An Integrated Taxonomic Approach Points towards a Single-Species Hypothesis for Santolina (Asteraceae) in Corsica and Sardinia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Material and Data
2.2. Molecular Analysis
| Marker | Name | Sequence 5′–3′ | Reference |
|---|---|---|---|
| ITS1-5.8S-ITS2 | JK14 | GGA GAA GTC GTA ACA AGG TTT CCG | [37] |
| JK12 | CCA AAC AAC CCG ACT CGT AGA CAG C | ||
| trnQ-rps16 | trnQ | GCG TGG CCA AGY GGT AAG GC | [38] |
| rps16 | GTT GCT TTY TAC CAC ATC GTT T | ||
| trnH-psbA | psbA | GTT ATG CAT GAA CGT AAT GCT C | [39] |
| trnH | CGC GCA TGG TGG ATT CAC AAT CC | ||
| trnL-trnF | trnF-IGS-f | GGT TCA AGT CCC TCT ATC CC | [39] |
| trnF-IGS-r | ATT TGA ACT GGT GAC ACG AG | ||
| trnS-trnG | trnS2-f | CGG TTT TCA AGA CCG GAG CTA TCA A | [40] |
| trnG2-r | CAT AAC CTT GAG GTC ACG GGT TCA AAT | ||
| psbM-trnD | psbM-f | TTT GAC TGA CTG TTT TTA CGT A | [40] |
| trnD-r | CAG AGC ACC GCC CTG TCA AG | ||
| rps15-ycf1 | rps15-IGSR | GCA ATT CTA AAT GTG AAG TAA G | [41] |
| ycf1-IGSR | ATT ATC GAT TAG AAG ATT TAG C |
| Locus | Denaturation (Start) | Denaturation | Annealing | Extension | Extension (End) |
|---|---|---|---|---|---|
| ×35 | |||||
| ITS1-5.8S-ITS2 | 94 °C 5′ | 94 °C 45″ | 55 °C 45″ | 72 °C 1′30″ | 72 °C 7′ |
| ×35 | |||||
| trnQ-rps16 | 94 °C 3′ | 94 °C 30″ | 53 °C 30″ | 72 °C 1′ | 72 °C 3′ |
| ×30 | |||||
| trnH-psbA | 95 °C 2′ | 95 °C 30″ | 55 °C 30″ | 72 °C 35″ | 72° C 1′ |
| ×30 | |||||
| trnL-trnF | 95 °C 2′ | 95 °C 30″ | 55 °C 30″ | 72 °C 35″ | 72 °C 1′ |
| ×35 | |||||
| trnS-trnG | 94 °C 3′ | 94 °C 30″ | 58 °C 30″ | 72 °C 1′30″ | 72 °C 5′ |
| ×35 | |||||
| psbM-trnD | 94 °C 3′ | 94 °C 30″ | 55 °C 30″ | 72 °C 1′30″ | 72 °C 5′ |
| ×35 | |||||
| rps15-ycf1 | 94 °C 3′ | 94 °C 30″ | 55 °C 30″ | 72 °C 1′30″ | 72 °C 5′ |
2.3. Cypsela Morpho-Colorimetric Analysis
2.4. Morphometric Analysis
2.5. Niche Analysis
3. Results
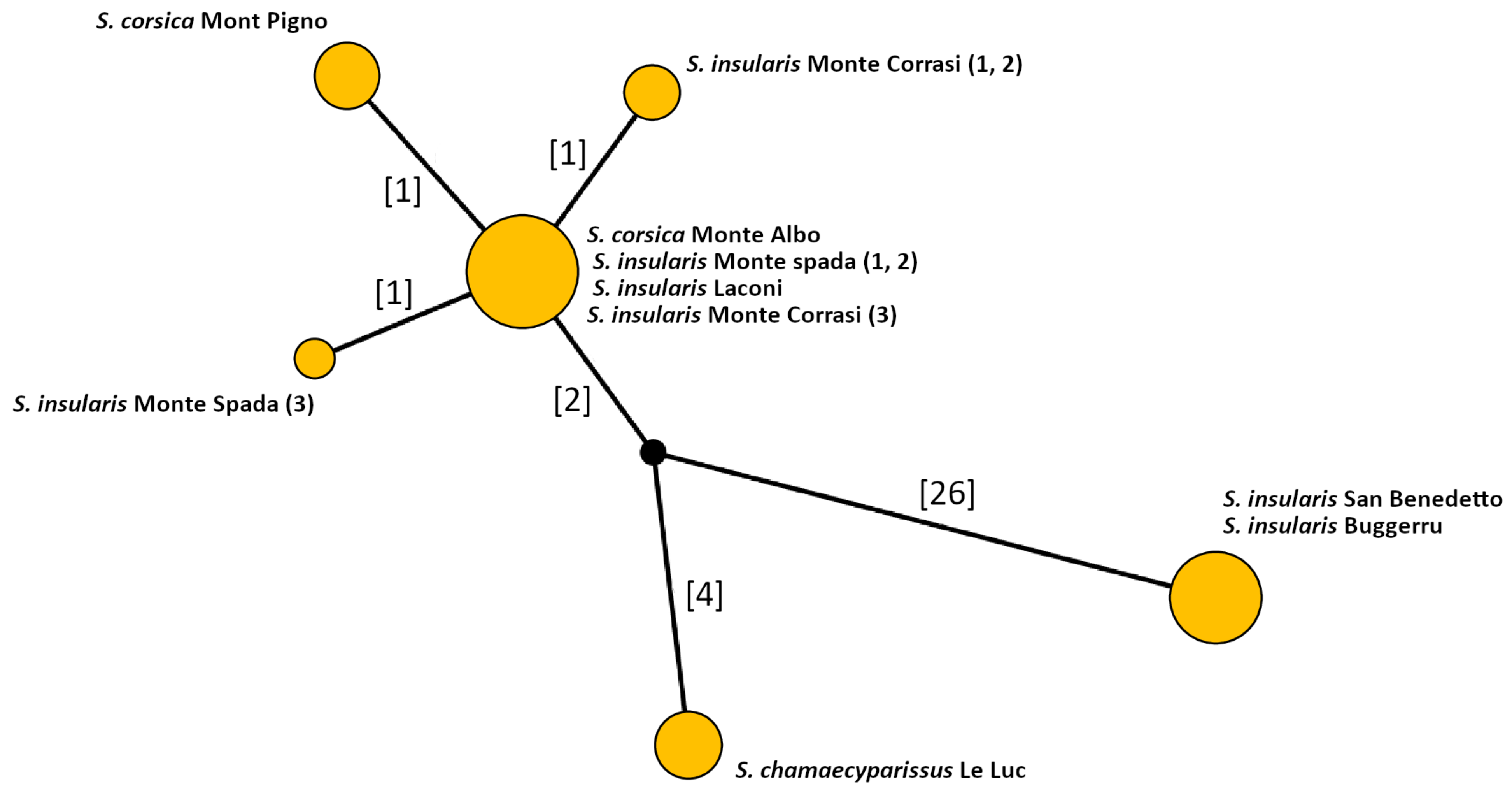
3.1. Molecular Analysis
3.2. Cypsela Morpho-Colorimetric Analysis
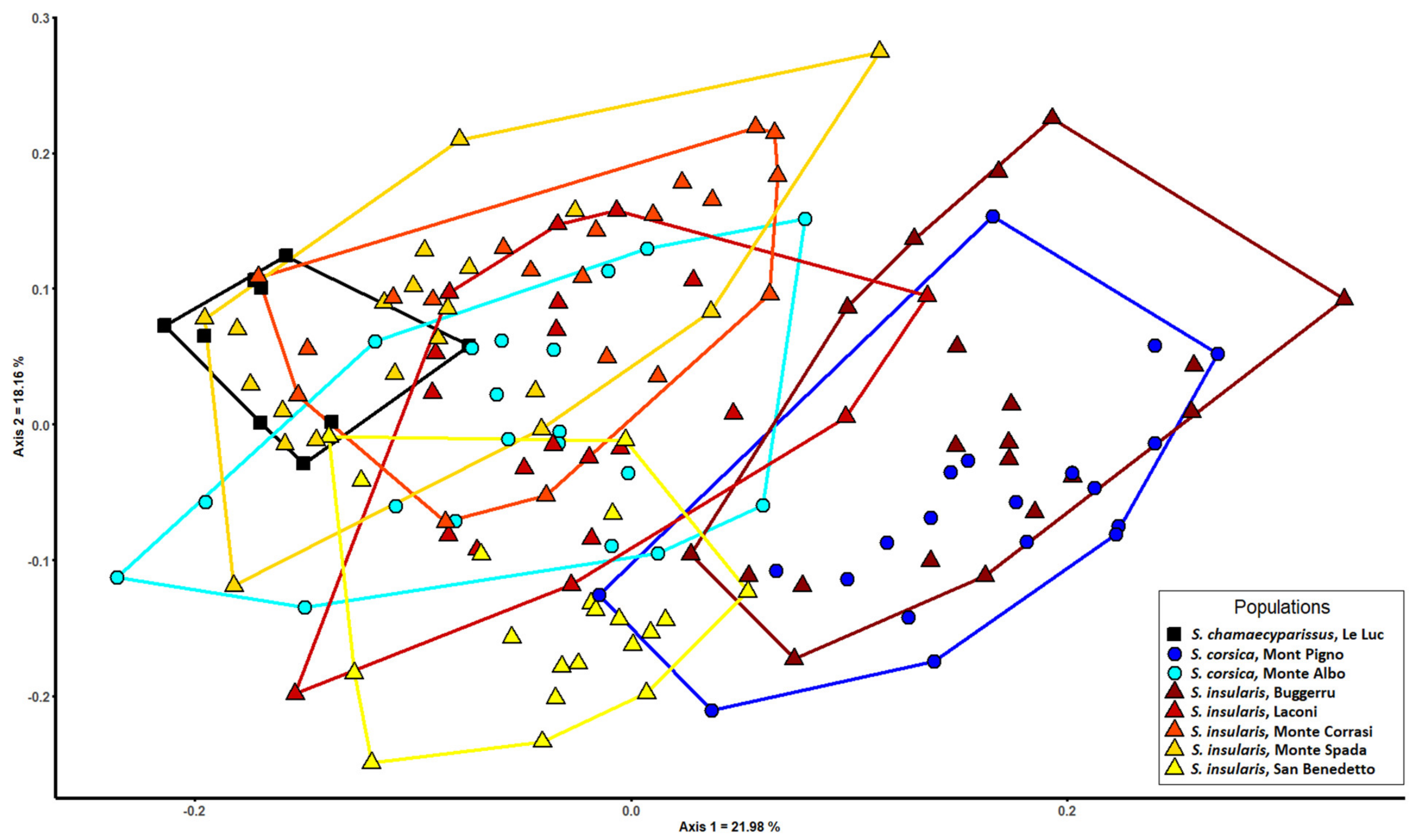
3.3. Morphometric Analysis
3.4. Niche Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittermeier, R.A.; Gil, P.R.; Hoffmann, M.; Pilgrim, J.; Brooks, T.; Mittermeier, C.G.; Lamoreux, J.; da Fonseca, G.A.B. Hotspots revisited. In Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Cañadas, E.M.; Fenu, G.; Peñas, J.; Lorite, J.; Mattana, E.; Bacchetta, G. Hotspots within hotspots: Endemic plant richness, environmental drivers, and implications for conservation. Biol. Conserv. 2014, 170, 282–291. [Google Scholar] [CrossRef]
- Greuter, W. Botanical diversity, endemism, rarity, and extinction in the Mediterranean area: An analysis based on the published volumes of Med-Checklist. Bot. Chron. 1991, 10, 63–79. [Google Scholar]
- Mansion, G.; Rosenbaum, G.; Schoenenberger, N.; Bacchetta, G.; Rosselló, J.A.; Conti, E. Phylogenetic analysis informed by geological history supports multiple, sequential invasions of the Mediterranean Basin by the angiosperm family Araceae. Syst. Biol. 2008, 57, 269–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grove, A.T.; Rackham, O. The Nature of Mediterranean Europe: An Ecological History; Yale University Press: New Haven, CT, USA, 2001. [Google Scholar]
- Jalut, G.; Dedoubat, J.J.; Fontugne, M.; Otto, T. Holocene circum-Mediterranean vegetation changes: Climate forcing and human impact. Quat. Int. 2009, 200, 4–18. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean: Insights for Conservation, 2nd ed.; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Médail, F.; Quézel, P. Hot-spots analysis for conservation of plant biodiversity in the Mediterranean Basin. Ann. Mo. Bot. Gard. 1997, 84, 112–127. [Google Scholar] [CrossRef]
- Bacchetta, G.; Pontecorvo, C. Contribution to the knowledge of the endemic vascular flora of Iglesiente (SW Sardinia-Italy). Candollea 2005, 60, 481–501. [Google Scholar]
- Véla, E.; Benhouhou, S. Évaluation d’un nouveau point chaud de biodiversité végétaledans le Bassin Méditerranéen (Afrique du Nord). Comptes Rendus Biol. 2007, 330, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Bartolucci, F.; Galasso, G.; Peruzzi, L.; Conti, F. Report 2020 on plant biodiversity in Italy: Native and alien vascular flora. Nat. Hist. Sci. 2021, 8, 41–54. [Google Scholar] [CrossRef]
- Jeanmonod, D.; Schlüssel, A.; Gamisans, J. Status and trends in the alien flora of Corsica. EPPO Bull. 2010, 41, 85–99. [Google Scholar] [CrossRef] [Green Version]
- Selvi, F.; Coppi, A.; Bigazzi, M. Karyotype variation, evolution and phylogeny in Borago (Boraginaceae), with emphasis on subgenus Buglossites in the Corso-Sardinian system. Ann. Bot. 2006, 98, 857–868. [Google Scholar] [CrossRef]
- Molins, A.; Bacchetta, G.; Rosato, M.; Rosselló, J.A.; Mayol, M. Molecular phylogeography of Thymus herba-barona (Lamiaceae): Insight into the evolutionary history of the flora of the western Mediterranean islands. Taxon 2011, 60, 1295–1305. [Google Scholar] [CrossRef]
- Dettori, C.A.; Sergi, S.; Tamburini, E.; Bacchetta, G. The genetic diversity and spatial genetic structure of the Corso-Sardinian endemic Ferula arrigonii Bocchieri (Apiaceae). Plant Biol. 2014, 16, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Meloni, M.; Dettori, C.A.; Reid, A.; Bacchetta, G.; Hugot, L.; Conti, E. High genetic diversity and presence of genetic structure characterise the Corso-Sardinian endemics Ruta corsica and Ruta lamarmorae (Rutaceae). Caryologia 2020, 73, 11–26. [Google Scholar] [CrossRef]
- Greuter, W.; Santolina, L. Med-checklist. In A Critical Inventory of Vascular Plants of the Circum-Mediterranean Countries. II. Dycotyledones (Compositae); Greuter, W., Raab-Straube von, E., Eds.; Organization for the Phyto-Taxonomic Investigation of the Mediterranean Area (OPTIMA): Genève, Switzerland, 2008; pp. 696–698. [Google Scholar]
- Carbajal, R.; Ortiz, S.; Sáez, L. Santolina, L. Flora Iberica; Castroviejo, S.B., Benedí, C., Buira, A., Rico, E., Crespo, M.B., Quintanar, A., Aedo, C., Eds.; Real Jardín Botánico, CSIC: Madrid, Spain, 2019; pp. 1938–1962. [Google Scholar]
- Giacò, A.; Astuti, G.; Peruzzi, L. Typification and nomenclature of the names in the Santolina chamaecyparissus species complex (Asteraceae). Taxon 2021, 70, 189–201. [Google Scholar] [CrossRef]
- Oberprieler, C. Temporal and spatial diversification of Circum-Mediterranean Compositae-Anthemideae. Taxon 2005, 54, 951–966. [Google Scholar] [CrossRef]
- Carbajal, R.; Serrano, M.; Ortiz, S.; Sáez, L. Two new combinations in Iberian Santolina (Compositae) based on morphology and molecular evidences. Phytotaxa 2017, 291, 217–223. [Google Scholar] [CrossRef]
- Arrigoni, P.V. Santolina, L. Flora D’Italia; Pignatti, S., Ed.; Edagricole: Milano, Italy, 2018; pp. 874–878. [Google Scholar]
- Tison, J.M.; de Foucault, B. Flora Gallica: Flore de France; Biotope Editions: Mèze, France, 2014. [Google Scholar]
- Giacò, A.; De Giorgi, P.; Astuti, G.; Varaldo, L.; Sáez, L.; Carbajal, R.; Serrano, M.; Casazza, G.; Bacchetta, G.; Caputo, P.; et al. Diploids and polyploids in the Santolina chamaecyparissus complex (Asteraceae) show different karyotype asymmetry. Plant Biosyst. 2022, in press. [Google Scholar] [CrossRef]
- Arrigoni, P.V. Santoline italiche nuove. Webbia 1977, 32, 129–134. [Google Scholar] [CrossRef]
- Marchi, P.; D’Amato, G. Numeri cromosomici per la flora italiana: 145–150. Inf. Bot. Ital. 1973, 5, 93–100. [Google Scholar]
- Fiori, A. Santolina. In Flora Analitica d’Italia; Fiori, A., Paoletti, G., Eds.; Tipografia del Seminario: Padova, Italy, 1903; pp. 269–270. [Google Scholar]
- Fiori, A. Nuova Flora Analitica d’Italia; Tipografia, M., Ed.; Ricci: Florence, Italy, 1927. [Google Scholar]
- Marchi, P.; Capineri, R.; D’Amato, G. Il cariotipo di Santolina corsica Jord. & Fourr. (Compositae) proveniente da Bastia (Corsica) ed altre osservazioni. Ann. Bot. 1979, 38, 1–13. [Google Scholar]
- Arrigoni, P.V.; Camarda, I.; Corrias, B.; Corrias, S.D.; Valsecchi, F. Le piante endemiche della Sardegna. Boll. Della Soc. Sarda Sci. Nat. 1982, 21, 338–348. [Google Scholar]
- Erst, A.S.; Sukhorukov, A.P.; Mitrenina, E.Y.; Skaptsov, M.V.; Kostikova, V.A.; Chernisheva, O.A.; Troshkina, V.; Kushunina, M.; Krivenko, D.A.; Ikeda, H.; et al. An integrative taxonomic approach reveals a new species of Eranthis (Ranunculaceae) in North Asia. PhytoKeys 2020, 140, 75–100. [Google Scholar] [CrossRef]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Bagella, S.; Filigheddu, R.; Peruzzi, L.; Bedini, G. Wikiplantbase #Sardegna v3.0. Available online: http://bot.biologia.unipi.it/wpb/sardegna/index.html (accessed on 4 December 2021).
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Aceto, S.; Caputo, P.; Cozzolino, S.; Gaudio, L.; Moretti, A. Phylogeny and evolution of Orchis and allied genera based on ITS DNA variation: Morphological gaps and molecular continuity. Mol. Phylogenetics Evol. 1999, 13, 67–76. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Prince, L.M. Plastid primers for angiosperm phylogenetics and phylogeography. Appl. Plant Sci. 2015, 3, 1400085. [Google Scholar] [CrossRef]
- Sarigu, M.; Grillo, O.; Lo Bianco, M.; Ucchesu, M.; d’Hallewin, G.; Loi, M.C.; Venora, G.; Bacchetta, G. Phenotypic identification of plum varieties (Prunus domestica L.) by endocarps morpho-colorimetric and textural descriptors. Comput. Electron. Agric. 2017, 136, 25–30. [Google Scholar] [CrossRef]
- Frigau, L.; Antoch, J.; Bacchetta, G.; Sarigu, M.; Ucchesu, M.; Zaratin Alves, C.; Mola, F. A statistical approach to the morphological classification of Prunus sp. seeds. Plant Biosyst. 2020, 154, 877–886. [Google Scholar] [CrossRef]
- Lo Bianco, M.; Grillo, O.; Escobar Garcia, P.; Mascia, F.; Venora, G.; Bacchetta, G. Morpho-colorimetric characterisation of Malva alliance taxa by seed image analysis. Plant Biol. 2017, 19, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Sarigu, M.; Porceddu, M.; Schmitt, E.; Camarda, I.; Bacchetta, G. Taxonomic discrimination of the Paeonia mascula group in the Tyrrhenian Islands by seed image analysis. Syst. Biodivers. 2019, 17, 801–810. [Google Scholar] [CrossRef]
- Farris, E.; Orrù, M.; Ucchesu, M.; Amadori, A.; Porceddu, M.; Bacchetta, G. Morpho-colorimetric characterization of the sardinian endemic taxa of the genus Anchusa L. by seed image analysis. Plants 2020, 9, 1321. [Google Scholar] [CrossRef] [PubMed]
- Shahin, M.A.; Symons, S.J. Color calibration of scanners for scanner-independent grain grading. Cereal Chem. 2003, 80, 285–289. [Google Scholar] [CrossRef]
- Landini, G. Quantitative analysis of the epithelial lining architecture in radicular cysts and odontogenic keratocysts. Head Face Med. 2006, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Terral, J.-F.; Tabard, E.; Bouby, L.; Ivorra, S.; Pastor, T.; Figueiral, I.; Picq, S.; Chevance, J.-B.; Jung, C.; Fabre, L.; et al. Evolution and history of grapevine (Vitis vinifera) under domestication: New morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann. Bot. 2010, 105, 443–455. [Google Scholar] [CrossRef]
- Orrù, M.; Grillo, O.; Lovicu, G.; Venora, G.; Bacchetta, G. Morphological characterisation of Vitis vinifera L. seeds by image analysis and comparison with archaeological remains. Veg. Hist. Archaeobotany 2013, 22, 231–242. [Google Scholar] [CrossRef]
- Morrison, D.F. Multivariate Statistical Methods; Duxbury Press: Belmont, CA, USA, 2004. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff; New York Botanical Garden: Bronx, NY, USA, 2021; Available online: http://sweetgum.nybg.org/ih/ (accessed on 4 December 2021).
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Shipunov, A.; Kosenko, Y.; Volkova, P. Floral polymorphism in common primrose (Primula vulgaris Huds., Primulaceae) of the Northeastern Black Sea coast. Plant Syst. Evol. 2011, 296, 167–178. [Google Scholar] [CrossRef]
- Skoracka, A.; Kuczyński, L.; Rector, B.; Amrine, J.W. Wheat curl mite and dry bulb mite: Untangling a taxonomic conundrum through a multidisciplinary approach. Biol. J. Linn. Soc. 2014, 111, 421–436. [Google Scholar] [CrossRef] [Green Version]
- Moffat, C.E.; Ensing, D.J.; Gaskin, J.F.; De Clerck-Floate, R.A.; Pither, J. Morphology delimits more species than molecular genetic clusters of invasive Pilosella. Am. J. Bot. 2015, 102, 1145–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobl, C.; Boulesteix, A.-L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touw, W.G.; Bayjanov, J.R.; Overmars, L.; Backus, L.; Boekhorst, J.; Wels, M.; van Hijum, S.A.F.T. Data mining in the Life Sciences with Random Forest: A walk in the park or lost in the jungle? Brief. Bioinform. 2013, 14, 315–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Broennimann, O.; Fitzpatrick, M.C.; Pearman, P.B.; Petitpierre, B.; Pellissier, L.; Yoccoz, N.G.; Thuiller, W.; Fortin, M.-J.; Randin, C.; Zimmermann, N.E.; et al. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob. Ecol. Biogeogr. 2012, 21, 481–497. [Google Scholar] [CrossRef] [Green Version]
- Schoener, T.W. Nonsynchronous spatial overlap of lizards in patchy habitats. Ecology 1970, 51, 408–418. [Google Scholar] [CrossRef] [Green Version]
- Broennimann, O.; Di Cola, V.; Petitpierre, B.; Breiner, F.; D’Amen, M.; Randin, C.; Engler, R.; Hordijk, W.; Pottier, J.; Di Febbraro, M.D.; et al. Ecospat: Spatial Ecology Miscellaneous Methods. Available online: https://cran.r-project.org/web/packages/ecospat/index.html (accessed on 4 December 2021).
- Angiolini, C.; Bacchetta, G. Analisi distributiva e studio fitosociologico delle comunità a Santolina insularis (Gennari ex Fiori) Arrigoni della Sardegna meridionale (Italia). Fitosociologia 2003, 40, 109–127. [Google Scholar]
- Cherchi, A.; Montadert, L. Oligo-Miocene rift of Sardinia and the early history of the Western Mediterranean Basin. Nature 1982, 298, 736–739. [Google Scholar] [CrossRef]
- Cocco, F.; Andreucci, S.; Sechi, D.; Cossu, G.; Funedda, A. Upper Pleistocene tectonics in western Sardinia (Italy): Insights from the Sinis peninsula structural high. Terra Nova 2019, 31, 485–493. [Google Scholar] [CrossRef]
- De Guia, A.P.O.; Saitoh, T. The gap between the concept and definitions in the Evolutionarily Significant Unit: The need to integrate neutral genetic variation and adaptive variation. Ecol. Res. 2007, 22, 604–612. [Google Scholar] [CrossRef]
- Ryder, O.A. Species conservation and systematics: The dilemma of subspecies. Trends Ecol. Evol. 1986, 1, 9–10. [Google Scholar] [CrossRef]
- Peruzzi, L.; Astuti, G.; Algisi, S.; Coppi, A. Genetic differentiation among populations of the threatened Bellevalia webbiana (Asparagaceae) and its consequence on conservation. Plant Biosyst. 2021, 155, 188–193. [Google Scholar] [CrossRef]
- Briquet, J. Etude carpologiques sur les genres de Composées Anthemis, Ormenis et Santolina. Annu. Du Conserv. Et Du Jard. Bot. De Genève 1916, 18–19, 257–313. [Google Scholar]
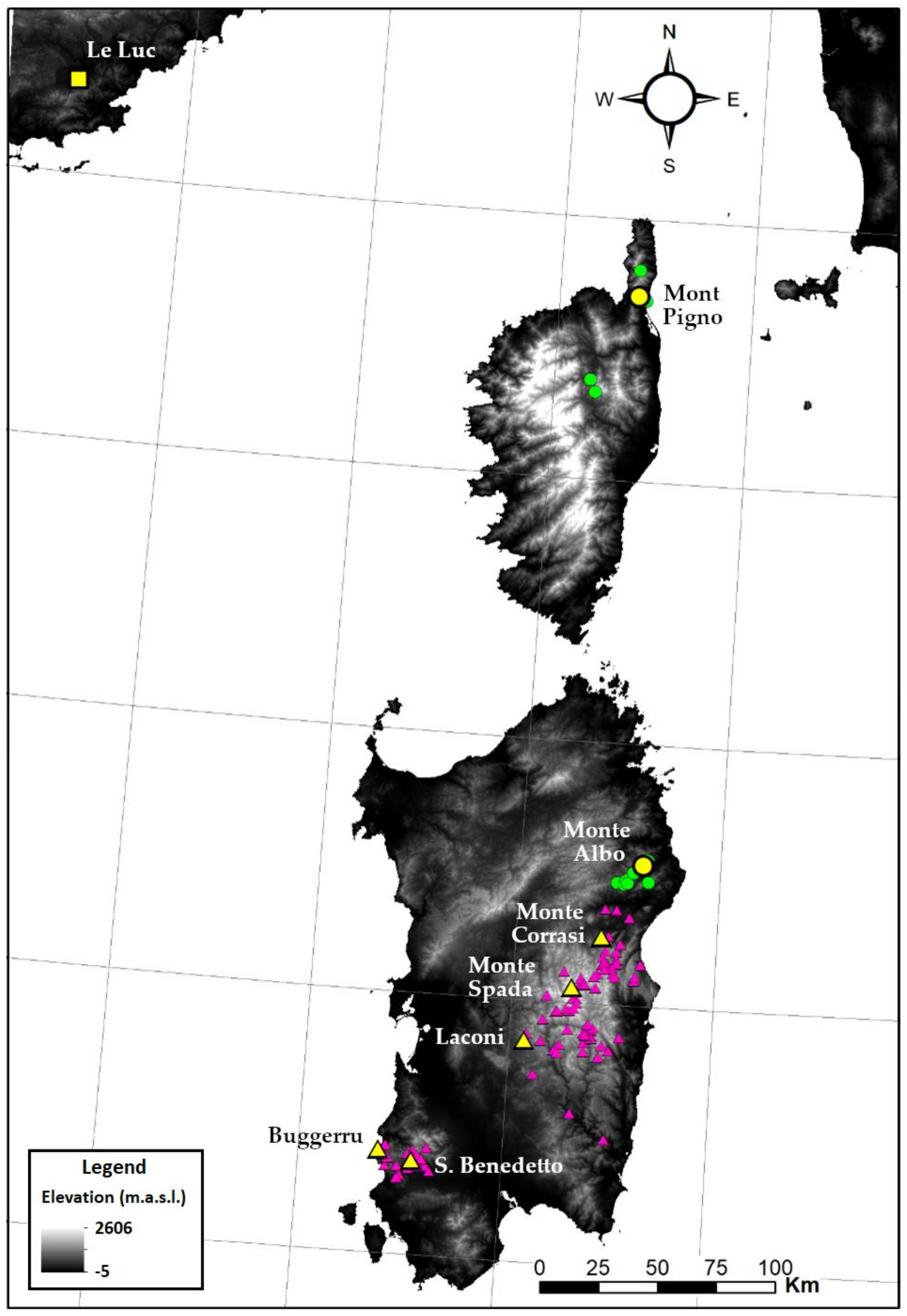
| Species | Population | Vouchers |
|---|---|---|
| S. chamaecyparissus | France, Provence-Alpes-Côte d’Azur, Le Luc [WGS84: 43.354166 N, 6.412222 E] | A. Giacò and L. Peruzzi, 30 June 2020, PI 034970–034974 |
| S. corsica | France, Corsica, Mont Pigno [WGS84: 42.7066667 N, 9.407777 E], type locality | A. Giacò and L. Peruzzi, 7 July 2020, PI 036636–036647 |
| S. corsica | Italy, Sardinia, Monte Albo [WGS84: 40.537853 N, 9.615131 E] | G. Calvia et al., 19 June 2020, PI 036122–036136 |
| S. insularis | Italy, Sardinia, Buggerru [WGS84: 39.393611 N, 8.391666 E] | G. Bacchetta et al., 14 June 2020, PI 036613–036625 |
| S. insularis | Italy, Sardinia, San Benedetto (Iglesias) [WGS84: 39.360311 N, 8.558333 E], type locality | G. Bacchetta et al., 14 June 2020, PI 036068–036085 |
| S. insularis | Italy, Sardinia, Laconi [WGS84: 39.847483 N, 9.071944 E] | G. Bacchetta et al., 15 June 2020, PI 036052–036067 |
| S. insularis | Italy, Sardinia, Monte Spada [WGS84: 40.058586 N, 9.293333 E] | G. Bacchetta et al., 14 June 2020, PI 036106–036121 |
| S. insularis | Italy, Sardinia, Monte Corrasi [WGS84: 40.256878 N, 9.426253 E] | G. Bacchetta et al., 14 June 2020, PI 036648–036663 |
| Code | Description of the Character | Type | Tool |
|---|---|---|---|
| Vegetative Parts | |||
| fs_length | Length of the fertile stem (cm) | QC | Ruler |
| br_ratio | Ratio between the highest ramification of the fertile stem and fs_length | QC | Ruler |
| dist_cap | Distance between the highest leaf on the stem and the floral head (mm) | QC | Caliper |
| fs_n_br | Number of branches of the fertile stem | QD | |
| br_type | Type of branch (no branch/parallel/or erect-patent) | CN | |
| fs_n_nodes | Number of nodes of the fertile stem | QD | |
| fs_node_length | Length of a random selected node of the fertile stem (mm) | QC | Caliper |
| fs_sterile_like | Fertile stem morphologically similar to sterile stems (Yes/No) | CB | |
| ss_length | Length of the sterile stem (cm) | QC | Ruler |
| ss_n_nodes | Number of nodes of the sterile stem | QD | |
| ss_node_length | Length of a random selected node of the sterile stem (mm) | QC | Caliper |
| ss_hair | Tomentosity of the sterile stem (hairless/slightly pubescent/pubescent/hairy/densely hairy) | CO | ImageJ |
| fs_hair | Degree of tomentosity of the fertile stem (%) | QC | ImageJ |
| fasc_type | Axillary leaves of the fertile stem (absent/fasciculate/ramified) | CN | |
| fsl_n_seg | Number of segments on the fertile stem leaf (the longest) | QD | |
| ssl_n_seg | Number of segments on the sterile stem leaf (the longest) | QD | |
| ssl_length | Length of the sterile stem leaf (mm) | QC | ImageJ |
| ssl_width | Width of the sterile stem leaf (mm) | QC | ImageJ |
| ssl_petiole_length | Length of the petiole of the sterile stem leaf (mm) | QC | ImageJ |
| ssl_seg_length | Length of the segment of the sterile stem leaf (mm) | QC | ImageJ |
| ssl_seg_width | Width of the segment of the sterile stem leaf (mm) | QC | ImageJ |
| ssl_seg_dist | Distance between the segments of the sterile stem leaf (mm) | QC | ImageJ |
| fsl_length | Length of the fertile stem leaf (mm) | QC | ImageJ |
| fsl_width | Width of the fertile stem leaf (mm) | QC | ImageJ |
| fsl_petiole_length | Length of the petiole of the fertile stem leaf (mm) | QC | ImageJ |
| fsl_seg_length | Length of the segment of the fertile stem leaf (mm) | QC | ImageJ |
| fsl_seg_width | Width of the segment of the fertile stem leaf (mm) | QC | ImageJ |
| fsl_seg_dist | Distance between the segments of the fertile stem leaf (mm) | QC | ImageJ |
| ssl_hair | Degree of tomentosity of the sterile stem leaf segment (%) | QC | ImageJ |
| fsl_hair | Degree of tomentosity of the fertile stem leaf segment (%) | QC | ImageJ |
| Floral head | |||
| cap_diam | Diameter of the floral head involucre (mm) | QC | Caliper |
| flowers_type | The flowers totally cover the involucre (Yes/No) | CB | |
| flower_length | Length of the floral tube (mm) | QC | ImageJ |
| flower_tooth_length | Length of the floral tooth (mm) | QC | ImageJ |
| sq_ext_length | Length of the external involucral bract (mm) | QC | ImageJ |
| sq_ext_width | Width of the external involucral bract (mm) | QC | ImageJ |
| sq_int_length | Length of the internal involucral bract (mm) | QC | ImageJ |
| sq_int_width | Width of the internal involucral bract (mm) | QC | ImageJ |
| sq_if_length | Length of the inter-floral bract (mm) | QC | ImageJ |
| sq_if_width | Width of the inter-floral bract (mm) | QC | ImageJ |
| sq_if_type | Tip of the inter-floral bract (rounded/truncate) | CB | |
| sq_if_n_hair | Tomentosity of the inter-floral bract (hairless/slightly pubescent/pubescent/hairy/densely hairy) | CO | ImageJ |
| sq_ext_hair | Tomentosity of the external involucral bract (hairless/only on the margin/everywhere) | CO | ImageJ |
| sq_int_hair | Tomentosity of the internal involucral bract (hairless/only on the margin/everywhere) | CO | ImageJ |
| Markers | Length (bp) | Consensus Length (bp) | Informative Sites |
|---|---|---|---|
| psbM-trnD | 708–716 | 717 | 5 |
| rps15-ycf1 | 495–510 | 517 | 7 |
| trnF-trnL | 381 | 381 | 1 |
| trnH-psbA | 422–457 | 458 | 9 |
| trnQ-rps16 | 885–895 | 905 | 10 |
| trnS-trnG | 420–425 | 425 | 2 |
| Concatenated matrix | --- | 3403 | 34 |
| Species | S. corsica | S. insularis |
|---|---|---|
| S. corsica | 54 | 46 |
| S. insularis | 12.4 | 87.6 |
| Populations | S. corsica Monte Albo | S. corsica Mont Pigno | S. insularis San Benedetto | S. insularis Buggerru | S. insularis Monte Spada | S. insularis Monte Corrasi | S. insularis Laconi |
|---|---|---|---|---|---|---|---|
| S. corsica Monte Albo | 53 | 19 | 15 | 10 | 2 | 1 | 0 |
| S. corsica Mont Pigno | 20 | 56 | 13 | 11 | 0 | 0 | 0 |
| S. insularis San Benedetto | 19 | 23 | 24 | 33 | 1 | 0 | 0 |
| S. insularis Buggerru | 12 | 17 | 22 | 48 | 1 | 0 | 0 |
| S. insularis Monte Spada | 0 | 0 | 2 | 0 | 67 | 20 | 11 |
| S. insularis Monte Corrasi | 0 | 0 | 0 | 0 | 23 | 63 | 14 |
| S. insularis Laconi | 0 | 0 | 0 | 0 | 26 | 21 | 53 |
| Species | S. chamaecyparissus | S. corsica | S. insularis |
|---|---|---|---|
| S. chamaecyparissus | 92 | 0 | 8 |
| S. corsica | 0 | 46 | 54 |
| S. insularis | 0 | 4 | 96 |
| Groups | S. chamaecyparissus | Group 1 | Group 2 |
|---|---|---|---|
| S. chamaecyparissus | 90 | 0 | 10 |
| Group 1 | 0 | 82 | 18 |
| Group 2 | 0 | 3 | 97 |
| Groups | S. chamaecyparissus | Group A | Group B |
|---|---|---|---|
| S. chamaecyparissus | 74 | 26 | 0 |
| Group A | 0 | 83 | 17 |
| Group B | 0 | 6 | 94 |
| Groups | S. chamaecyparissus | Group X | Group Y |
|---|---|---|---|
| S. chamaecyparissus | 78 | 0 | 22 |
| Group X | 0 | 57 | 43 |
| Group Y | 0 | 1 | 99 |
| Character | S. chamaecyparissus Le Luc | S. corsica Monte Albo | S. corsica Mont Pigno | S. insularis San Benedetto | S. insularis Buggerru | S. insularis Monte Spada | S. insularis Monte Corrasi | S. insularis Laconi |
|---|---|---|---|---|---|---|---|---|
| fs_length (cm) | 16.7 (± 2.6) | 15.7 (± 3.5) | 12.6 (± 3.9) | 20.4 (± 5.2) | 11.3 (± 4.4) | 15.7 (± 4.9) | 14.3 (± 4.7) | 16.4 (± 3.8) |
| br_ratio | 0.1 (± 0.1) | 0.2 (± 0.3) | 0.6 (± 0.2) | 0.6 (± 0.3) | 0.7 (± 0.3) | 0.1 (± 0.2) | 0.1 (± 0.2) | 0.3 (± 0.3) |
| dist_cap_lf (mm) | 41.9 (± 11.6) | 24.7 (± 12.4) | 20.4 (± 7.3) | 34.0 (± 13.8) | 11.5 (± 6.4) | 36.4 (± 22.5) | 35.9 (± 19.2) | 27.5 (± 11.6) |
| fs_n_br | 1.0 (± 1.2) | 0.7 (± 1.4) | 4.0 (± 2.9) | 4.0 (± 2.8) | 5.1 (± 5.3) | 1.0 (± 1.6) | 0.4 (± 0.7) | 1.5 (± 1.6) |
| fs_n_nodes | 12.9 (± 2.5) | 14.8 (± 2.5) | 15.0 (± 2.4) | 17.3 (± 3.9) | 19.9 (± 6.0) | 14.4 (± 2.9) | 11.5 (± 2.2) | 16.3 (± 3.5) |
| fs_node_length (mm) | 13.7 (± 7.1) | 9.8 (± 4.1) | 8.6 (± 4.2) | 12.0 (± 6.4) | 6.2 (± 3.5) | 7.5 (± 3.2) | 9.5 (± 4.9) | 9.1 (± 5.6) |
| ss_length (cm) | 10.4 (± 2.8) | 9.3 (± 2.2) | 6.7 (± 3.6) | 11.9 (± 3.8) | 5.9 (± 3.2) | 10.3 (± 3.7) | 7.1 (± 3.2) | 10.9 (± 2.9) |
| ss_n_nodes | 16.0 (± 3.2) | 17.1 (± 3.2) | 15.4 (± 3.1) | 18.4 (± 3.1) | 16.1 (± 5.2) | 16.5 (± 3.3) | 12.7 (± 2.8) | 17.1 (± 4.8) |
| ss_node_length (mm) | 6.8 (± 2.3) | 6.9 (± 2.2) | 5.3 (± 2.2) | 6.9 (± 2.0) | 4.5 (± 2.6) | 7.0 (± 3.4) | 6.5 (± 3.7) | 8.8 (± 3.7) |
| fs_hair | 0.2 (± 0.1) | 0.3 (± 0.2) | 0.5 (± 0.1) | 0.4 (± 0.2) | 0.7 (± 0.2) | 0.3 (± 0.1) | 0.3 (± 0.1) | 0.3 (± 0.1) |
| fsl_n_seg | 14.3 (± 3.5) | 59.7 (± 17.3) | 90.2 (± 28.6) | 82.3 (± 15.4) | 105.2 (± 32.1) | 48.8 (± 9.2) | 52.4 (± 16.4) | 48.7 (± 15.0) |
| ssl_n_seg | 45.6 (± 6.4) | 102.9 (± 20.9) | 135.9 (± 37.7) | 109.5 (± 18.7) | 121.6 (± 31.3) | 72.5 (± 12.0) | 83.7 (± 14.8) | 73.5 (± 16.8) |
| ssl_length (mm) | 23.0 (± 3.4) | 36.5 (± 10.4) | 33.1 (± 9.3) | 38.5 (± 8.8) | 24.9 (± 8.3) | 23.9 (± 3.5) | 26.2 (± 6.8) | 25.1 (± 4.5) |
| ssl_width (mm) | 0.9 (± 0.2) | 0.8 (± 0.1) | 0.8 (± 0.2) | 0.8 (± 0.2) | 0.8 (± 0.2) | 0.9 (± 0.2) | 0.8 (± 0.2) | 0.8 (± 0.2) |
| ssl_petiole_length (mm) | 3.0 (± 0.8) | 4.3 (± 2.2) | 3.2 (± 1.6) | 4.5 (± 1.8) | 3.3 (± 1.5) | 2.2 (± 1.0) | 2.9 (± 1.0) | 3.2 (± 1.8) |
| ssl_seg_length (mm) | 1.5 (± 0.2) | 1.2 (± 0.6) | 1.5 (± 0.3) | 1.4 (± 0.2) | 1.0 (± 0.4) | 1.3 (± 8.3) | 1.3 (± 0.3) | 1.3 (± 0.4) |
| ssl_seg_width (mm) | 0.8 (± 0.1) | 0.6 (± 0.1) | 0.4 (± 0.1) | 0.6 (± 0.1) | 0.6 (± 0.1) | 0.6 (± 0.1) | 0.6 (± 0.1) | 0.6 (± 0.1) |
| ssl_seg_dist (mm) | 0.8 (± 0.2) | 0.9 (± 0.4) | 0.9 (± 0.4) | 0.7 (± 0.4) | 0.4 (± 0.3) | 0.8 (± 0.3) | 0.9 (± 0.4) | 0.7 (± 0.3) |
| fsl_length (mm) | 13.6 (± 2.2) | 21.8 (± 8.2) | 22.4 (± 5.5) | 31.1 (± 8.5) | 21.8 (± 8.7) | 16.3 (± 4.1) | 16.1 (± 4.5) | 15.8 (± 5.3) |
| fsl_width (mm) | 1.0 (± 0.2) | 0.8 (± 0.4) | 0.7 (± 0.2) | 0.7 (± 0.1) | 0.8 (± 0.2) | 0.9 (± 0.2) | 1.1 (± 0.7) | 0.8 (± 0.2) |
| fsl_petiole_length (mm) | 5.4 (± 1.2) | 4.0 (± 1.6) | 2.3 (± 1.0) | 4.8 (± 1.5) | 2.5 (± 1.1) | 2.9 (± 1.2) | 2.4 (± 1.2) | 2.9 (± 1.7) |
| fsl_seg_length (mm) | 1.2 (± 0.2) | 0.7 (± 0.3) | 1.3 (± 0.4) | 1.1 (± 0.3) | 0.8 (± 0.2) | 1.1 (± 0.4) | 0.9 (± 0.2) | 1.0 (± 0.4) |
| fsl_seg_width (mm) | 0.6 (± 0.1) | 0.5 (± 0.1) | 0.4 (± 0.1) | 0.6 (± 0.1) | 0.6 (± 0.2) | 0.6 (± 0.3) | 0.5 (± 0.1) | 0.5 (± 0.2) |
| fsl_seg_dist (mm) | 0.4 (± 0.3) | 0.7 (± 0.4) | 0.7 (± 0.2) | 0.6 (± 0.3) | 0.4 (± 0.3) | 0.3 (± 0.3) | 0.6 (± 0.3) | 0.3 (± 0.3) |
| ssl_hair | 0.9 (± 0.1) | 0.7 (± 0.2) | 0.8 (± 0.1) | 0.8 (± 0.1) | 0.8 (± 0.2) | 0.7 (± 0.2) | 0.9 (± 0.1) | 0.6 (± 0.2) |
| fsl_hair | 0.4 (± 0.1) | 0.4 (± 0.2) | 0.7 (± 0.2) | 0.5 (± 0.1) | 0.6 (± 0.2) | 0.2 (± 0.1) | 0.5 (± 0.2) | 0.3 (± 0.1) |
| cap_diam (mm) | 7.0 (± 0.4) | 7.1 (± 1.6) | 5.1 (± 0.7) | 6.8 (± 1.1) | 5.6 (± 0.8) | 6.6 (± 1.1) | 6.5 (± 0.9) | 6.8 (± 1.3) |
| flower_length (mm) | 4.3 (± 0.3) | 3.3 (± 0.4) | 2.8 (± 0.3) | 3.3 (± 0.4) | 3.1 (± 3.1) | 3.3 (± 0.4) | 3.5 (± 0.3) | 2.9 (± 0.5) |
| flower_tooth_length (mm) | 0.6 (± 0.1) | 0.8 (± 0.1) | 0.7 (± 0.1) | 0.7 (± 0.1) | 0.6 (± 0.1) | 0.7 (± 0.1) | 0.8 (± 0.1) | 0.7 (± 0.2) |
| sq_ext_length (mm) | 3.3 (± 0.4) | 3.2 (± 0.6) | 2.9 (± 0.4) | 3.6 (± 0.5) | 3.3 (± 0.5) | 3.6 (± 0.6) | 3.2 (± 0.4) | 3.3 (± 0.6) |
| sq_ext_width (mm) | 1.6 (± 0.2) | 1.3 (± 0.2) | 1.2 (± 0.2) | 1.3 (± 0.3) | 1.3 (± 0.3) | 1.4 (± 0.2) | 1.3 (± 0.2) | 1.3 (± 0.3) |
| sq_int_length (mm) | 4.1 (± 0.3) | 3.3 (± 0.4) | 2.9 (± 0.3) | 3.5 (± 0.5) | 3.4 (± 0.4) | 4.0 (± 0.5) | 3.2 (± 0.6) | 3.3 (± 0.5) |
| sq_int_width (mm) | 1.7 (± 0.2) | 1.4 (± 0.2) | 1.2 (± 0.1) | 1.5 (± 0.2) | 1.3 (± 0.2) | 1.5 (± 0.4) | 1.4 (± 0.2) | 1.4 (± 0.3) |
| sq_if_length (mm) | 3.7 (± 0.4) | 3.4 (± 0.5) | 3.1 (± 0.4) | 3.3 (± 0.3) | 2.9 (± 0.3) | 3.4 (± 0.5) | 3.9 (± 0.5) | 3.1 (± 0.3) |
| sq_if_width (mm) | 1.2 (± 0.2) | 1.0 (± 0.2) | 0.9 (± 0.2) | 1.0 (± 0.2) | 1.0 (± 0.2) | 1.1 (± 0.2) | 1.0 (± 0.3) | 1.0 (± 0.2) |
| Current Taxonomic Hypothesis | |||
|---|---|---|---|
| Background | Niche overlap | S. corsica vs S. insularis | S. insularis vs S. corsica |
| 5 km | 0.34 | more | ns |
| 10 km | 0.36 | more | ns |
| 15 km | 0.26 | more | ns |
| “Cypsela Morpho-Colorimetric” Grouping | |||
| Background | Niche overlap | Group A vs Group B | Group B vs Group A |
| 5 km | 0.09 | ns | more |
| 10 km | 0.16 | ns | more |
| 15 km | 0.25 | ns | more |
| “Molecular” Grouping | |||
| Background | Niche overlap | Group x vs Group y | Group y vs Group x |
| 5 km | 0.04 | ns | ns |
| 10 km | 0.03 | ns | ns |
| 15 km | 0.05 | ns | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Giorgi, P.; Giacò, A.; Astuti, G.; Minuto, L.; Varaldo, L.; De Luca, D.; De Rosa, A.; Bacchetta, G.; Sarigu, M.; Peruzzi, L. An Integrated Taxonomic Approach Points towards a Single-Species Hypothesis for Santolina (Asteraceae) in Corsica and Sardinia. Biology 2022, 11, 356. https://doi.org/10.3390/biology11030356
De Giorgi P, Giacò A, Astuti G, Minuto L, Varaldo L, De Luca D, De Rosa A, Bacchetta G, Sarigu M, Peruzzi L. An Integrated Taxonomic Approach Points towards a Single-Species Hypothesis for Santolina (Asteraceae) in Corsica and Sardinia. Biology. 2022; 11(3):356. https://doi.org/10.3390/biology11030356
Chicago/Turabian StyleDe Giorgi, Paola, Antonio Giacò, Giovanni Astuti, Luigi Minuto, Lucia Varaldo, Daniele De Luca, Alessandro De Rosa, Gianluigi Bacchetta, Marco Sarigu, and Lorenzo Peruzzi. 2022. "An Integrated Taxonomic Approach Points towards a Single-Species Hypothesis for Santolina (Asteraceae) in Corsica and Sardinia" Biology 11, no. 3: 356. https://doi.org/10.3390/biology11030356
APA StyleDe Giorgi, P., Giacò, A., Astuti, G., Minuto, L., Varaldo, L., De Luca, D., De Rosa, A., Bacchetta, G., Sarigu, M., & Peruzzi, L. (2022). An Integrated Taxonomic Approach Points towards a Single-Species Hypothesis for Santolina (Asteraceae) in Corsica and Sardinia. Biology, 11(3), 356. https://doi.org/10.3390/biology11030356










