ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. Donor Plasmid Construction
2.3. Synthesis of gRNA, Pre-crRNA, and mRNAs Encoding Cas9, ErCas12a, and T5exo-ErCas12a
2.4. Microinjection of Zebrafish Embryos
2.5. Target Site Efficiency Evaluation
2.6. Junction PCR and Sanger Sequencing
2.7. Imaging and Processing
2.8. Statistics
3. Results
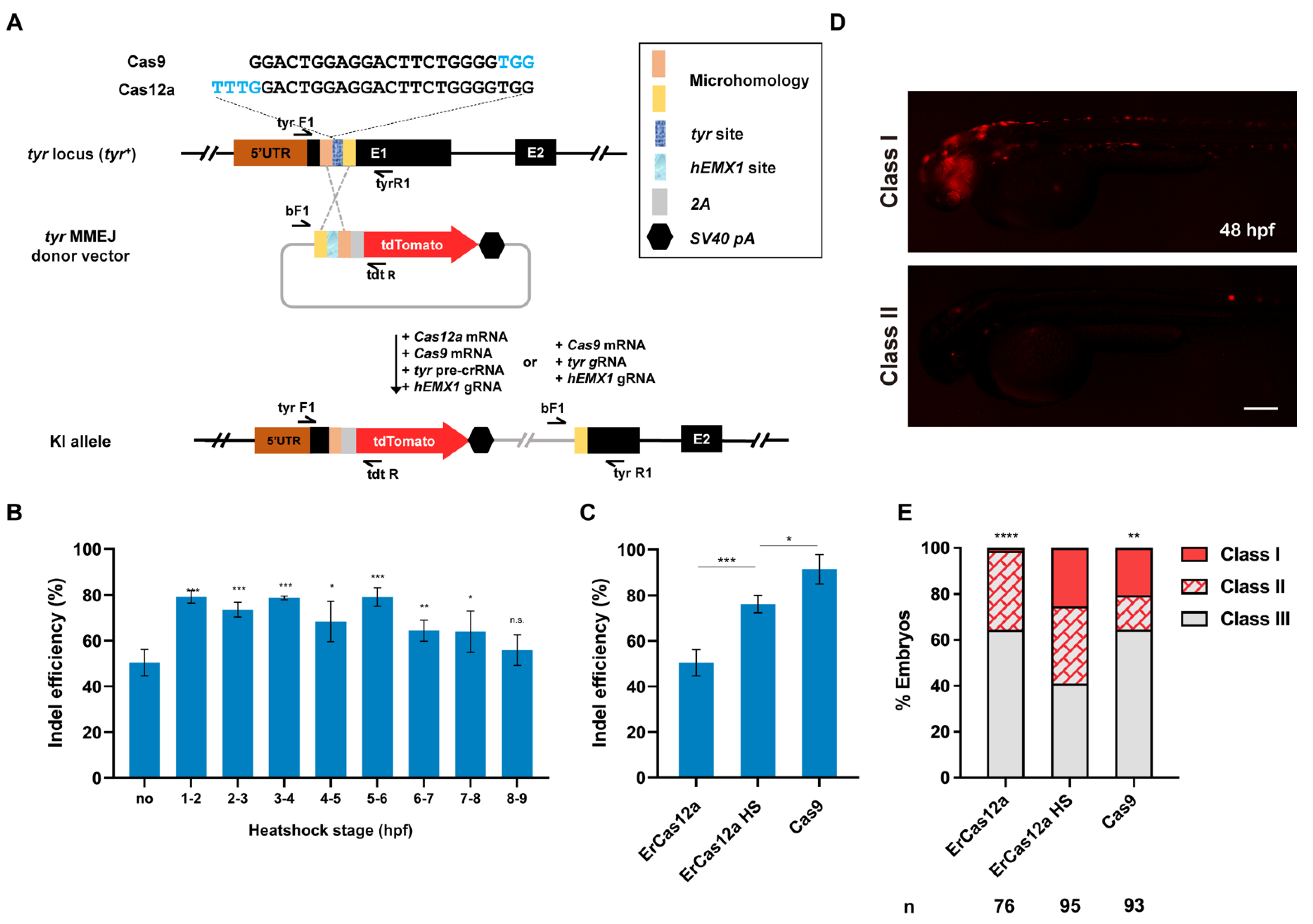
3.1. Optimization of Heatshock Conditions for the Application of ErCas12a mRNA in Zebrafish
3.2. ErCas12a Could Achieve Efficient MMEJ-Mediated Knockin in Zebrafish
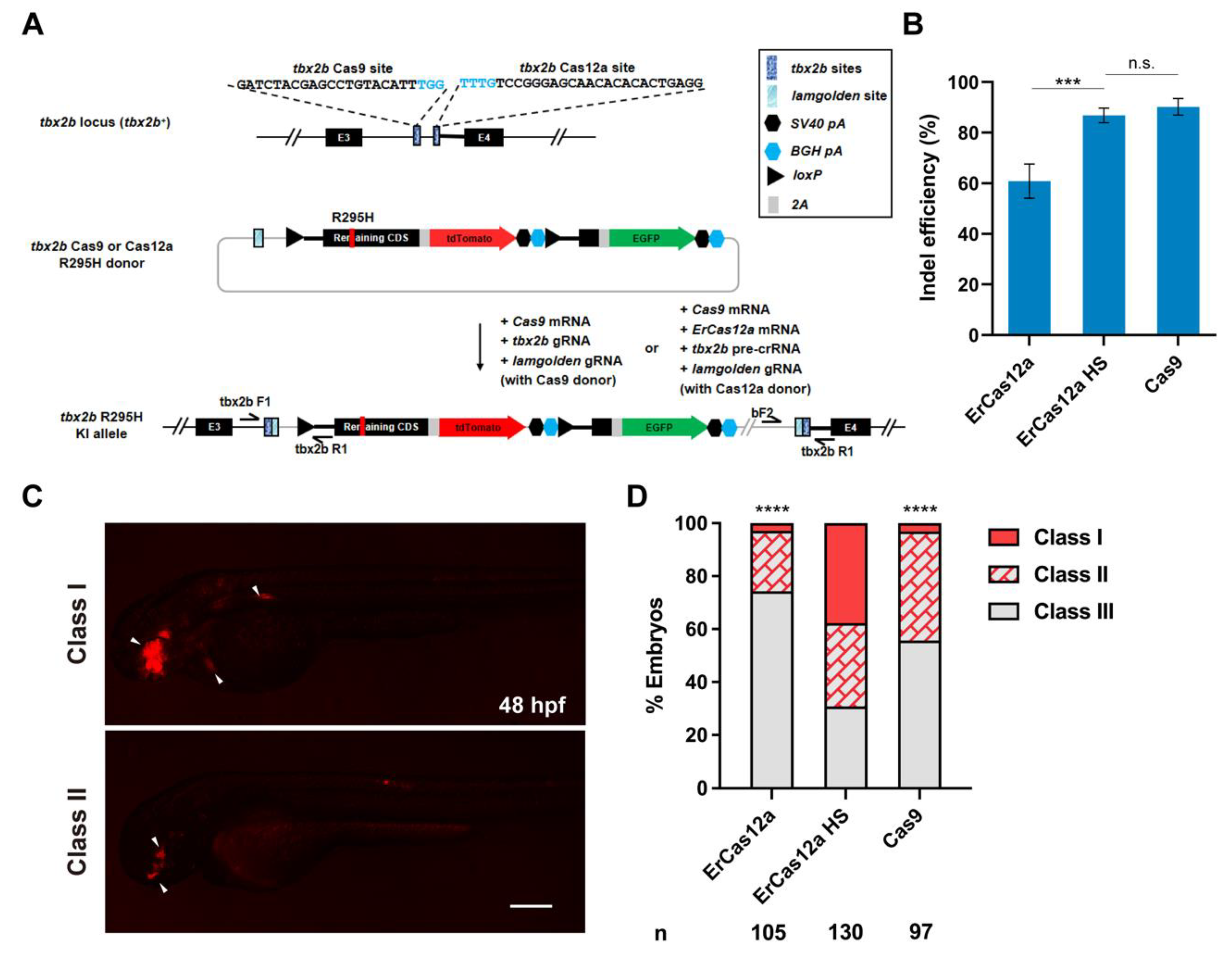
3.3. ErCas12a Could Efficiently Induce NHEJ-Based Knockin in Zebrafish
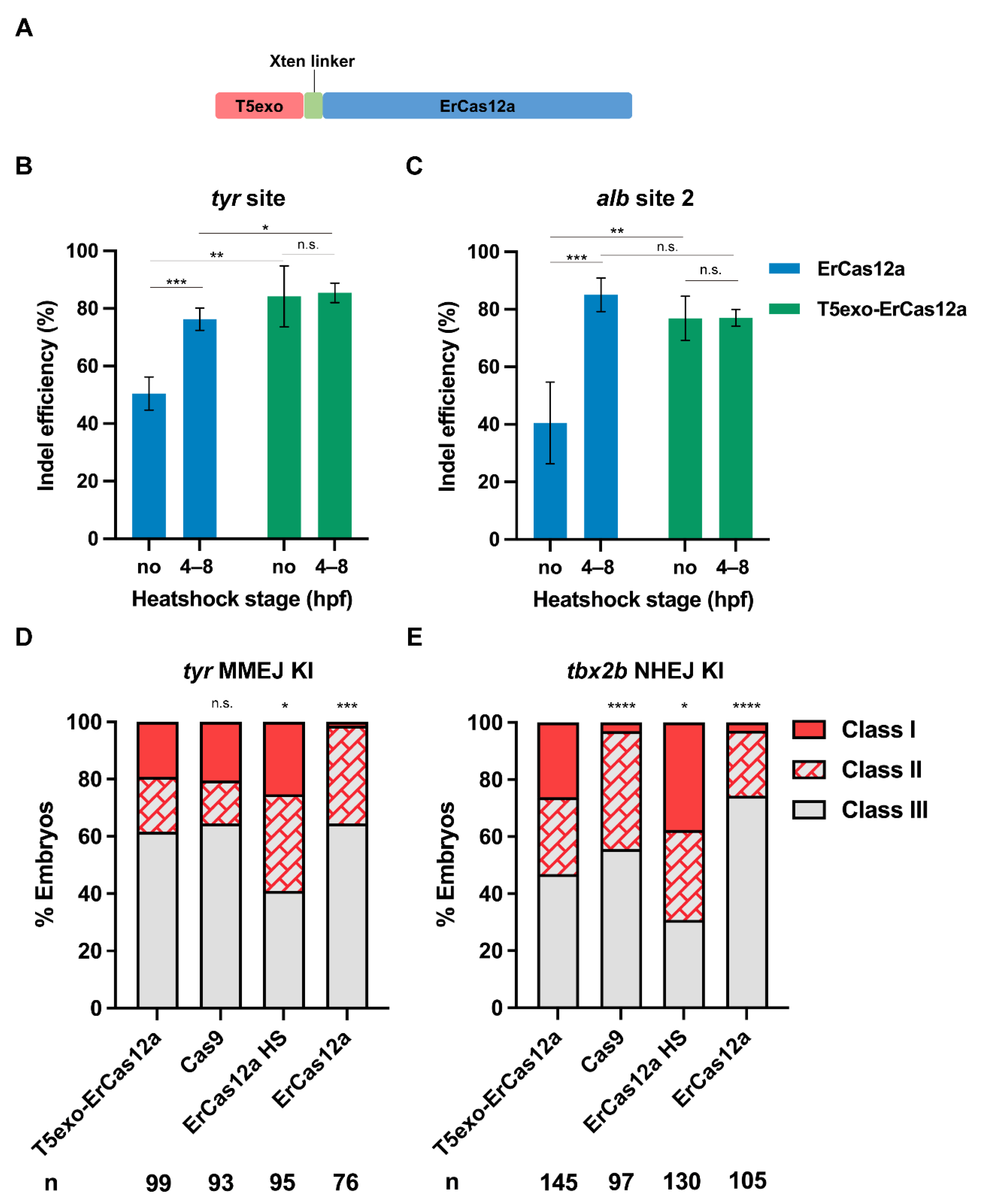
3.4. T5exo-ErCas12a Efficiently Induces Indel Mutation and Promotes Knockin while Avoiding Heatshock Treatment in Zebrafish
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.H.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hur, J.K.; Kim, K.; Been, K.W.; Baek, G.; Ye, S.; Hur, J.W.; Ryu, S.M.; Lee, Y.S.; Kim, J.S. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 2016, 34, 807–808. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef]
- Kim, Y.; Cheong, S.A.; Lee, J.G.; Lee, S.W.; Lee, M.S.; Baek, I.J.; Sung, Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016, 34, 808–810. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. [Google Scholar] [CrossRef]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [Green Version]
- Wierson, W.A.; Simone, B.W.; WareJoncas, Z.; Mann, C.; Welker, J.M.; Kar, B.; Emch, M.J.; Friedberg, I.; Gendron, W.A.C.; Barry, M.A.; et al. Expanding the CRISPR Toolbox with ErCas12a in Zebrafish and Human Cells. CRISPR J. 2019, 2, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Auer, T.O.; Duroure, K.; De Cian, A.; Concordet, J.P.; Del Bene, F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014, 24, 142–153. [Google Scholar] [CrossRef] [Green Version]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisano, Y.; Sakuma, T.; Nakade, S.; Ohga, R.; Ota, S.; Okamoto, H.; Yamamoto, T.; Kawahara, A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015, 5, 8841. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.Y.; Fu, Y.F.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Li, J.; Li, H.Y.; Gu, S.Y.; Zi, H.X.; Jiang, L.; Du, J.L. One-step generation of zebrafish carrying a conditional knockout-knockin visible switch via CRISPR/Cas9-mediated intron targeting. Sci. China Life Sci. 2020, 63, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, B.B.; Ren, Y.G.; Gu, S.Y.; Xiang, Y.H.; Du, J.L. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Res. 2015, 25, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Z.X.; Xiao, A.; Zhang, Y.T.; Li, W.Y.; Zu, Y.; Yao, S.H.; Lin, S.; Zhang, B. Efficient Gene Targeting in Zebrafish Mediated by a Zebrafish-Codon-Optimized Cas9 and Evaluation of Off-Targeting Effect. J. Genet. Genom. 2014, 41, 43–46. [Google Scholar] [CrossRef]
- Luo, J.J.; Bian, W.P.; Liu, Y.; Huang, H.Y.; Yin, Q.; Yang, X.J.; Pei, D.S. CRISPR/Cas9-based genome engineering of zebrafish using a seamless integration strategy. FASEB J. 2018, 32, 5132–5142. [Google Scholar] [CrossRef] [Green Version]
- Wierson, W.A.; Welker, J.M.; Almeida, M.P.; Mann, C.M.; Webster, D.A.; Torrie, M.E.; Weiss, T.J.; Kambakam, S.; Vollbrecht, M.K.; Lan, M.; et al. Efficient targeted integration directed by short homology in zebrafish and mammalian cells. Elife 2020, 9, e53968. [Google Scholar] [CrossRef]
- Liu, P.; Luk, K.; Shin, M.; Idrizi, F.; Kwok, S.; Roscoe, B.; Mintzer, E.; Suresh, S.; Morrison, K.; Frazao, J.B.; et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019, 47, 4169–4180. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Zhang, Y.; Bi, X.; Zhou, Y.; Krueger, C.J.; Hu, X.; Zhu, Z.; Tong, X.; Zhang, B. Bi-FoRe: An efficient bidirectional knockin strategy to generate pairwise conditional alleles with fluorescent indicators. Protein Cell 2021, 12, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Zhang, Y.G.; Han, B.Z.; Li, L.Y.; Li, M.H.; Lu, X.C.; Chen, C.; Lu, M.J.; Zhang, Y.J.; Jia, X.F.; et al. One-step efficient generation of dual-function conditional knockout geno-tagging alleles in zebrafish. Elife 2019, 8, e48081. [Google Scholar] [CrossRef]
- Burger, A.; Lindsay, H.; Felker, A.; Hess, C.; Anders, C.; Chiavacci, E.; Zaugg, J.; Weber, L.M.; Catena, R.; Jinek, M.; et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 2016, 143, 2025–2037. [Google Scholar] [CrossRef] [Green Version]
- Hoshijima, K.; Jurynec, M.J.; Klatt Shaw, D.; Jacobi, A.M.; Behlke, M.A.; Grunwald, D.J. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell 2019, 51, 645–657.e644. [Google Scholar] [CrossRef]
- Kotani, H.; Taimatsu, K.; Ohga, R.; Ota, S.; Kawahara, A. Efficient Multiple Genome Modifications Induced by the crRNAs, tracrRNA and Cas9 Protein Complex in Zebrafish. PLoS ONE 2015, 10, e0128319. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.; Cheng, Z.; Kong, L.; Zhu, Z.; Lin, S.; Gao, G.; Zhang, B. CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 2014, 30, 1180–1182. [Google Scholar] [CrossRef] [Green Version]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, E.K.; Kousholt, A.N.; Harmsen, T.; Leemans, C.; Chen, T.; Jonkers, J.; van Steensel, B. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 2018, 46, e58. [Google Scholar] [CrossRef]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [Google Scholar] [CrossRef]
- Chan, S.H.; Yu, A.M.; McVey, M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010, 6, e1001005. [Google Scholar] [CrossRef] [Green Version]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Lazzerini-Denchi, E.; Sfeir, A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, R.D.; Doublie, S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair 2016, 44, 22–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yumlu, S.; Bashir, S.; Stumm, J.; Kuhn, R. Efficient Gene Editing of Human Induced Pluripotent Stem Cells Using CRISPR/Cas9. Methods Mol. Biol 2019, 1961, 137–151. [Google Scholar] [PubMed]
- Wu, Y.; Yuan, Q.; Zhu, Y.; Gao, X.; Song, J.; Yin, Z. Improving FnCas12a Genome Editing by Exonuclease Fusion. CRISPR J. 2020, 3, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yin, K.; Liu, G.; Li, S.; Li, M.; Qiu, J.L. Fusing T5 exonuclease with Cas9 and Cas12a increases the frequency and size of deletion at target sites. Sci. China Life Sci. 2020, 63, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Schiel, J.A.; Maksimova, E.; Strezoska, Z.; Zhao, G.; Anderson, E.M.; Wu, Y.; Warren, J.; Bartels, A.; van Brabant Smith, A.; et al. ErCas12a CRISPR-MAD7 for Model Generation in Human Cells, Mice, and Rats. CRISPR J. 2020, 3, 97–108. [Google Scholar] [CrossRef]
- Chaurasia, A.; Tarallo, A.; Berna, L.; Yagi, M.; Agnisola, C.; D’Onofrio, G. Length and GC content variability of introns among teleostean genomes in the light of the metabolic rate hypothesis. PLoS ONE 2014, 9, e103889. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Akhmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hans, S.; Zoller, D.; Hammer, J.; Stucke, J.; Spiess, S.; Kesavan, G.; Kroehne, V.; Eguiguren, J.S.; Ezhkova, D.; Petzold, A.; et al. Cre-Controlled CRISPR mutagenesis provides fast and easy conditional gene inactivation in zebrafish. Nat. Commun. 2021, 12, 1125. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Wang, I.J. Heat-shock-induced tyrosinase gene ablation with CRISPR in zebrafish. Mol. Genet. Genom. 2020, 295, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Marques, J.; Espinosa-Medina, I.; Ku, K.Y.; Yang, C.P.; Koyama, M.; Yu, H.H.; Lee, T. A programmable sequence of reporters for lineage analysis. Nat. Neurosci. 2020, 23, 1618–1628. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef]


| Gene | F0 | Sex | Number of tdTomato-Positive F1 Embryos | Number of Total F1 Progeny | F0 Germ Cell Mosaicism (Ratio of Positive F1) |
|---|---|---|---|---|---|
| tyr | #3 | Male | 35 | 103 | 34.0% |
| #5 | Male | 27 | 82 | 33.0% | |
| #7 | Male | 4 | 26 | 15.4% | |
| tbx2b | #1 | Male | 6 | 34 | 17.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, B.; Zhang, Y.; Zhou, Y.; Zhang, B.; Krueger, C.J.; Bi, X.; Zhu, Z.; Tong, X.; Zhang, B. ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish. Biology 2022, 11, 411. https://doi.org/10.3390/biology11030411
Han B, Zhang Y, Zhou Y, Zhang B, Krueger CJ, Bi X, Zhu Z, Tong X, Zhang B. ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish. Biology. 2022; 11(3):411. https://doi.org/10.3390/biology11030411
Chicago/Turabian StyleHan, Bingzhou, Yage Zhang, Yang Zhou, Biao Zhang, Christopher J. Krueger, Xuetong Bi, Zuoyan Zhu, Xiangjun Tong, and Bo Zhang. 2022. "ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish" Biology 11, no. 3: 411. https://doi.org/10.3390/biology11030411
APA StyleHan, B., Zhang, Y., Zhou, Y., Zhang, B., Krueger, C. J., Bi, X., Zhu, Z., Tong, X., & Zhang, B. (2022). ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish. Biology, 11(3), 411. https://doi.org/10.3390/biology11030411






