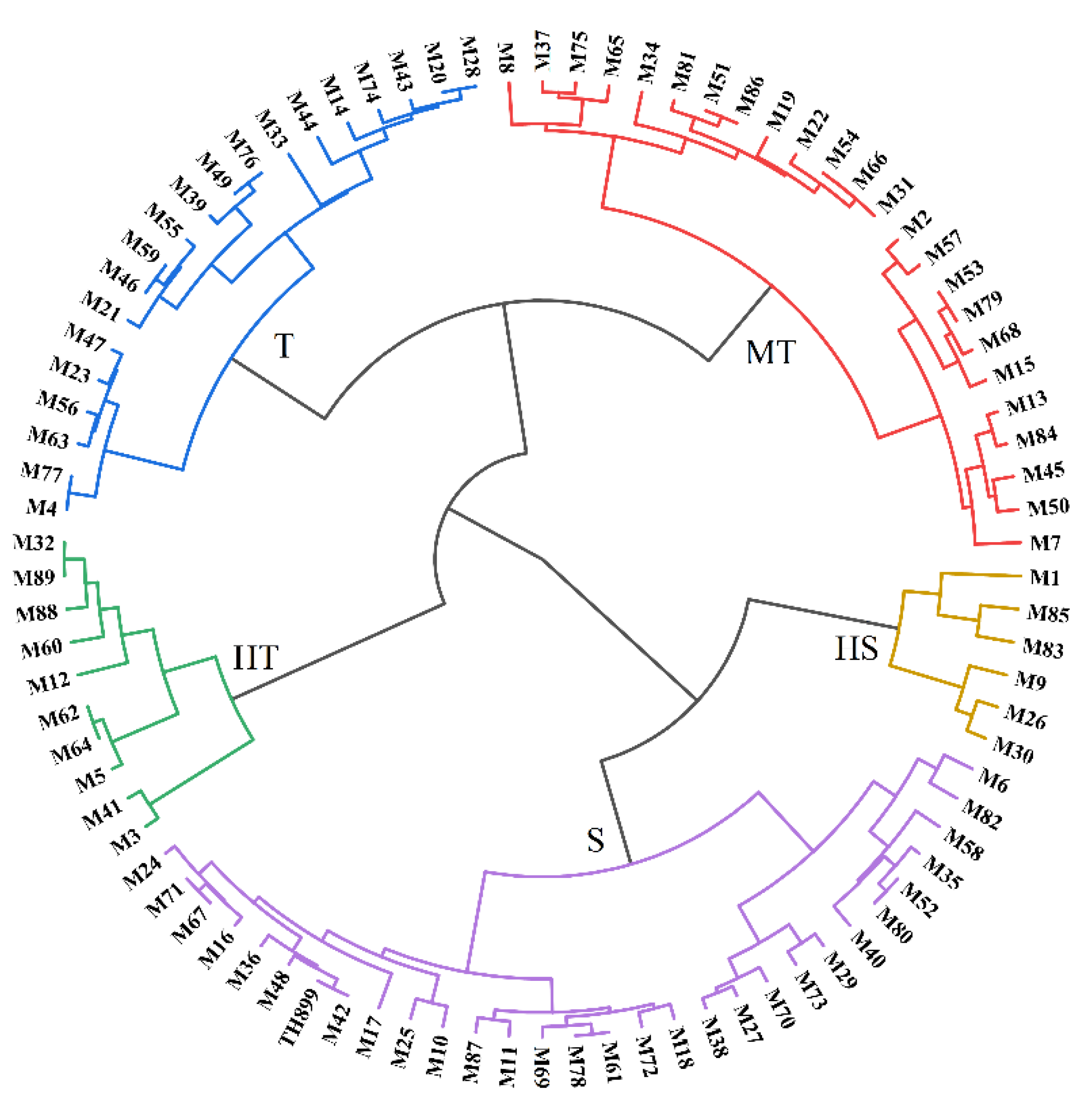
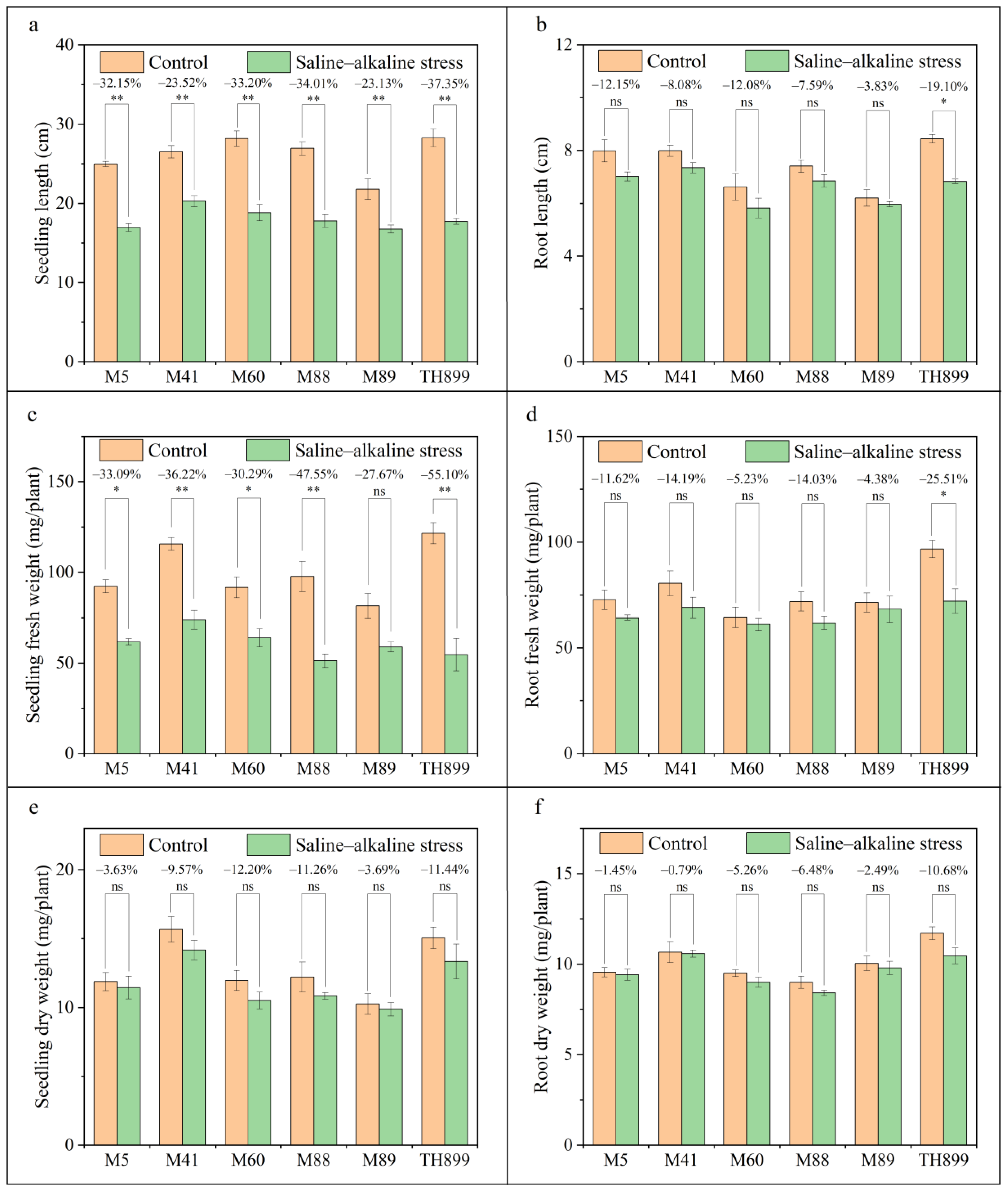
3.2. Seedling Growth of Mutant Lines under Saline–Alkaline Stress
3.3. K+ and Na+ Content in Mutant Lines under Saline–Alkaline Stress
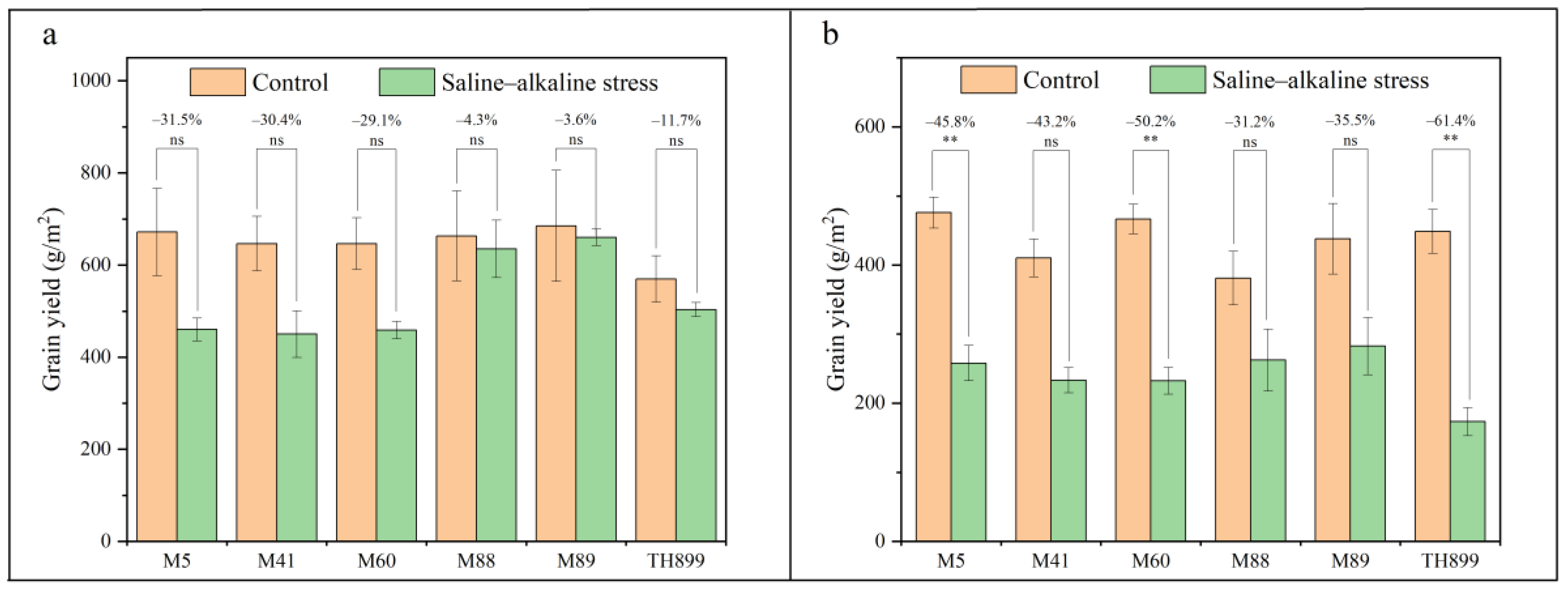
3.4. Grain Yield and Yield Components of Mutant Lines under Saline–Alkaline Stress
3.5. Field Performance of M89
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Amini, S.; Ghadiri, H.; Chen, C.; Marschner, P. Salt-affected soils, reclamation, carbon dynamics, and biochar: A review. J. Soils Sediments 2016, 16, 939–953. [Google Scholar] [CrossRef]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil Salinity: Historical Perspectives and a World Overview of the Problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Zaman, M., Shahid, S.A., Heng, L., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 43–53. [Google Scholar]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 2008, 56, 179–190. [Google Scholar] [CrossRef]
- Zhang, X.G.; Huang, B.; Liang, Z.W.; Zhao, Y.C.; Sun, W.X.; Hu, W.Y. Study on salinization characteristics of surface soil in Western Songnen Plain. Soils 2013, 45, e338. [Google Scholar] [CrossRef]
- Ma, H.; Liang, Z. Effects of different soil pH and soil extracts on the germination and seedling growth of Leymus chinensis. Chin. Bull. Bot. 2007, 24, 181. [Google Scholar]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Pan, T.; Allakhverdiev, S.I.; Yu, M.; Shabala, S. Crop halophytism: An environmentally sustainable solution for global food security. Trends Plant Sci. 2020, 25, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [Green Version]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef] [Green Version]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Haque, M.A.; Rafii, M.Y.; Yusoff, M.M.; Ali, N.S.; Yusuff, O.; Datta, D.R.; Anisuzzaman, M.; Ikbal, M.F. Advanced Breeding Strategies and Future Perspectives of Salinity Tolerance in Rice. Agronomy 2021, 11, 1631. [Google Scholar] [CrossRef]
- Jenks, M.A.; Hasegawa, P.M.; Jain, S.M. Advances in Molecular Breeding toward Drought and Salt Tolerant Crops; Springer: New York, NY, USA, 2007. [Google Scholar]
- Sun, M.F.; Yan, G.H.; Wang, A.M.; Zhu, G.Y.; Tang, H.S.; He, C.X.; Ren, Z.L.; Liu, K.; Zhang, G.Y.; Shi, W. Research progress on the breeding of salt-tolerant rice varieties. Barley Cereals Sci. 2017, 34, 1–9. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Zhao, L.; Lu, K.; Zhu, Z.; Chen, T.; Zhao, Q.; Yao, S.; Zhou, L.; Zhao, C. Research status, problems and suggestions on salt-alkali tolerant rice. China Rice 2019, 25, 1–6. [Google Scholar] [CrossRef]
- Kotula, L.; Garcia Caparros, P.; Zörb, C.; Colmer, T.D.; Flowers, T.J. Improving crop salt tolerance using transgenic approaches: An update and physiological analysis. Plant Cell Environ. 2020, 43, 2932–2956. [Google Scholar] [CrossRef] [PubMed]
- Pental, D. When scientists turn against science: Exceptionally flawed analysis of plant breeding technologies. Curr. Sci. 2019, 117, 932–939. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Parry, M.A.J.; Madgwick, P.J.; Bayon, C.; Tearall, K.; Hernandez-Lopez, A.; Baudo, M.; Rakszegi, M.; Hamada, W.; Al-Yassin, A.; Ouabbou, H. Mutation discovery for crop improvement. J. Exp. Bot. 2009, 60, 2817–2825. [Google Scholar] [CrossRef] [Green Version]
- Ahloowalia, B.S.; Maluszynski, M.; Nichterlein, K. Global impact of mutation-derived varieties. Euphytica 2004, 135, 187–204. [Google Scholar] [CrossRef]
- Masoabi, M.; Lloyd, J.; Kossmann, J.; van der Vyver, C. Ethyl methanesulfonate mutagenesis and in vitro polyethylene glycol selection for drought tolerance in sugarcane (Saccharum spp.). Sugar Tech 2018, 20, 50–59. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Tah, J. Mutagenesis—A Potential Approach for Crop Improvement. In Crop Improvement: New Approaches and Modern Techniques; Hakeem, K.R., Ahmad, P., Ozturk, M., Eds.; Springer: Boston, MA, USA, 2013; pp. 149–187. [Google Scholar]
- Serrat, X.; Esteban, R.; Guibourt, N.; Moysset, L.; Nogués, S.; Lalanne, E. EMS mutagenesis in mature seed-derived rice calli as a new method for rapidly obtaining TILLING mutant populations. Plant Methods 2014, 10, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.-w.; Luo, H.-c.; Li, J.-j.; Yu, X.-q. Molecular Variation and Application from Aerospace Mutagenesis in Upland Rice Huhan 3 and Huhan 7. Rice Sci. 2013, 20, 249–258. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, S.; Huang, J.; Fu, H.; Zhou, L.; Furusawa, Y.; Shu, Q. Mutagenic Effect of Three Ion Beams on Rice and Identification of Heritable Mutations by Whole Genome Sequencing. Plants 2020, 9, 551. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shimizu, A.; Nishio, T.; Tsutsumi, N.; Kato, H. Comparison and characterization of mutations induced by gamma-ray and carbon-ion irradiation in rice (Oryza sativa L.) using whole-genome resequencing. G3 Genes Genomes Genet. 2019, 9, 3743–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Luo, S.; Yu, L.; Cui, T.; Chen, X.; Yang, J.; Li, X.; Li, W.; Wang, J.; Zhou, L. Strategies for identification of mutations induced by carbon-ion beam irradiation in Arabidopsis thaliana by whole genome re-sequencing. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2018, 807, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Hisamura, A.; Mine, D.; Takebe, T.; Abe, T.; Hayashi, Y.; Hirano, T. Breeding of summer-autumn flowering chrysanthemum cv. Hakuryo with a little generation of malformed flower. RIKEN Accel. Prog. Rep. 2016, 49, 24. [Google Scholar]
- Okamura, M.; Nakayama, M.; Umemoto, N.; Cano, E.A.; Hase, Y.; Nishizaki, Y.; Sasaki, N.; Ozeki, Y. Crossbreeding of a metallic color carnation and diversification of the peculiar coloration by ion-beam irradiation. Euphytica 2013, 191, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Kazama, Y.; Fujiwara, M.T.; Takehisa, H.; Ohbu, S.; Saito, H.; Ichida, H.; Hayashi, Y.; Abe, T. Characterization of a heavy-ion induced white flower mutant of allotetraploid Nicotiana tabacum. Plant Cell Rep. 2013, 32, 11–19. [Google Scholar] [CrossRef]
- Duan, H.; Li, X.; Zhou, C.; Zhou, Y. Effects of low energy ion beam on fruit qualities of tomato. Guizhou Agric. Sci. 2014, 42, 168–171. [Google Scholar]
- Dong, X.; Li, W. Biological features of an early-maturity mutant of sweet sorghum induced by carbon ions irradiation and its genetic polymorphism. Adv. Space Res. 2012, 50, 496–501. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.-K.; Tu, B.-J.; Li, Y.-S.; Zhang, Q.-Y.; Liu, X.-B. Effects of Carbon Ion Beam Irradiation on Phenotypic Variations and Biochemical Parameters in Early Generations of Soybean Plants. Agriculture 2021, 11, 98. [Google Scholar] [CrossRef]
- Li, Y.; Meng, L.; Jia, Y.; Han, K.; Shi, Y.; Jin, D. Breeding Report on the New Rice Variety Tonghe-899. North Rice 2016, 46, 54–55. [Google Scholar] [CrossRef]
- Lv, B.S.; Ma, H.Y.; Li, X.W.; Wei, L.X.; Lv, H.Y.; Yang, H.Y.; Jiang, C.J.; Liang, Z.W. Proline Accumulation Is Not Correlated with Saline-Alkaline Stress Tolerance in Rice Seedlings. Agron. J. 2015, 107, 51–60. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, K. Comparison of extractive methods of Na and K in wheat leaves. Plant Physiol. Commun. 1995, 31, 50–52. [Google Scholar] [CrossRef]
- Yoshida, S. Fundamentals of Rice Crop Science; Int. Rice Res. Inst.: Los Banos, Philippines, 1981. [Google Scholar]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 2020, 11, 833. [Google Scholar] [CrossRef]
- Ahloowalia, B.S.; Maluszynski, M. Induced mutations–A new paradigm in plant breeding. Euphytica 2001, 118, 167–173. [Google Scholar] [CrossRef]
- Gottschalk, W.; Wolff, G. Induced Mutations in Plant Breeding; Springer Science & Business Media: New York, NY, USA, 2012; Volume 7. [Google Scholar]
- Song, J.Y.; Kim, D.S.; Lee, M.C.; Lee, K.J.; Kim, J.B.; Kim, S.H.; Ha, B.K.; Yun, S.J.; Kang, S.Y. Physiological characterization of gamma-ray induced salt tolerant rice mutants. Aust. J. Crop Sci. 2012, 6, 421–429. [Google Scholar] [CrossRef]
- Rasel, M.; Tahjib-Ul-Arif, M.; Hossain, M.A.; Hassan, L.; Farzana, S.; Brestic, M. Screening of salt-tolerant rice landraces by seedling stage phenotyping and dissecting biochemical determinants of tolerance mechanism. J. Plant Growth Regul. 2021, 40, 1853–1868. [Google Scholar] [CrossRef]
- Peng, Z.; He, S.; Sun, J.; Pan, Z.; Gong, W.; Lu, Y.; Du, X. Na+ compartmentalization related to salinity stress tolerance in upland cotton (Gossypium hirsutum) seedlings. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.-H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef] [PubMed]
- Nakhoda, B.; Leung, H.; Mendioro, M.S.; Mohammadi-nejad, G.; Ismail, A.M. Isolation, characterization, and field evaluation of rice (Oryza sativa L., Var. IR64) mutants with altered responses to salt stress. Field Crops Res. 2012, 127, 191–202. [Google Scholar] [CrossRef]
- Singh, R.K.; Redoña, E.; Refuerzo, L. Varietal improvement for abiotic stress tolerance in crop plants: Special reference to salinity in rice. In Abiotic Stress Adaptation in Plants; Springer: New York, NY, USA, 2009; pp. 387–415. [Google Scholar]
- Domingo, C.; Lalanne, E.; Catalá, M.M.; Pla, E.; Reig-Valiente, J.L.; Talón, M. Physiological basis and transcriptional profiling of three salt-tolerant mutant lines of rice. Front. Plant Sci. 2016, 7, 1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Type | pH | EC (dS/m) | CEC (cmol/kg) | Na+ (cmol/kg) | ESP (%) |
|---|---|---|---|---|---|
| Field 1 | 8.3 | 0.31 | 37.41 | 3.85 | 10.6 |
| Field 2 | 9.2 | 0.48 | 27.66 | 9.15 | 32.8 |
| G | GP (%) | G | GP (%) | G | GP (%) | |||
|---|---|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Control | Stress | |||
| M1 | 96.7 | 54 | M31 | 100 | 48.7 | M61 | 100 | 39.3 |
| M2 | 100 | 48.7 | M32 | 100 | 81.3 | M62 | 100 | 76 |
| M3 | 99.3 | 36 | M33 | 100 | 66 | M63 | 100 | 68.7 |
| M4 | 100 | 81.3 | M34 | 100 | 46 | M64 | 100 | 76.7 |
| M5 | 99.3 | 88.7 | M35 | 98.7 | 27.3 | M65 | 100 | 44.7 |
| M6 | 100 | 19.3 | M36 | 100 | 37.3 | M66 | 100 | 48.7 |
| M7 | 98.7 | 53.3 | M37 | 97.3 | 45.3 | M67 | 100 | 35.3 |
| M8 | 99.3 | 61.3 | M38 | 100 | 30.7 | M68 | 100 | 55.3 |
| M9 | 100 | 15.3 | M39 | 99.3 | 64 | M69 | 99.3 | 40 |
| M10 | 99.3 | 41.3 | M40 | 100 | 26 | M70 | 100 | 28.7 |
| M11 | 98.7 | 40 | M41 | 100 | 88 | M71 | 100 | 36 |
| M12 | 100 | 76 | M42 | 98 | 38 | M72 | 99.3 | 38.7 |
| M13 | 100 | 56.7 | M43 | 100 | 60.7 | M73 | 99.3 | 29.3 |
| M14 | 100 | 59.3 | M44 | 96 | 62 | M74 | 100 | 62 |
| M15 | 100 | 56 | M45 | 100 | 54 | M75 | 100 | 45.3 |
| M16 | 99.3 | 37.3 | M46 | 99.3 | 63.3 | M76 | 100 | 66 |
| M17 | 98.7 | 34.7 | M47 | 99.3 | 70 | M77 | 100 | 68.7 |
| M18 | 100 | 31.3 | M48 | 100 | 38 | M78 | 99.3 | 40 |
| M19 | 100 | 47.3 | M49 | 99.3 | 66 | M79 | 100 | 55.3 |
| M20 | 99.3 | 60.7 | M50 | 98.7 | 52 | M80 | 100 | 26 |
| M21 | 100 | 70 | M51 | 100 | 46.7 | M81 | 99.3 | 50 |
| M22 | 98.7 | 48.7 | M52 | 100 | 26.7 | M82 | 100 | 22 |
| M23 | 99.3 | 70.7 | M53 | 99.3 | 58 | M83 | 98.7 | 13.3 |
| M24 | 100 | 36 | M54 | 100 | 48 | M84 | 100 | 53.3 |
| M25 | 98.7 | 33.3 | M55 | 100 | 64 | M85 | 100 | 9.3 |
| M26 | 98 | 21.3 | M56 | 100 | 70.7 | M86 | 100 | 47.3 |
| M27 | 100 | 32 | M57 | 98.7 | 57 | M87 | 100 | 40.7 |
| M28 | 99.3 | 61.3 | M58 | 100 | 24 | M88 | 98 | 80.7 |
| M29 | 98.7 | 25.3 | M59 | 100 | 64 | M89 | 100 | 80 |
| M30 | 98.7 | 19.3 | M60 | 99.3 | 84 | TH899 | 100. | 38 |
| Genotype | K+ Content (mM/g) | Na+ Content (mM/g) | K+/Na+ | |||
|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Control | Stress | |
| M5 | 55.5 ± 1.1 ab | 16.5 ± 0.7 ac | 10.6 ± 0.1 b | 25.5 ± 1.0 ab | 5.2 ± 0.0 b | 0.65 ± 0.1 ab |
| M41 | 55.1 ± 4.2 ab | 21.2 ± 0.8 a | 11.2 ± 0.5 b | 27.9 ± 0.8 ab | 4.9 ± 0.3 b | 0.8 ± 0.0 a |
| M60 | 45.7 ± 4.1 b | 15.1 ± 1.2 bc | 11.7 ± 0.5 b | 26.9 ± 1.8 ab | 4.0 ± 0.5 b | 0.6 ± 0.1 ab |
| M88 | 65.7 ± 3.1 a | 16.4 ± 0.5 ac | 11.9 ± 0.8 ab | 27.5 ± 0.9 ab | 5.6 ± 0.5 ab | 0.6 ± 0.0 ab |
| M89 | 52.6 ± 0.7 ab | 18.1 ± 0.9 ab | 11.1 ± 0.6 b | 24.2 ± 0.9 b | 4.8 ± 0.2 b | 0.8 ± 0.1 a |
| TH899 | 54.1 ± 1.0 ab | 13.0 ± 1.0 c | 14.9 ± 0.5 a | 32.7 ± 1.6 a | 3.7 ± 0.2 a | 0.4 ± 0.0 b |
| G | PH (cm) | PL (cm) | PN | SP | 1000-GW (g) | PSF (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Control | Stress | Control | Stress | Control | Stress | Control | Stress | |
| M5 | 101.3 ± 3.9 a | 98.3 ± 4.0 a | 19.5 ± 0.6 ab | 20.2 ± 0.2 a | 14 ± 1.0 a | 10 ± 0.7 a | 152 ± 5.7 a | 130 ± 0.8 ab | 25.05 ± 0.4 ab | 23.60 ± 0.1 bc | 94.5 ± 0.3 ab | 93.8 ± 0.9 a |
| M41 | 108.3 ± 3.3 a | 103.7 ± 1.1 a | 16.8 ± 1.0 b | 19.7 ± 0.4 a | 16 ± 2.5 a | 12 ± 0.4 a | 98 ± 6.6 b | 109 ± 7.7 b | 26.18 ± 0.1 a | 24.95 ± 0.1 ab | 95.7 ± 0.5 ab | 92.9 ± 2.0 a |
| M60 | 115.7 ± 4.0 a | 102.7 ± 1.9 a | 21.4 ± 0.2 a | 21.3 ± 0.1 a | 15 ± 1.5 a | 11 ± 0.6 a | 158 ± 9.6 a | 152 ± 7.7 a | 23.35 ± 0.6 b | 20.83 ± 0.4 d | 90.6 ± 2.1 b | 91.9 ± 0.3 a |
| M88 | 106.0 ± 2.6 a | 100.0 ± 1.7 a | 21.1 ± 0.7 a | 20.0 ± 0.2 a | 14 ± 0.5 a | 13 ± 1.0 a | 164 ± 12.8 a | 150 ± 2.6 a | 23.30 ± 0.2 b | 23.73 ± 0.3 abc | 96.9 ± 0.6 a | 97.7 ± 0.5 a |
| M89 | 107.0 ± 2.1 a | 104.3 ± 2.6 a | 20.2 ± 0.5 ab | 19.4 ± 0.9 a | 12 ± 0.9 a | 12 ± 0.7 a | 150 ± 10.5 a | 120 ± 10.3 ab | 26.12 ± 0.2 a | 25.32 ± 0.3 a | 96.8 ± 0.3 a | 97.2 ± 0.3 a |
| TH899 | 105.7 ± 2.6 a | 97.7 ± 1.8 a | 19.8 ± 0.1 ab | 18.5 ± 1.0 a | 13 ± 0.9 a | 11 ± 0.4 a | 153 ± 0.7 a | 137 ± 8.1 ab | 23.63 ± 0.5 b | 22.85 ± 0.4 c | 95.4 ± 1.2 ab | 94.7 ± 1.9 a |
| G | PH (cm) | PL (cm) | PN | SP | 1000-GW (g) | PSF (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Control | Stress | Control | Stress | Control | Stress | Control | Stress | |
| M5 | 96.1 ± 1.6 c | 84.8 ± 0.6 c | 20.8 ± 0.2 a | 18.3 ± 0.4 ab | 10 ± 0.4 a | 8 ± 0.6 a | 149 ± 9.5 a | 112 ± 3.2 ab | 25.89 ± 0.5 a | 23.99 ± 0.1 ab | 97.1 ± 1.0 a | 96.2 ± 1.4 a |
| M41 | 101.6 ± 1.3 ac | 90.5 ± 1.2 bc | 19.7 ± 0.4 a | 17.7 ± 0.4 b | 9 ± 0.7 a | 7 ± 0.4 a | 119 ± 4.8 a | 103 ± 5.8 b | 27.04 ± 0.6 a | 25.30 ± 0.4 a | 93.4 ± 0.9 a | 91.45 ± 3.7 a |
| M60 | 107.1 ± 2.3 ab | 95.0 ± 1.3 ab | 21.4 ± 0.3 a | 19.8 ± 0.1 a | 11 ± 0.4 a | 7 ± 0.7 a | 172 ± 9.1 a | 133 ± 2.5 a | 23.11 ± 0.3 b | 22.02 ± 0.6 bc | 92.9 ± 1.4 a | 95.4 ± 0.8 a |
| M88 | 94.2 ± 1.8 c | 85.3 ± 2.7 c | 20.3 ± 0.8 a | 18.4 ± 0.4 ab | 9 ± 0.4 a | 7 ± 0.5 a | 162 ± 15.1 a | 132 ± 8.5 a | 22.21 ± 0.4 b | 22.18 ± 0.3 bc | 92.4 ± 1.1 a | 96.4 ± 0.7 a |
| M89 | 98.3 ± 3.1 bc | 89.3 ± 0.3 bc | 19.2 ± 0.5 a | 17.5 ± 0.3 b | 10 ± 0.4 a | 8 ± 0.9 a | 136 ± 15.4 a | 108 ± 4.2 ab | 25.49 ± 0.1 a | 23.81 ± 0.9 ab | 96.3 ± 0.9 a | 93.2 ± 1.5 a |
| TH899 | 110.9 ± 0.9 a | 98.5 ± 3.3 a | 16.7 ± 0.2 b | 15.3 ± 0.3 c | 9 ± 0.9 a | 6 ± 0.6 a | 166 ± 9.4 a | 114 ± 3.9 ab | 22.32 ± 0.2 b | 20.40 ± 0.1 c | 95.9 ± 0.9 a | 97.3 ± 0.3 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yang, F.; Ma, H.; Li, J. Evaluation of the Saline–Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis. Biology 2022, 11, 126. https://doi.org/10.3390/biology11010126
Zhang X, Yang F, Ma H, Li J. Evaluation of the Saline–Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis. Biology. 2022; 11(1):126. https://doi.org/10.3390/biology11010126
Chicago/Turabian StyleZhang, Xin, Fu Yang, Hongyuan Ma, and Jingpeng Li. 2022. "Evaluation of the Saline–Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis" Biology 11, no. 1: 126. https://doi.org/10.3390/biology11010126
APA StyleZhang, X., Yang, F., Ma, H., & Li, J. (2022). Evaluation of the Saline–Alkaline Tolerance of Rice (Oryza sativa L.) Mutants Induced by Heavy-Ion Beam Mutagenesis. Biology, 11(1), 126. https://doi.org/10.3390/biology11010126





