A Probabilistic Structural Equation Model to Evaluate Links between Gut Microbiota and Body Weights of Chicken Fed or Not Fed Insect Larvae
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bird and Insect Management
2.2. 16S rDNA Detection and Analyses
2.3. Data Processing and Modeling
3. Results
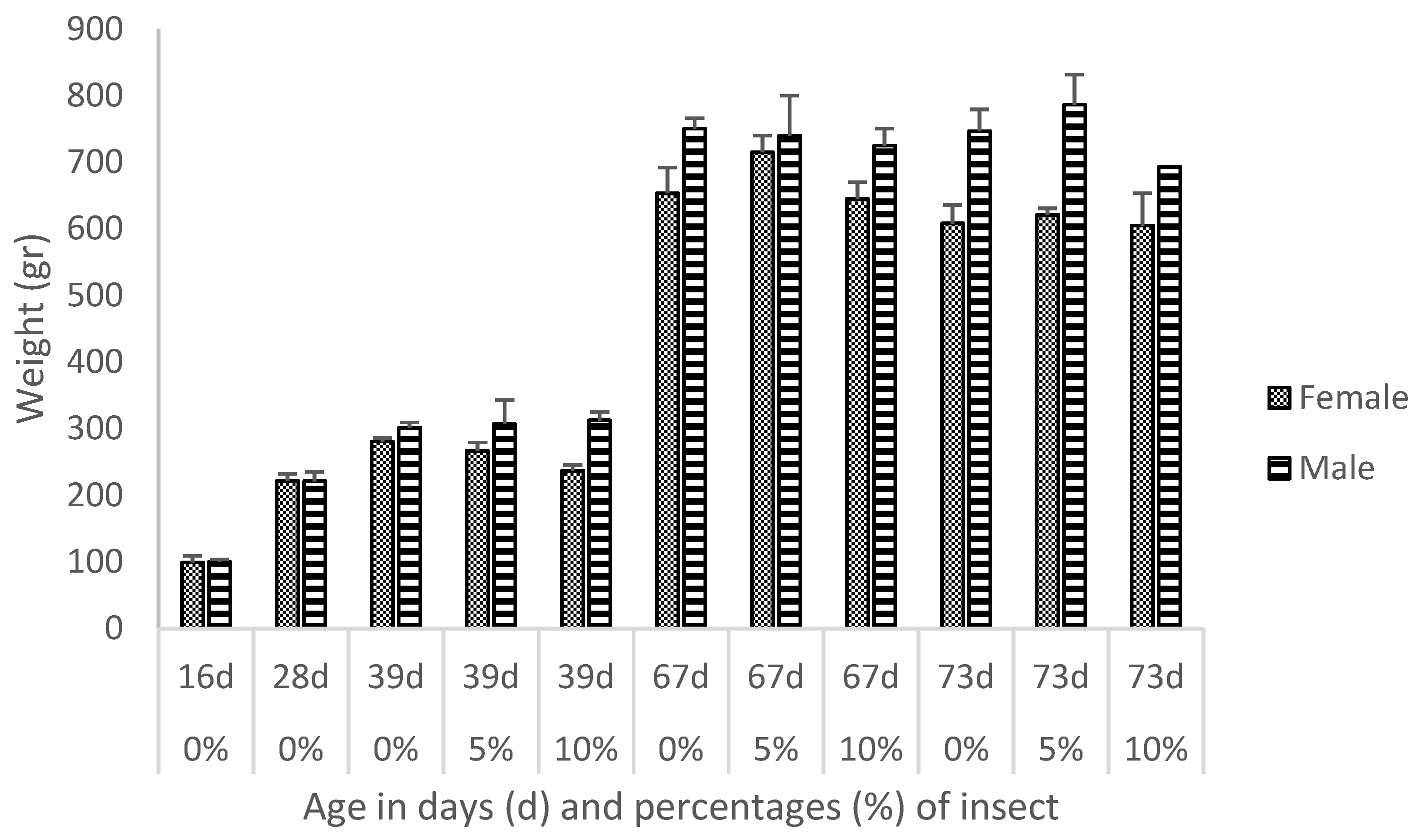
3.1. Descriptive Analytics
3.2. Discretization
3.3. Network Construction
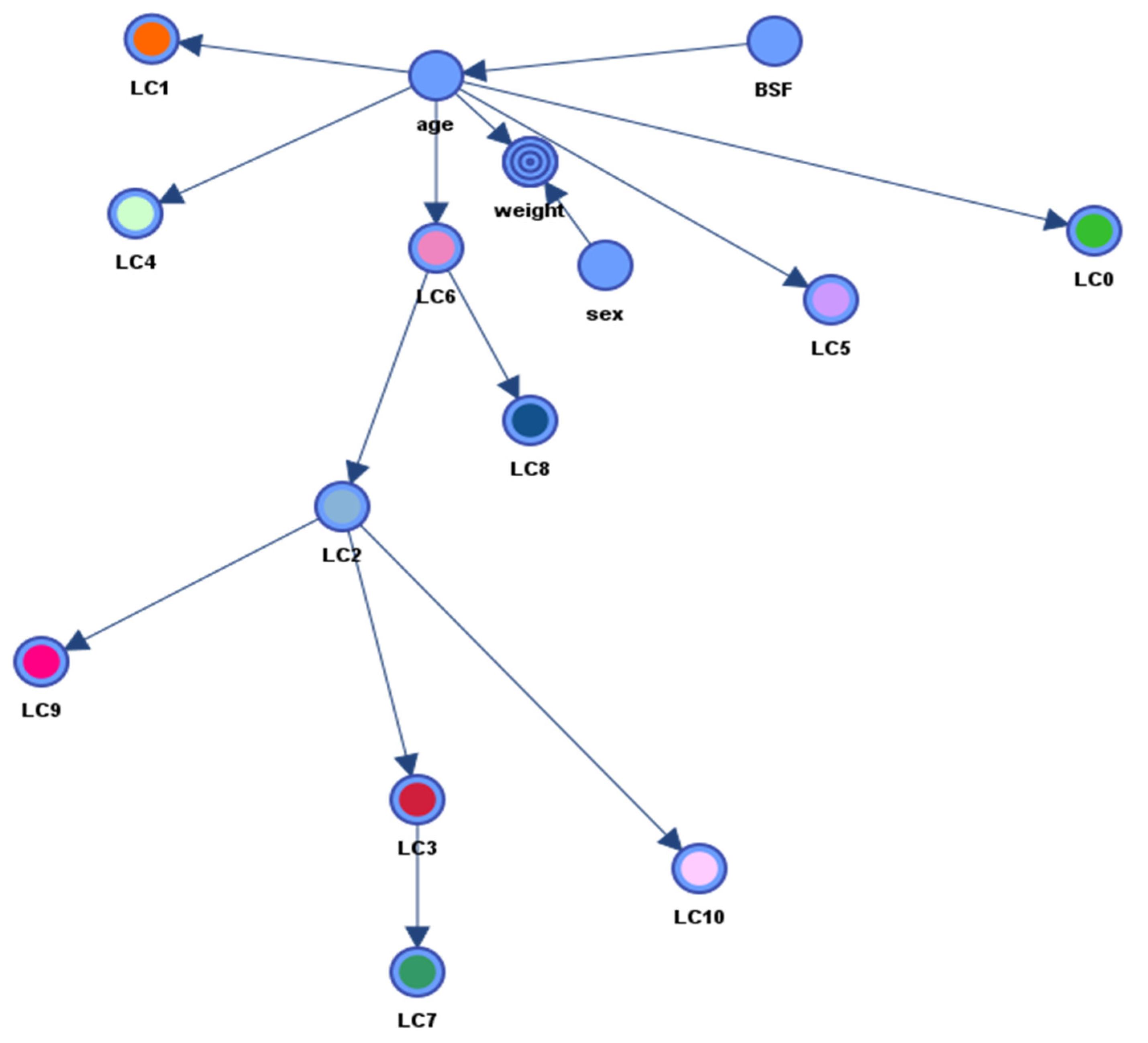
3.4. Measurement Part of the pSEM
3.5. Structural Part of the pSEM
4. Discussion
4.1. Descriptive Analytics
4.2. Measurement Part of the pSEM
4.3. Structural Part of the pSEM
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Huis, A.; Oonincx, D.G.A.B. The environmental sustainability of insects as food and feed. A review. Agron. Sustain. Dev. 2017, 37, 43. [Google Scholar] [CrossRef] [Green Version]
- Moula, N.; Detilleux, J. A meta-analysis of the effects of insects in feed on poultry growth performances. Animals 2019, 9, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz Carrasco, J.M.; Casanova, N.A.; Fernández Miyakawa, M.E. Microbiota, gut health and chicken productivity: What is the connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, P.; Fothergill, J.; Bernardeau, M.; Wigley, P. Development of the caecal microbiota in three broiler breeds. Front. Vet. Sci. 2019, 6, 201. [Google Scholar] [CrossRef]
- Jozefiak, D.; Rutkowski, A.; Martin, S.A. Carbohydrate fermentation in the avian ceca: A review. Anim. Feed Sci. Technol. 2004, 133, 1–15. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Chia, N.; Jeraldo, P.; Sipos, M.; Goldenfeld, N.D.; White, B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 2012, 13, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Ocejo, M.; Oporto, B.; Hurtado, A. 16S rRNA amplicon sequencing characterization of caecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci. Rep. 2019, 9, 2506. [Google Scholar] [CrossRef] [Green Version]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Stegeman, J.A.; Smidt, H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018, 9, 235. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Y.; Li, J.; Xing, T.; Jiang, Y.; Zhang, L.; Gao, F. Dietary resistant starch modifies the composition and function of caecal microbiota of broilers. J. Sci. Food Agric. 2020, 100, 1274–1284. [Google Scholar] [CrossRef]
- Biasato, I.; Ferrocino, I.; Dabbou, S.; Evangelista, R.; Gai, F.; Gasco, L.; Cocolin, L.; Capucchio, M.T.; Schiavone, A. Black soldier fly and gut health in broiler chickens: Insights into the relationship between cecal microbiota and intestinal mucin composition. J. Anim. Sci. Biotechnol. 2020, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Borrelli, L.; Coretti, L.; Dipineto, L.; Bovera, F.; Menna, F.; Chiariotti, L.; Nizza, A.; Lembo, F.; Fioretti, A. Insect-based diet, a promising nutritional source, modulates gut microbiota composition and SCFAs production in laying hens. Sci. Rep. 2017, 7, 16269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biasato, I.; Ferrocino, I.; Grego, E.; Dabbou, S.; Gai, F.; Gasco, L.; Cocolin, L.; Capucchio, M.T.; Schiavone, A. Yellow mealworm inclusion in diets for heavy-size broiler chickens: Implications for intestinal microbiota and mucin dynamics. Animals 2020, 10, 1909. [Google Scholar] [CrossRef] [PubMed]
- Moula, N.; Hornick, J.L.; Cabaraux, J.F.; Korsak, N.; Dawans, E.; Antoine, N.; Taminiau, B.; Detilleux, J. Effects of dietary black soldier fly larvae on performance of broilers mediated or not through changes in microbiota. J. Insects Food Feed. 2017, 4, 31–42. [Google Scholar] [CrossRef]
- Mahmood, T.; Guo, Y. Dietary fiber and chicken microbiome interaction: Where will it lead to? Anim. Nutr. 2020, 202, 1–8. [Google Scholar] [CrossRef]
- Tran, G.; Gnaedinger, C.; Mélin, C. Feedipedia, an Online Encyclopedia of Animla Feeds. Available online: https://www.feedipedia.org/node/16388 (accessed on 25 January 2022).
- Dörper, A.; Veldkamp, T.; Dicke, M. Use of black soldier fly and house fly in feed to promote sustainable poultry production. JIFF 2020, 7, 761–780. [Google Scholar] [CrossRef]
- Allali, I.; Arnold, J.W.; Roach, J.; Cadenas, M.B.; Butz, N.; Hassan, H.M.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R.; et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017, 17, 194. [Google Scholar] [CrossRef]
- Xia, Y.; Sun, J. Hypothesis testing and statistical analysis of microbiome. Genes Dis. 2017, 4, 138–148. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [Green Version]
- Holmes, I.; Harris, K.; Quince, C. Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLoS ONE 2012, 7, e30126. [Google Scholar]
- Egozcue, J.J.; Graffelman, J.; Ortego, M.I.; Pawlowsky-Glahn, V. Some thoughts on counts in sequencing studies. NAR Genom. Bioinform. 2020, 2, lqaa094. [Google Scholar]
- Quinn, T.P.; Erb, I.; Gloor, G.; Notredame, C.; Richardson, M.F.; Crowley, T.M. A field guide for the compositional analysis of any-omics data. Gigascience 2019, 8, giz107. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Vatcheva, K.P.; Lee, M.; McCormick, J.B.; Rahbar, M.H. Multicollinearity in regression analyses conducted in epidemiologic studies. Epidemiology 2016, 6, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Dohlman, A.B.; Shen, X. Mapping the microbial interactome: Statistical and experimental approaches for microbiome network inference. Exp. Biol Med. 2019, 244, 445–458. [Google Scholar] [CrossRef]
- Pearl, J. Causality: Models, Reasoning, and Inference, 1st ed.; Cambridge University Press: Cambridge, UK, 2000; pp. 1–384. [Google Scholar]
- Scanagatta, M.; Salmerón, A.; Stella, F. A survey on Bayesian network structure learning from data. Prog. Artif. Intell. 2019, 8, 425–439. [Google Scholar] [CrossRef]
- Grunwald, P. Minimum description length tutorial. In Advances in Minimum Description Length. Theory and Applications, 1st ed.; Grunwald, P.D., Myung, J.I., Pitt, M.A., Eds.; MIT Press: Cambridge, MA, USA, 2005; pp. 23–81. [Google Scholar]
- Ji, Z.; Xia, Q.; Meng, G. A review of parameter learning methods in Bayesian network. In Advanced Intelligent Computing Theories and Applications, 1st ed.; Huang, D.S., Han, K., Eds.; Springer: London, UK, 2015; Volume 3, pp. 3–12. [Google Scholar]
- Saborío-Montero, A.; Gutiérrez-Rivas, M.; García-Rodríguez, A.; Atxaerandio, R.; Goiri, I.; López de Maturana, E.; Jiménez-Montero, J.A.; Alenda, R.; González-Recio, O. Structural equation models to disentangle the biological relationship between microbiota and complex traits: Methane production in dairy cattle as a case of study. J. Anim Breed. Genet. 2020, 137, 36–48. [Google Scholar] [CrossRef]
- Tiezzi, F.; Fix, J.; Schwab, C.; Shull, C.; Maltecca, C. Gut microbiome mediates host genomic effects on phenotypes: A case study with fat deposition in pigs. Comput. Struct. Biotechnol. J. 2020, 19, 530–544. [Google Scholar] [CrossRef]
- Haenlein, M.; Kaplan, A.M. A beginner’s guide to partial least squares analysis. Underst. Statistics 2004, 3, 283–297. [Google Scholar] [CrossRef]
- Detilleux, J.; Theron, L.; Duprez, J.N.; Reding, E.; Humblet, M.F.; Planchon, V.; Delfosse, C.; Bertozzi, C.; Mainil, J.; Hanzen, C. Structural equation models to estimate risk of infection and tolerance to bovine mastitis. Genet. Sel. Evol. 2013, 45, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Waal, A.; Yoo, K. Latent variable Bayesian networks constructed using structural equation modelling. In Proceedings of the 21st International Conference on Information Fusion, Cambridge, UK, 10–13 July 2018; pp. 688–695. [Google Scholar]
- Detilleux, J. Tolerance to bovine clinical mastitis: Total, direct, and indirect milk losses. J. Dairy Sci. 2018, 10, 3334–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunzler, D.; Chen, T.; Wu, P.; Zhang, H. Introduction to mediation analysis with structural equation modeling. Shanghai Arch. Psychiatry 2013, 25, 390–394. [Google Scholar] [PubMed]
- Hoc, B.; Noël, G.; Carpentier, J.; Francis, F.; Caparros, M.; Megido, R. Optimization of black soldier fly (Hermetia illucens) artificial reproduction. PLoS ONE 2019, 30, e0216160. [Google Scholar] [CrossRef] [Green Version]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Conrady, S.; Jouffe, L. Bayesian Networks & BayesiaLab: A Practical Introduction for Researchers, 1st ed.; Bayesia: Franklin, TN, USA, 2015; pp. 1–371. [Google Scholar]
- Ryynänen, O.P.; Leppänen, T.; Kekolahti, P.; Mervaala, E.; Töyräs, J. Bayesian network model to evaluate the effectiveness of continuous positive airway pressure treatment of sleep apnea. Healthcare Inform. Res. 2018, 24, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Chernick, M.R. The jackknife: A resampling method with connections to the bootstrap. Wiley Interdiscip. Rev. Comput. Stat. 2012, 4, 224–226. [Google Scholar] [CrossRef]
- Rychlik, I. Composition and function of chicken gut microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chica Cardenas, L.A.; Clavijo, V.; Vives, M.; Reyes, A. Bacterial meta-analysis of chicken cecal microbiota. PeerJ 2021, 9, e10571. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Geier, M.S.; Chen, H.; Hughes, R.J.; Moore, R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015, 15, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Videvall, E.; Strandh, M.; Engelbrecht, A.; Cloete, S.; Cornwallis, C.K. Measuring the gut microbiome in birds: Comparison of faecal and cloacal sampling. Mol. Ecol. Resour. 2018, 18, 424–434. [Google Scholar] [CrossRef]
- Lee, S.J.; Cho, S.; La, T.M.; Lee, H.J.; Lee, J.B.; Park, S.Y.; Song, C.S.; Choi, I.S.; Lee, S.W. Comparison of microbiota in the cloaca, colon, and magnum of layer chicken. PLoS ONE 2020, 15, e0237108. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Larivière, J.M.; Michaux, C.; Farnir, F.; Detilleux, J.; Verleyen, V.; Leroy, P. Reproductive performance of the Ardennaise chicken breed under traditional and modern breeding management systems. Int. J. Poultry Sci. 2009, 8, 446–451. [Google Scholar] [CrossRef] [Green Version]
- Moula, N.; Antoine-Moussiaux, N.; Farnir, F.; Philippart de Foy, M.; Leroy, P. Performances zootechniques de la poule Ardennaise, une race ancienne pour le futur? Ann. Méd. Vét. 2009, 153, 66–75. [Google Scholar]
- Kaakoush, N.O.; Sodhi, N.; Chenu, J.W.; Cox, J.M.; Riordan, S.M.; Mitchell, H.M. The interplay between Campylobacter and Helicobacter species and other gastrointestinal microbiota of commercial broiler chickens. Gut Pathog. 2014, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.S.; Mi, J.D.; Mei, L.; Liang, J.; Feng, K.X.; Wu, Y.B.; Liao, X.D.; Wang, Y. Microbial diversity and community variation in the intestines of layer chickens. Animals 2021, 11, 840. [Google Scholar] [CrossRef] [PubMed]
- Zou, A.; Sharif, S.; Parkinson, J. Lactobacillus elicits a ‘Marmite effect’ on the chicken cecal microbiome. Jpn. Biofilms. Microbiomes. 2018, 4, 27. [Google Scholar] [CrossRef] [Green Version]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef] [PubMed]
- Svihus, B.; Choct, M.; Classen, H.L. Function and nutritional roles of the avian caeca: A review. World Poultry Sci. J. 2013, 69, 249–263. [Google Scholar] [CrossRef]
- Karasawa, Y. Significant role of the nitrogen recycling system through the ceca occurs in protein-depleted chickens. J. Exp. Zool. 1999, 283, 418–425. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Van Immerseel, F.; Croubels, S.; De Baere, S.; Haesebrouck, F.; Ducatelle, R.; Louis, P.; Vandamme, P. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb. Biotechnol. 2011, 4, 503–512. [Google Scholar] [CrossRef] [Green Version]
- Polansky, O.; Sekelova, Z.; Faldynova, M.; Sebkova, A.; Sisak, F.; Rychlik, I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 2015, 82, 1569–1576. [Google Scholar] [CrossRef] [Green Version]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- De Maesschalck, C.; Eeckhaut, V.; Maertens, L.; De Lange, L.; Marchal, L.; Daube, G.; Dewulf, J.; Haesebrouck, F.; Ducatelle, R.; Taminau, B.; et al. Amorphous cellulose feed supplement alters the broiler caecal microbiome. PoultSci 2019, 98, 3811–3817. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.Y.; Zhang, H.L.; Zhao, F.J.; Wang, S.Q.; Wang, Z.X.; Wei, Z.Y. Modulation of gut microbiota, short-chain fatty acid production, and inflammatory cytokine expression in the cecum of porcine Deltacoronavirus-infected chicks. Front. Microbiol. 2020, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.C.; Kil, D.Y.; Sul, W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017, 55, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Amaretti, A.; Gozzoli, C.; Simone, M.; Raimondi, S.; Righini, L.; Pérez-Brocal, V.; García-López, R.; Moya, A.; Rossi, M. Profiling of protein degraders in cultures of human gut microbiota. Front. Microbiol. 2019, 10, 2614. [Google Scholar] [CrossRef]
- Slobodkin, A. The Family Peptostreptococcaceae. In The Prokaryotes, 1st ed.; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrand, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014; pp. 291–302. [Google Scholar]
- Zhou, C.; Chen, L.L.; Lu, R.Q.; Ma, W.W.; Xiao, R. Alteration of intestinal microbiota composition in oral sensitized C3H/HeJ mice is associated with changes in dendritic cells and T cells in mesenteric lymph nodes. Front. Immunol. 2021, 12, 631494. [Google Scholar] [CrossRef]
- Hedblom, G.A.; Reiland, H.A.; Sylte, M.J.; Johnson, T.J.; Baumler, D.J. Segmented filamentous bacteria - Metabolism meets immunity. Front. Microbiol. 2018, 9, 1991. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012, 3, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Awad, W.A.; Mann, E.; Dzieciol, M.; Hess, C.; Schmitz-Esser, S.; Wagner, M.; Hess, M. Age-related differences in the luminal and mucosa-associated gut microbiome of broiler chickens and shifts associated with Campylobacter jejuni infection. Front. Cell Infect. Microbiol. 2016, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Stamilla, A.; Ruiz-Ruiz, S.; Artacho, A.; Pons, J.; Messina, A.; Lucia Randazzo, C.; Caggia, C.; Lanza, M.; Moya, A. Analysis of the microbial intestinal tract in broiler chickens during the rearing period. Biology 2021, 10, 942. [Google Scholar] [CrossRef]
- Donaldson, E.E.; Stanley, D.; Hughes, R.J.; Moore, R.J. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ 2017, 5, e3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vilela Souza, J.V.; Kheravii, S.K.; Bajagai, Y.S.; Kolakshyapati, M.; Wu, S.; Ruhnke, I. Inclusion, of black soldier fly larvae in a meat chicken diet has minor effect on caeca microbiota. In Proceedings of the 32nd Annual Australian Poultry Science Symposium; University of Sydney: Sydney, Australia, 2021; Volume 32, p. 146. [Google Scholar]
- Dabbou, S.; Lauwaerts, A.; Ferrocino, I.; Biasato, I.; Sirri, F.; Zampiga, M.; Bergagna, S.; Pagliasso, G.; Gariglio, M.; Colombino, E.; et al. Modified black soldier fly larva fat in broiler diet: Effects on performance, carcass traits, blood parameters, histomorphological features and gut microbiota. Animals 2021, 11, 1837. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, H.; Zhang, R.; Cao, G.; Li, Q.; Zhang, B.; Wang, Y.; Yang, C. Serum metabolome and gut microbiome alterations in broiler chickens supplemented with lauric acid. Poult. Sci. 2021, 100, 101315. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Hack, M.E.; Shafi, M.E.; Alghamdi, W.Y.; Abdelnour, S.A.; Shehata, A.M.; Noreldin, A.E.; Ashour, E.A.; Swelum, A.A.; Al-Sagan, A.A.; Alkhateeb, M.; et al. Black soldier fly (Hermetia illucens) meal as a promising feed ingredient for poultry: A comprehensive review. Agriculture 2020, 10, 339. [Google Scholar] [CrossRef]
- Weththasinghe, P.; Hansen, J.Ø.; Mydland, L.T.; Øverland, M. A systematic meta-analysis based review on black soldier fly (Hermetia illucens) as a novel protein source for salmonids. Rev. Aquac. 2021. [Google Scholar] [CrossRef]
- Shumo, M.; Osuga, I.M.; Khamis, F.M.; Tanga, C.M.; Fiaboe, K.K.M.; Subramanian, S.; Ekesi, S.; van Huis, A.; Borgemeister, C. The nutritive value of black soldier fly larvae reared on common organic waste streams in Kenya. Sci. Rep. 2019, 9, 10110. [Google Scholar] [CrossRef]
- DiGiacomo, K.; Leury, B. Review: Insect meal: A future source of protein feed for pigs? Animal 2019, 13, 3022–3030. [Google Scholar] [CrossRef] [Green Version]
- Dabbou, S.; Gai, F.; Biasato, I.; Capucchio, M.T.; Biasibetti, E.; Dezzutto, D.; Meneguz, M.; Plachà, I.; Gasco, L.; Schiavone, A. Black soldier fly defatted meal as a dietary protein source for broiler chickens: Effects on growth performance, blood traits, gut morphology and histological features. J. Anim. Sci. Biotechnol. 2018, 9, 49. [Google Scholar] [CrossRef]
- Marmion, M.; Ferone, M.T.; Whyte, P.; Scannell, A.G.M. The changing microbiome of poultry meat; from farm to fridge. Food Microbiol. 2021, 99, 103823. [Google Scholar] [CrossRef]
- Moula, N.; Scippo, M.L.; Douny, C.; Degand, G.; Dawans, E.; Cabaraux, J.F.; Hornick, J.L.; Medigo, R.C.; Leroy, P.; Francis, F.; et al. Performances of local poultry breed fed black soldier fly larvae reared on horse manure. Anim Nutr. 2018, 4, 73–78. [Google Scholar] [CrossRef]
- Grünwald, P.D. Introducing the minimum description length principle. In Advances in Minimum Description Length. Theory and Applications, 1st ed.; Grünwald, P.D., Myung, I.J., Pitt, M.A., Eds.; MIT Press: London, UK, 2004; pp. 3–23. [Google Scholar]
- Lehmann, D.R.; Hulbert, J. Are three-point scales always good enough? J. Marketing Res. 1972, 9, 444–446. [Google Scholar] [CrossRef]
- Kim, S.K.; Frisby, C.L. Gaining from discretization of continuous data: The correspondence analysis biplot approach. Behav. Res. 2019, 51, 589–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safari, S.; Baratloo, A.; Elfil, M.; Negida, A. Evidence based emergency medicine; Part 5 Receiver Operating Curve and Area under the Curve. Emergency 2016, 4, 111–113. [Google Scholar] [PubMed]
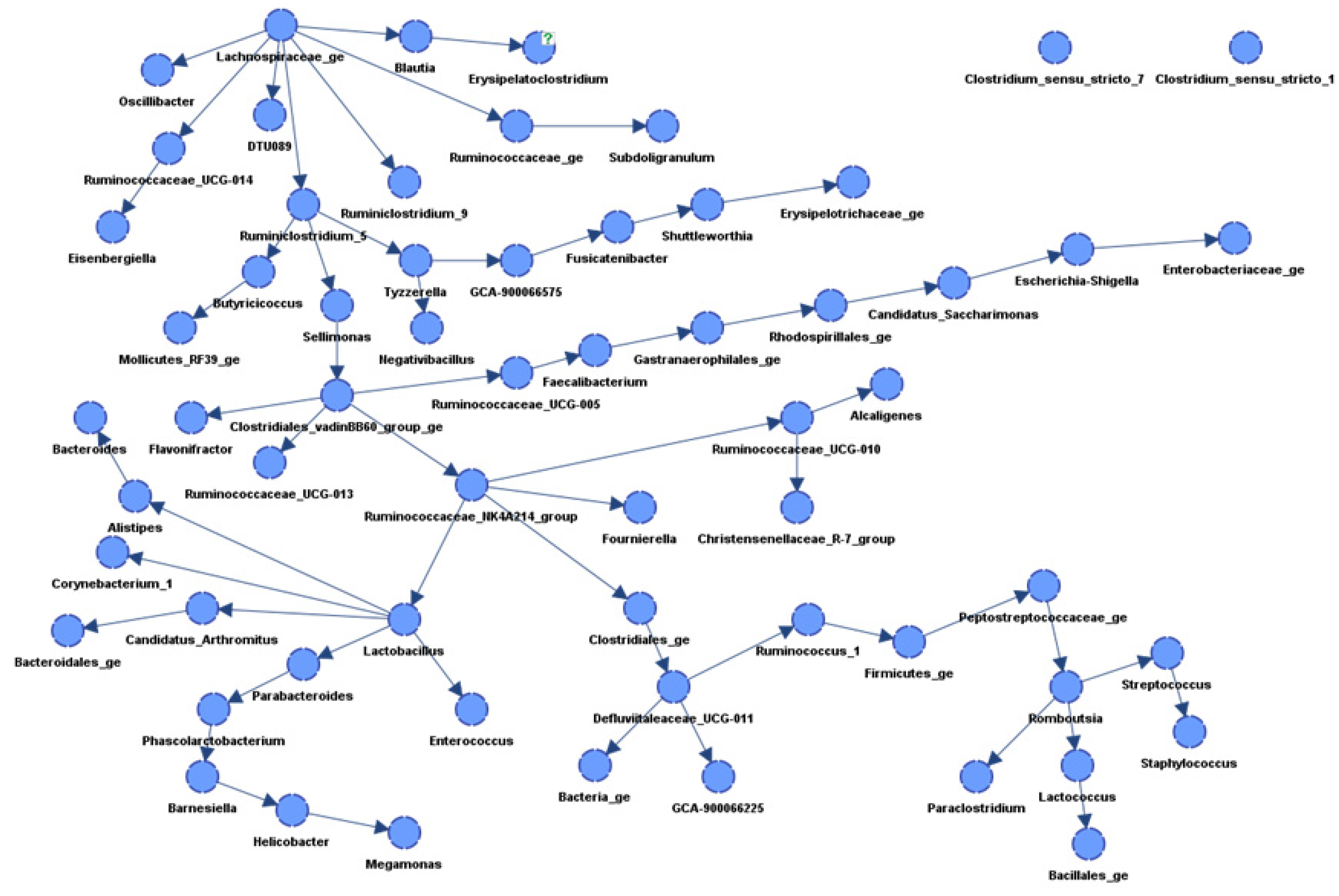
| Parent Node | Child Node | KL | Corr |
|---|---|---|---|
| Ruminococcaceae NK4A214 group | Ruminococcaceae UCG-010 | 0.77 | 0.83 |
| Lachnospiraceae | Ruminococcaceae | 0.76 | 0.94 |
| Ruminiclostridium 5 | Butyricicoccus | 0.76 | 0.88 |
| Tyzzerella | Negativibacillus | 0.75 | 0.86 |
| Ruminococcaceae UCG-010 | Christensenellaceae R-7 group | 0.73 | 0.81 |
| Fusicatenibacter | Shuttleworthia | 0.73 | 0.71 |
| Tyzzerella | GCA-900066575 | 0.72 | 0.85 |
| Clostridiales vadinBB60 group | Flavonifractor | 0.72 | 0.76 |
| Clostridiales vadinBB60 group | Ruminococcaceae NK4A214 group | 0.72 | 0.79 |
| Ruminiclostridium 5 | Sellimonas | 0.69 | 0.83 |
| Ruminococcaceae UCG-014 | Eisenbergiella | 0.69 | 0.84 |
| GCA-900066575 | Fusicatenibacter | 0.68 | 0.83 |
| Ruminiclostridium 5 | Tyzzerella | 0.68 | 0.82 |
| Lachnospiraceae | Ruminococcaceae UCG-014 | 0.67 | 0.88 |
| Lachnospiraceae | Ruminiclostridium 5 | 0.67 | 0.86 |
| Lachnospiraceae | Ruminiclostridium 9 | 0.66 | 0.86 |
| Ruminococcaceae NK4A214 group | Clostridiales | 0.65 | 0.77 |
| Lachnospiraceae | Oscillibacter | 0.65 | 0.84 |
| Clostridiales | Defluviitaleaceae UCG-011 | 0.65 | 0.71 |
| Escherichia-Shigella | Enterobacteriaceae | 0.63 | 0.73 |
| Sellimonas | Clostridiales vadinBB60 group | 0.62 | 0.75 |
| Ruminococcaceae | Subdoligranulum | 0.61 | 0.85 |
| Alistipes | Bacteroides | 0.59 | 0.69 |
| Clostridiales vadinBB60 group | Ruminococcaceae UCG-013 | 0.58 | 0.70 |
| Ruminococcaceae NK4A214 group | Lactobacillus | 0.57 | −0.81 |
| Ruminococcaceae UCG-005 | Faecalibacterium | 0.57 | 0.78 |
| Lachnospiraceae | DTU089 | 0.56 | 0.78 |
| Rhodospirillales | Candidatus Saccharimonas | 0.54 | 0.58 |
| Defluviitaleaceae UCG-011 | GCA-900066225 | 0.54 | 0.70 |
| Lactobacillus | Alistipes | 0.54 | −0.74 |
| Shuttleworthia | Erysipelotrichaceae | 0.53 | 0.64 |
| Clostridiales vadinBB60 group | Ruminococcaceae UCG-005 | 0.51 | 0.72 |
| Lachnospiraceae | Blautia | 0.48 | 0.57 |
| Barnesiella | Helicobacter | 0.48 | 0.63 |
| Ruminococcus 1 | Firmicutes | 0.48 | 0.26 |
| Firmicutes | Peptostreptococcaceae | 0.46 | 0.36 |
| Lactobacillus | Candidatus Arthromitus | 0.45 | 0.75 |
| Faecalibacterium | Gastranaerophilales | 0.45 | 0.62 |
| Defluviitaleaceae UCG-011 | Ruminococcus 1 | 0.45 | 0.69 |
| Butyricicoccus | Mollicutes RF39 | 0.44 | 0.66 |
| Ruminococcaceae NK4A214 group | Fournierella | 0.43 | 0.51 |
| Defluviitaleaceae UCG-011 | Bacteria | 0.40 | 0.53 |
| Romboutsia | Paraclostridium | 0.39 | 0.53 |
| Peptostreptococcaceae | Romboutsia | 0.39 | 0.73 |
| Blautia | Erysipelatoclostridium | 0.38 | 0.59 |
| Helicobacter | Megamonas | 0.36 | 0.68 |
| Romboutsia | Lactococcus | 0.36 | 0.64 |
| Lactobacillus | Corynebacterium 1 | 0.36 | 0.28 |
| Parabacteroides | Phascolarctobacterium | 0.35 | 0.64 |
| Lactobacillus | Enterococcus | 0.33 | 0.39 |
| Lactococcus | Bacillales | 0.33 | 0.63 |
| Gastranaerophilales | Rhodospirillales | 0.32 | 0.52 |
| Ruminococcaceae UCG-010 | Alcaligenes | 0.30 | −0.34 |
| Streptococcus | Staphylococcus | 0.30 | 0.62 |
| Romboutsia | Streptococcus | 0.30 | 0.42 |
| Phascolarctobacterium | Barnesiella | 0.27 | −0.02 |
| Candidatus Saccharimonas | Escherichia-Shigella | 0.27 | −0.30 |
| Lactobacillus | Parabacteroides | 0.23 | −0.36 |
| Candidatus Arthromitus | Bacteroidales | 0.14 | −0.24 |
| Cluster | Purity (%) | CTF (%) |
|---|---|---|
| 0 | 99.65 | 79.32 |
| 1 | 98.91 | 61.79 |
| 2 | 98.46 | 80.41 |
| 3 | 98.23 | 88.48 |
| 4 | 97.98 | 76.14 |
| 5 | 99.57 | 80.59 |
| 6 | 98.98 | 87.11 |
| 7 | 98.66 | 87.84 |
| 8 | 96.82 | 87.85 |
| 9 | 97.28 | 96.10 |
| 10 | 96.43 | 98.02 |
| Parent Node | Child Node | KL Divergence | Correlation |
|---|---|---|---|
| age | weight | 1.51 | 0.94 |
| age | LC1 | 1.34 | −0.24 |
| LC6 | LC2 | 0.95 | 0.62 |
| age | LC4 | 0.92 | 0.53 |
| LC6 | LC8 | 0.92 | 0.56 |
| LC2 | LC10 | 0.87 | 0.84 |
| LC2 | LC3 | 0.82 | 0.86 |
| age | LC6 | 0.78 | 0.37 |
| LC3 | LC7 | 0.78 | 0.79 |
| age | LC0 | 0.68 | −0.70 |
| age | LC5 | 0.68 | 0.53 |
| LC2 | LC9 | 0.41 | 0.68 |
| BSF | age | 0.34 | 0.44 |
| sex | weight | 0.18 | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detilleux, J.; Moula, N.; Dawans, E.; Taminiau, B.; Daube, G.; Leroy, P. A Probabilistic Structural Equation Model to Evaluate Links between Gut Microbiota and Body Weights of Chicken Fed or Not Fed Insect Larvae. Biology 2022, 11, 357. https://doi.org/10.3390/biology11030357
Detilleux J, Moula N, Dawans E, Taminiau B, Daube G, Leroy P. A Probabilistic Structural Equation Model to Evaluate Links between Gut Microbiota and Body Weights of Chicken Fed or Not Fed Insect Larvae. Biology. 2022; 11(3):357. https://doi.org/10.3390/biology11030357
Chicago/Turabian StyleDetilleux, Johann, Nassim Moula, Edwin Dawans, Bernard Taminiau, Georges Daube, and Pascal Leroy. 2022. "A Probabilistic Structural Equation Model to Evaluate Links between Gut Microbiota and Body Weights of Chicken Fed or Not Fed Insect Larvae" Biology 11, no. 3: 357. https://doi.org/10.3390/biology11030357
APA StyleDetilleux, J., Moula, N., Dawans, E., Taminiau, B., Daube, G., & Leroy, P. (2022). A Probabilistic Structural Equation Model to Evaluate Links between Gut Microbiota and Body Weights of Chicken Fed or Not Fed Insect Larvae. Biology, 11(3), 357. https://doi.org/10.3390/biology11030357






