Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
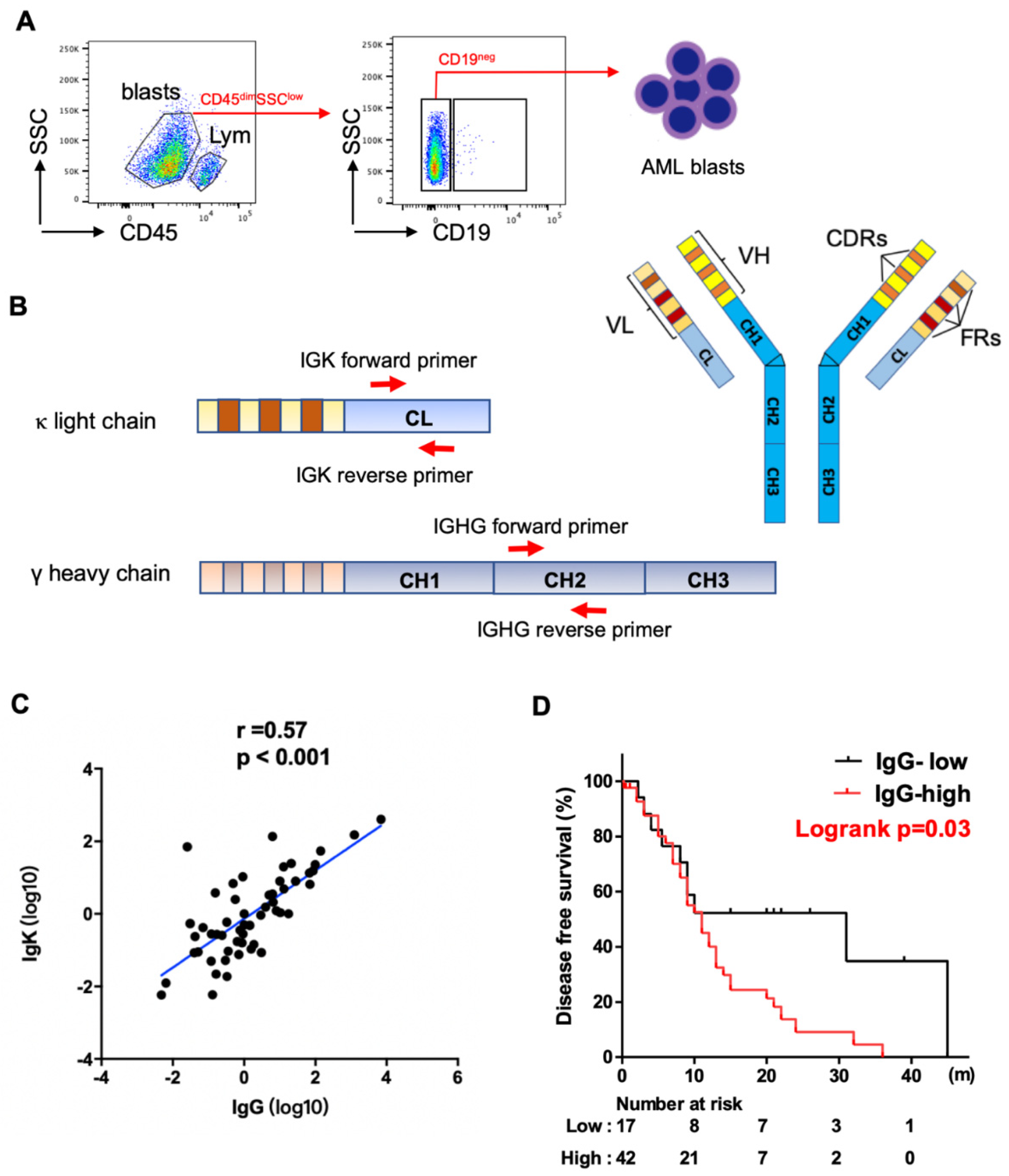
2.2. Fluorescence-Activated Cell Sorting (FACS)
2.3. Quantitative Real-Time Polymerase Chain Reaction (PCR)
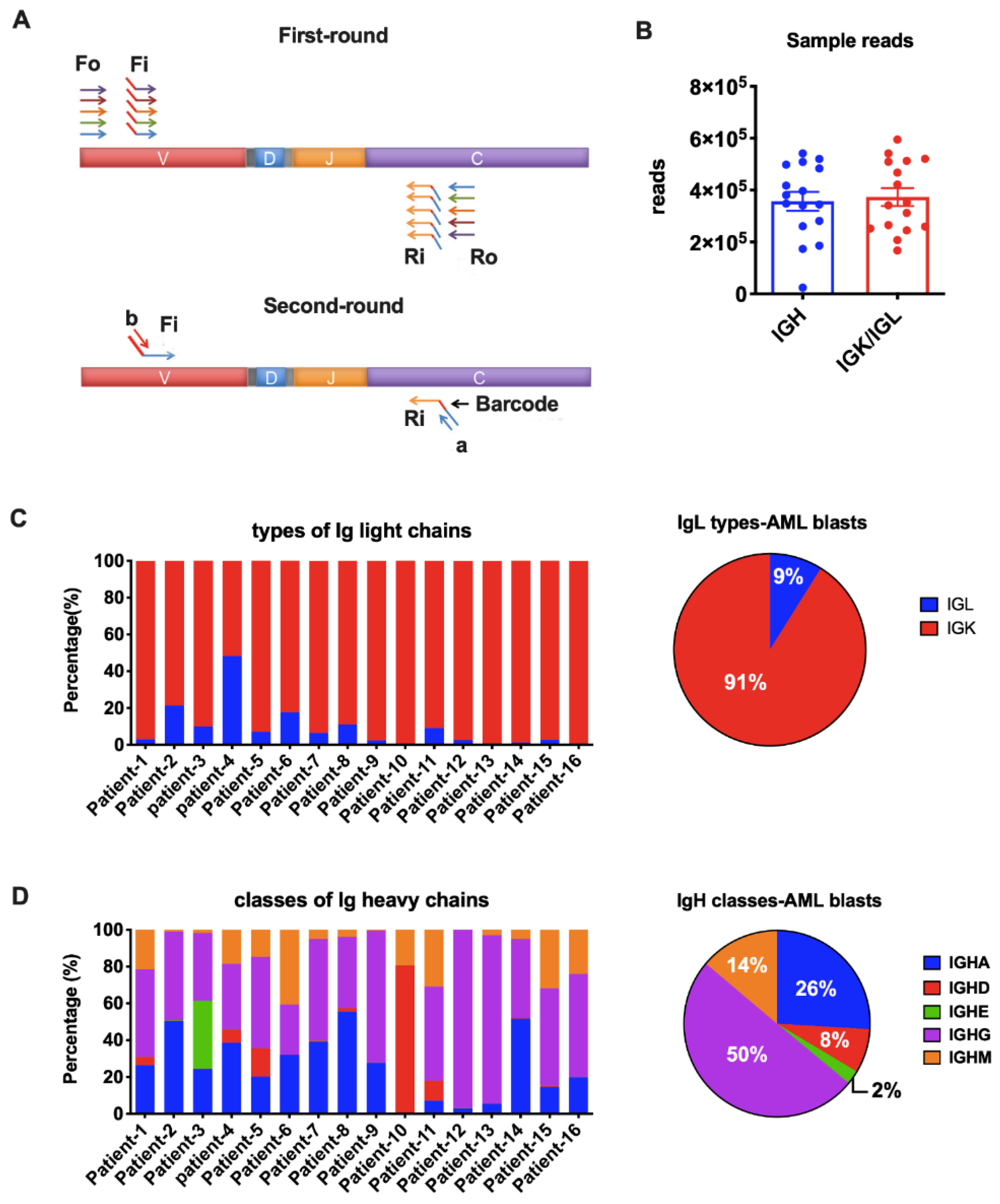
2.4. Next-Generation Sequencing (NGS)
2.5. Data Analysis
2.6. Statistical Analysis
3. Results
3.1. Correlation between Levels of Ig Expression and Clinicopathologic Features
3.2. Detecting AML-Derived Ig Repertoire by NGS
3.3. Biased Ig Rearrangements in AML Blasts
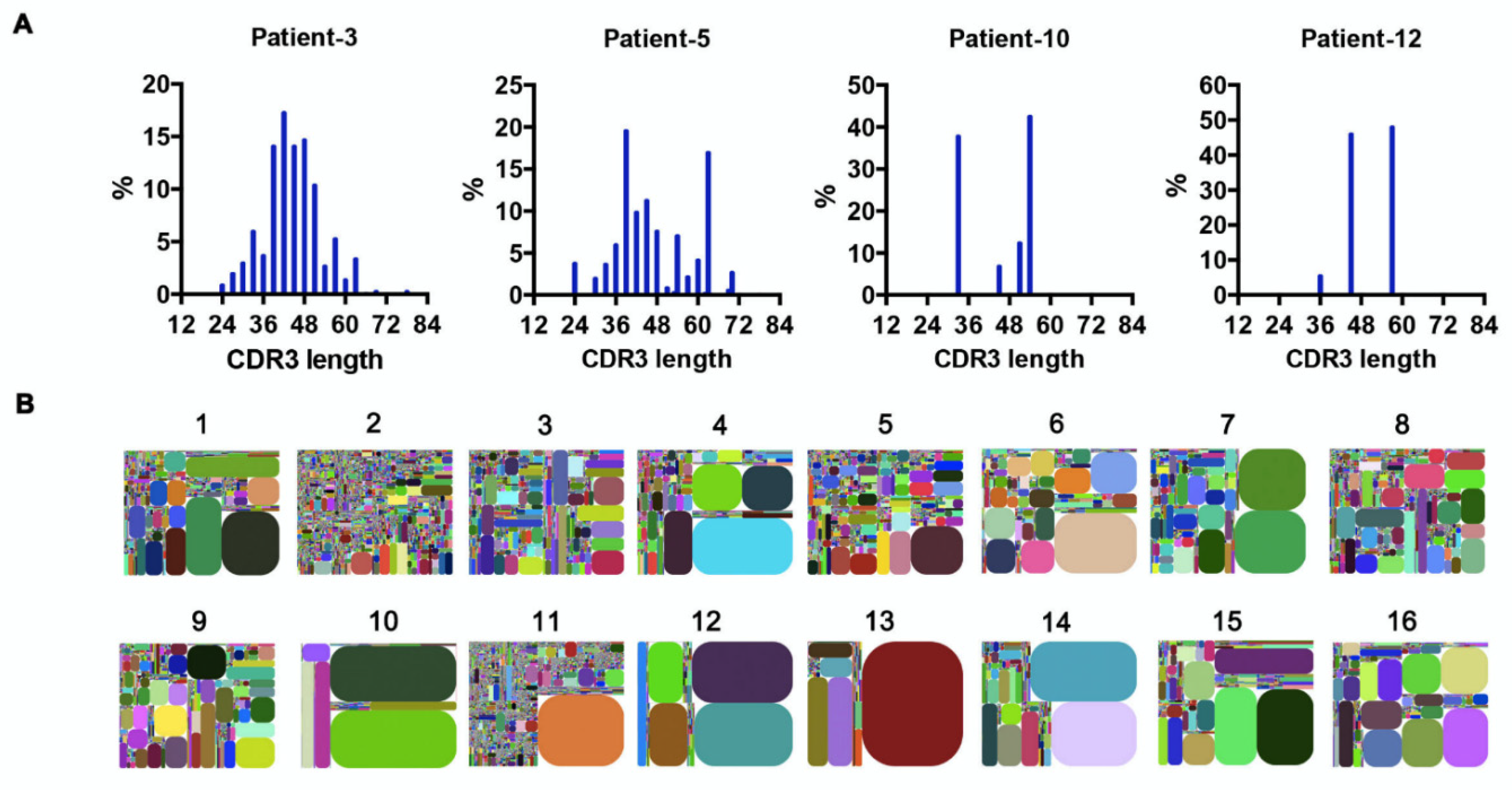
3.4. Recurring CDR3s Revealed Clonal Expansion in AML
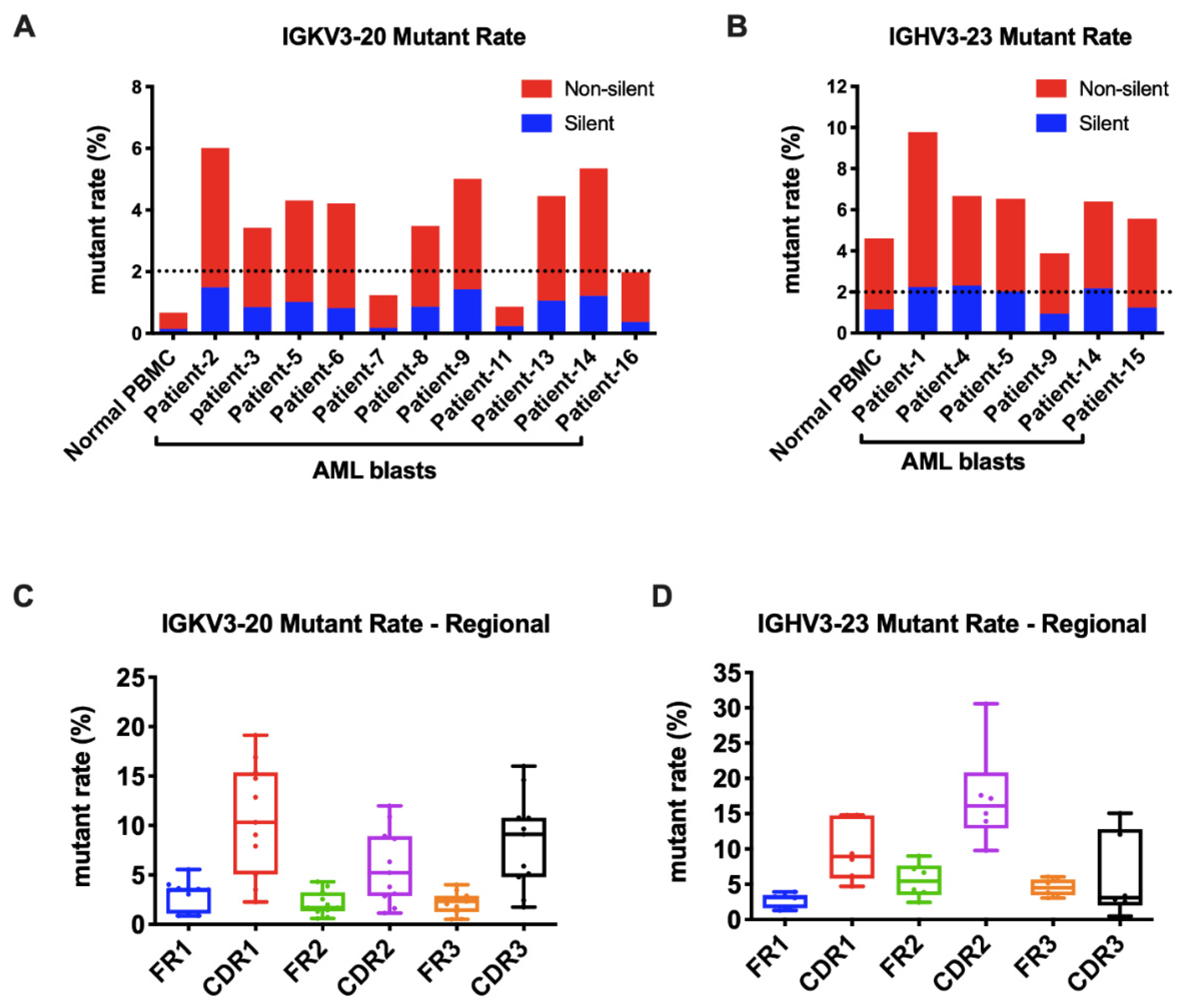
3.5. Frequent Somatic Hypermutations Occurred in AML-Derived Igs
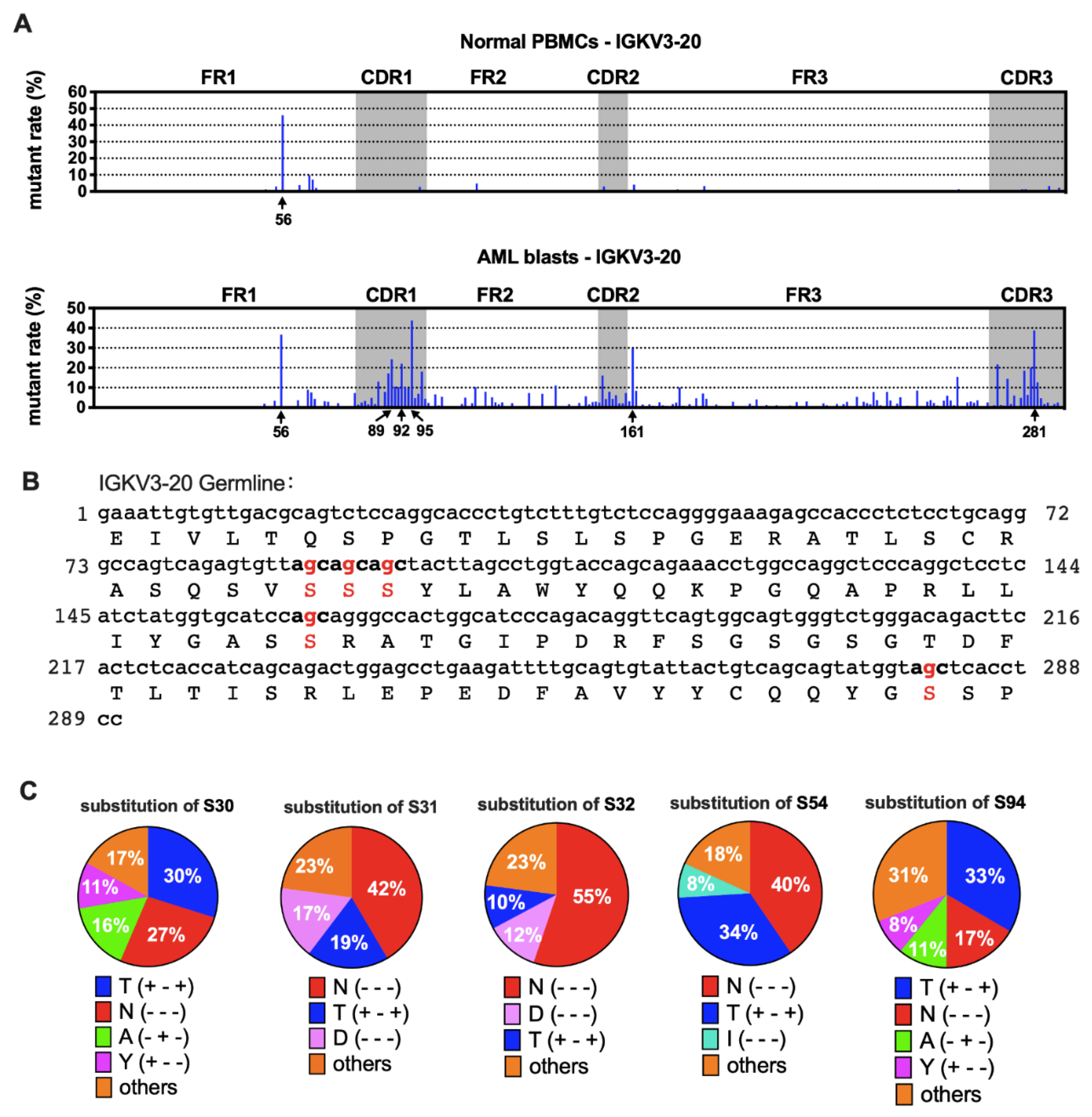
3.6. AML-Derived IGKV3-20 Displayed Specific Mutation Hotspots at the Serine Codons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [Green Version]
- Yohe, S. Molecular Genetic Markers in Acute Myeloid Leukemia. J. Clin. Med. 2015, 4, 460–478. [Google Scholar] [CrossRef] [Green Version]
- Kumar, C.C. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer 2011, 2, 95–107. [Google Scholar] [CrossRef]
- De Kouchkovsky, I.; Abdul-Hay, M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016, 6, e441. [Google Scholar] [CrossRef]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- Bagg, A.; Braziel, R.M.; Arber, D.A.; Bijwaard, K.E.; Chu, A.Y. Immunoglobulin heavy chain gene analysis in lymphomas: A multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J. Mol. Diagn 2002, 4, 81–89. [Google Scholar] [CrossRef]
- Cui, M.; Huang, J.; Zhang, S.; Liu, Q.; Liao, Q.; Qiu, X. Immunoglobulin Expression in Cancer Cells and Its Critical Roles in Tumorigenesis. Front. Immunol. 2021, 12, 613530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mao, Y.; Huang, J.; Ma, T.; Zhang, L.; Zhu, X.; Zheng, J.; Wu, L.; Yin, C.C.; Qiu, X. Immunoglobulin gene locus events in epithelial cells of lactating mouse mammary glands. Cell Mol. Life Sci. 2010, 67, 985–994. [Google Scholar] [CrossRef]
- Kang, B.Y.; Hu, C.; Prayaga, S.; Khaidakov, M.; Sawamura, T.; Seung, K.B.; Mehta, J.L. LOX-1 dependent overexpression of immunoglobulins in cardiomyocytes in response to angiotensin II. Biochem. Biophys Res. Commun. 2009, 379, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wang, X.; Liu, Y.; Tian, X.; Deng, S.; Sun, Y.; Wang, S.; Zheng, D.; Cui, Z.; Pan, Y.; et al. Single-cell RNA sequencing confirms IgG transcription and limited diversity of VHDJH rearrangements in proximal tubular epithelial cells. Sci. Rep. 2020, 10, 19657. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Ge, J.; Liao, Q.; Ma, J.; Liu, Y.; Huang, J.; Wang, C.; Xu, W.; Zheng, J.; Shao, W.; et al. IgG and IgA with potential microbial-binding activity are expressed by normal human skin epidermal cells. Int. J. Mol. Sci. 2015, 16, 2574–2590. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Zhang, L.; Zheng, J.; Zhao, L.; Huang, J.; Shao, W.; Liao, Q.; Ma, T.; Geng, L.; Yin, C.C.; et al. Spontaneous production of immunoglobulin M in human epithelial cancer cells. PLoS ONE 2012, 7, e51423. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhu, X.; Zhang, L.; Mao, Y.; Zhang, J.; Hao, P.; Li, G.; Lv, P.; Li, Z.; Sun, X.; et al. Human epithelial cancers secrete immunoglobulin g with unidentified specificity to promote growth and survival of tumor cells. Cancer Res. 2003, 63, 6488–6495. [Google Scholar]
- Liu, J.; Xia, M.; Wang, P.; Wang, C.; Geng, Z.; Cameron Yin, C.; Zhang, C.; Qiu, X. Immunoglobulin gene expression in umbilical cord blood-derived CD34(+) hematopoietic stem/progenitor cells. Gene 2016, 575, 108–117. [Google Scholar] [CrossRef]
- Qiu, X.; Sun, X.; He, Z.; Huang, J.; Hu, F.; Chen, L.; Lin, P.; You, M.J.; Medeiros, L.J.; Yin, C.C. Immunoglobulin gamma heavy chain gene with somatic hypermutation is frequently expressed in acute myeloid leukemia. Leukemia 2013, 27, 92–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Sun, X.; Gong, X.; He, Z.; Chen, L.; Qiu, X.; Yin, C.C. Rearrangement and expression of the immunoglobulin mu-chain gene in human myeloid cells. Cell Mol. Immunol. 2014, 11, 94–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Xia, M.; Sun, X.; He, Z.; Hu, F.; Chen, L.; Bueso-Ramos, C.E.; Qiu, X.; Yin, C.C. IGK with conserved IGKappaV/IGKappaJ repertoire is expressed in acute myeloid leukemia and promotes leukemic cell migration. Oncotarget 2015, 6, 39062–39072. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Xia, M.; Sun, X.; Han, X.; Zu, Y.; Jabbour, E.J.; You, M.J.; Lin, P.; Li, S.; Xu, J.; et al. High levels of immunoglobulin expression predict shorter overall survival in patients with acute myeloid leukemia. Eur. J. Haematol. 2020, 105, 449–459. [Google Scholar] [CrossRef]
- Gearhart, P.J. Generation of immunoglobulin variable gene diversity. Immunol. Today 1982, 3, 107–112. [Google Scholar] [CrossRef]
- Alamyar, E.; Duroux, P.; Lefranc, M.P.; Giudicelli, V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012, 882, 569–604. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.R.; Hayakawa, K. B cell development pathways. Annu. Rev. Immunol. 2001, 19, 595–621. [Google Scholar] [CrossRef]
- Santos, P.; Arumemi, F.; Park, K.S.; Borghesi, L.; Milcarek, C. Transcriptional and epigenetic regulation of B cell development. Immunol. Res. 2011, 50, 105–112. [Google Scholar] [CrossRef]
- Deneys, V.; Mazzon, A.M.; Marques, J.L.; Benoit, H.; De Bruyere, M. Reference values for peripheral blood B-lymphocyte subpopulations: A basis for multiparametric immunophenotyping of abnormal lymphocytes. J. Immunol. Methods 2001, 253, 23–36. [Google Scholar] [CrossRef]
- Marti, G.E.; Rawstron, A.C.; Ghia, P.; Hillmen, P.; Houlston, R.S.; Kay, N.; Schleinitz, T.A.; Caporaso, N.; International Familial, C.L.L.C. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br. J. Haematol. 2005, 130, 325–332. [Google Scholar] [CrossRef]
- Xu, J.L.; Davis, M.M. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000, 13, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Miqueu, P.; Guillet, M.; Degauque, N.; Dore, J.C.; Soulillou, J.P.; Brouard, S. Statistical analysis of CDR3 length distributions for the assessment of T and B cell repertoire biases. Mol. Immunol. 2007, 44, 1057–1064. [Google Scholar] [CrossRef]
- Darzentas, N.; Stamatopoulos, K. Stereotyped B cell receptors in B cell leukemias and lymphomas. Methods Mol. Biol. 2013, 971, 135–148. [Google Scholar] [CrossRef]
- Pommie, C.; Levadoux, S.; Sabatier, R.; Lefranc, G.; Lefranc, M.P. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004, 17, 17–32. [Google Scholar] [CrossRef]
- Ostronoff, F.; Othus, M.; Lazenby, M.; Estey, E.; Appelbaum, F.R.; Evans, A.; Godwin, J.; Gilkes, A.; Kopecky, K.J.; Burnett, A.; et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: A SWOG and UK National Cancer Research Institute/Medical Research Council report. J. Clin. Oncol. 2015, 33, 1157–1164. [Google Scholar] [CrossRef] [Green Version]
- Falini, B.; Bolli, N.; Liso, A.; Martelli, M.P.; Mannucci, R.; Pileri, S.; Nicoletti, I. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: Molecular basis and clinical implications. Leukemia 2009, 23, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, M.; Ren, W.; Zeng, L.; Liu, H.D.; Hu, D.; Deng, X.; Tang, M.; Shi, Y.; Gong, J.; et al. Expression and secretion of immunoglobulin alpha heavy chain with diverse VDJ recombinations by human epithelial cancer cells. Mol. Immunol. 2007, 44, 2221–2227. [Google Scholar] [CrossRef]
- Zheng, H.; Li, M.; Liu, H.; Ren, W.; Hu, D.S.; Shi, Y.; Tang, M.; Cao, Y. Immunoglobulin alpha heavy chain derived from human epithelial cancer cells promotes the access of S phase and growth of cancer cells. Cell Biol. Int. 2007, 31, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, C.; Sun, X.; Mao, Y.; Li, G.; Liu, X.; Zhang, Y.; Qiu, X. Immunoglobulin mRNA and protein expression in human oral epithelial tumor cells. Appl. Immunohistochem. Mol. Morphol. 2008, 16, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.J.; Brezinschek, H.P.; Brezinschek, R.I.; Lipsky, P.E. Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J. Clin. Invest. 1997, 99, 1614–1627. [Google Scholar] [CrossRef]
- Juul, L.; Hougs, L.; Andersen, V.; Svejgaard, A.; Barington, T. The normally expressed kappa immunoglobulin light chain gene repertoire and somatic mutations studied by single-sided specific polymerase chain reaction (PCR); frequent occurrence of features often assigned to autoimmunity. Clin. Exp. Immunol. 1997, 109, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Feeney, A.J.; Lugo, G.; Escuro, G. Human cord blood kappa repertoire. J. Immunol. 1997, 158, 3761–3768. [Google Scholar]
- Girschick, H.J.; Lipsky, P.E. The kappa gene repertoire of human neonatal B cells. Mol. Immunol. 2002, 38, 1113–1127. [Google Scholar] [CrossRef]
- Briney, B.; Inderbitzin, A.; Joyce, C.; Burton, D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 2019, 566, 393–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Huang, J.; Mao, Y.; Liu, S.; Sun, X.; Zhu, X.; Ma, T.; Zhang, L.; Ji, J.; Zhang, Y.; et al. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. J. Biol. Chem. 2009, 284, 13610–13619. [Google Scholar] [CrossRef] [Green Version]
- Chiorazzi, N.; Ferrarini, M. B cell chronic lymphocytic leukemia: Lessons learned from studies of the B cell antigen receptor. Annu. Rev. Immunol. 2003, 21, 841–894. [Google Scholar] [CrossRef] [Green Version]
- Dighiero, G. Unsolved issues in CLL biology and management. Leukemia 2003, 17, 2385–2391. [Google Scholar] [CrossRef] [PubMed]
- Ferrarini, M.; Chiorazzi, N. Recent advances in the molecular biology and immunobiology of chronic lymphocytic leukemia. Semin. Hematol. 2004, 41, 207–223. [Google Scholar] [CrossRef]
- Stevenson, F.K.; Caligaris-Cappio, F. Chronic lymphocytic leukemia: Revelations from the B-cell receptor. Blood 2004, 103, 4389–4395. [Google Scholar] [CrossRef]
- Fais, F.; Ghiotto, F.; Hashimoto, S.; Sellars, B.; Valetto, A.; Allen, S.L.; Schulman, P.; Vinciguerra, V.P.; Rai, K.; Rassenti, L.Z.; et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 1998, 102, 1515–1525. [Google Scholar] [CrossRef]
- Tobin, G.; Thunberg, U.; Johnson, A.; Thorn, I.; Soderberg, O.; Hultdin, M.; Botling, J.; Enblad, G.; Sallstrom, J.; Sundstrom, C.; et al. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood 2002, 99, 2262–2264. [Google Scholar] [CrossRef]
- Tobin, G.; Thunberg, U.; Johnson, A.; Eriksson, I.; Soderberg, O.; Karlsson, K.; Merup, M.; Juliusson, G.; Vilpo, J.; Enblad, G.; et al. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vlambda2-14 gene use and homologous CDR3s: Implicating recognition of a common antigen epitope. Blood 2003, 101, 4952–4957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghia, P.; Stamatopoulos, K.; Belessi, C.; Moreno, C.; Stella, S.; Guida, G.; Michel, A.; Crespo, M.; Laoutaris, N.; Montserrat, E.; et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: The lesson of the IGHV3-21 gene. Blood 2005, 105, 1678–1685. [Google Scholar] [CrossRef]
- Muramatsu, M.; Kinoshita, K.; Fagarasan, S.; Yamada, S.; Shinkai, Y.; Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000, 102, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Revy, P.; Muto, T.; Levy, Y.; Geissmann, F.; Plebani, A.; Sanal, O.; Catalan, N.; Forveille, M.; Dufourcq-Labelouse, R.; Gennery, A.; et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 2000, 102, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Reynaud, C.A.; Aoufouchi, S.; Faili, A.; Weill, J.C. What role for AID: Mutator, or assembler of the immunoglobulin mutasome? Nat. Immunol. 2003, 4, 631–638. [Google Scholar] [CrossRef]
- Kiyoshi, M.; Tsumoto, K.; Ishii-Watabe, A.; Caaveiro, J.M.M. Glycosylation of IgG-Fc: A molecular perspective. Int. Immunol. 2017, 29, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Zhu, L.P.; Wei, Q. IgG-Fc N-glycosylation at Asn297 and IgA O-glycosylation in the hinge region in health and disease. Glycoconj. J. 2013, 30, 735–745. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, J.; Liu, Y.; Liao, Q.; Huang, J.; Geng, Z.; Xu, W.; Sheng, Z.; Lee, G.; Zhang, Y.; et al. Lung squamous cell carcinoma cells express non-canonically glycosylated IgG that activates integrin-FAK signaling. Cancer Lett. 2018, 430, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Niwa, R.; Natsume, A.; Uehara, A.; Wakitani, M.; Iida, S.; Uchida, K.; Satoh, M.; Shitara, K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J. Immunol. Methods 2005, 306, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Geng, Z.; Shao, W.; Liu, E.; Zhang, J.; Tang, J.; Wang, P.; Sun, X.; Xiao, L.; Xu, W.; et al. Cancer-derived sialylated IgG promotes tumor immune escape by binding to Siglecs on effector T cells. Cell Mol. Immunol. 2020, 17, 1148–1162. [Google Scholar] [CrossRef]

| PID * | Vκ | Jκ | Utilization (%) | PID * | Vκ | Jκ | Utilization (%) |
|---|---|---|---|---|---|---|---|
| IGKV2-30*02 | IGKJ5*01 | 39.4 | IGKV2D-28*01 | IGKJ1*01 | 27.3 | ||
| 1 | IGKV4-1*01 | IGKJ1*01 | 7.8 | 9 | IGKV2D-28*01 | IGKJ3*01 | 10.5 |
| IGKV3-15*01 | IGKJ2*01 | 6.8 | IGKV3-20*01 | IGKJ2*01 | 7.7 | ||
| IGKV3-20*01 | IGKJ1*01 | 14.4 | IGKV3-11*01 | IGKJ4*01 | 56.4 | ||
| 2 | IGKV3-20*01 | IGKJ3*01 | 6.5 | 10 | IGKV2D-28*01 | IGKJ2*01 | 18.2 |
| IGKV2D-28*01 | IGKJ5*01 | 6.0 | IGKV1-5*03 | IGKJ2*01 | 9.1 | ||
| IGKV2-24*01 | IGKJ1*01 | 20.4 | IGKV2D-28*01 | IGKJ1*01 | 11.6 | ||
| 3 | IGKV3-20*01 | IGKJ4*01 | 10.3 | 11 | IGKV3-20*01 | IGKJ1*01 | 11.4 |
| IGKV4-1*01 | IGKJ2*01 | 9.8 | IGKV4-1*01 | IGKJ1*01 | 8.7 | ||
| IGKV3-11*01 | IGKJ4*01 | 55.5 | IGKV1-16*02 | IGKJ1*01 | 55.7 | ||
| 4 | IGKV1-5*03 | IGKJ1*01 | 3.9 | 12 | IGKV1D-39*01 | IGKJ2*01 | 31.9 |
| IGKV4-1*01 | IGKJ1*01 | 3.6 | IGKV1D-39*01 | IGKJ1*01 | 5.8 | ||
| IGKV2-24*01 | IGKJ1*01 | 28.1 | IGKV3-20*01 | IGKJ4*01 | 14.5 | ||
| 5 | IGKV2D-28*01 | IGKJ1*01 | 6.1 | 13 | IGKV2D-28*01 | IGKJ1*01 | 13.5 |
| IGKV3-20*01 | IGKJ1*01 | 4.3 | IGKV3-11*01 | IGKJ5*01 | 8.5 | ||
| IGKV2-30*02 | IGKJ4*01 | 19.7 | IGKV2D-28*01 | IGKJ1*01 | 60.9 | ||
| 6 | IGKV3-20*01 | IGKJ1*01 | 10.0 | 14 | IGKV3-20*01 | IGKJ5*01 | 16.0 |
| IGKV3-11*01 | IGKJ4*01 | 9.8 | IGKV4-1*01 | IGKJ1*01 | 13.0 | ||
| IGKV2D-28*01 | IGKJ2*01 | 24.9 | IGKV2D-28*01 | IGKJ2*01 | 33.2 | ||
| 7 | IGKV3-20*01 | IGKJ2*01 | 20.4 | 15 | IGKV4-1*01 | IGKJ1*01 | 28.8 |
| IGKV2D-28*01 | IGKJ3*01 | 8.2 | IGKV2-30*01 | IGKJ1*01 | 14.9 | ||
| IGKV3-20*01 | IGKJ4*01 | 20.0 | IGKV3-11*01 | IGKJ1*01 | 18.1 | ||
| 8 | IGKV3-20*01 | IGKJ1*01 | 17.6 | 16 | IGKV3-20*01 | IGKJ1*01 | 16.6 |
| IGKV3-20*01 | IGKJ2*01 | 14.7 | IGKV2-30*02 | IGKJ1*01 | 12.0 |
| Patient ID | VH | D | JH | Utilization (%) |
|---|---|---|---|---|
| IGHV3-23*04 | IGHD3-3*02 | IGHJ6*03 | 19.3 | |
| 1 | IGHV4-59*01 | IGHD2-15*01 | IGHJ3*02 | 14.7 |
| IGHV1-2*02 | IGHD7-27*01 | IGHJ3*02 | 10.0 | |
| IGHV1-18*01 | IGHD3-3*02 | IGHJ6*02 | 4.7 | |
| 2 | IGHV3-74*03 | IGHD1-7*01 | IGHJ4*02 | 2.4 |
| IGHV1-18*01 | IGHD3-10*01 | IGHJ6*03 | 1.7 | |
| IGHV4-61*08 | IGHD6-13*01 | IGHJ4*02 | 4.4 | |
| 3 | IGHV4-4*07 | IGHD3-10*02 | IGHJ3*02 | 4.4 |
| IGHV3-9*01 | IGHD5-5*01 | IGHJ4*02 | 4.3 | |
| IGHV2-5*10 | IGHD3-9*01 | IGHJ5*02 | 32.0 | |
| 4 | IGHV7-4-1*02 | IGHD6-19*01 | IGHJ4*02 | 12.1 |
| IGHV3-23*04 | IGHD3-16*02 | IGHJ4*02 | 11.9 | |
| IGHV3-23*04 | IGHD1-7*01 | IGHJ4*02 | 13.2 | |
| 5 | IGHV1-2*02 | IGHD1-7*01 | IGHJ4*02 | 2.9 |
| IGHV1-46*01 | IGHD6-13*01 | IGHJ4*02 | 2.7 | |
| IGHV3-48*03 | IGHD3-16*02 | IGHJ5*02 | 25.2 | |
| 6 | IGHV1-2*02 | IGHD3-10*01 | IGHJ4*02 | 10.2 |
| IGHV3-73*02 | - | IGHJ6*02 | 5.8 | |
| IGHV4-59*01 | IGHD3-16*01 | IGHJ5*02 | 21.0 | |
| 7 | IGHV4-b*01 | IGHD5-5*01 | IGHJ4*01 | 19.0 |
| IGHV3-30*02 | IGHD6-13*01 | IGHJ6*03 | 5.7 | |
| IGHV1-46*03 | IGHD2-2*03 | IGHJ4*02 | 7.5 | |
| 8 | IGHV4-59*08 | IGHD3-3*02 | IGHJ3*02 | 4.8 |
| IGHV2-5*10 | IGHD3-22*01 | IGHJ4*02 | 4.3 | |
| IGHV3-23*04 | IGHD6-19*01 | IGHJ4*02 | 8.4 | |
| 9 | IGHV4-31*03 | IGHD3-16*02 | IGHJ4*02 | 6.1 |
| IGHV3-74*02 | IGHD3-22*01 | IGHJ4*02 | 5.4 | |
| IGHV3-48*02 | IGHD1-26*01 | IGHJ3*02 | 38.6 | |
| 10 | IGHV2-5*10 | IGHD5-5*01 | IGHJ5*02 | 37.4 |
| IGHV3-30*18 | IGHD4-17*01 | IGHJ3*02 | 8.4 | |
| IGHV7-4-1*02 | IGHD3-22*01 | IGHJ4*02 | 31.9 | |
| 11 | IGHV3-33*01 | IGHD1-20*01 | IGHJ4*02 | 1.3 |
| IGHV3-33*01 | IGHD3-22*01 | IGHJ3*02 | 1.2 | |
| IGHV3-48*03 | IGHD6-13*01 | IGHJ6*02 | 34.6 | |
| 12 | IGHV3-11*01 | IGHD6-6*01 | IGHJ5*02 | 33.8 |
| IGHV3-11*01 | IGHD6-13*01 | IGHJ6*02 | 13.4 | |
| IGHV1-18*01 | IGHD6-19*01 | IGHJ6*02 | 65.8 | |
| 13 | IGHV1-24*01 | IGHD3-9*01 | IGHJ6*02 | 11.1 |
| IGHV1-24*01 | IGHD3-10*01 | IGHJ6*02 | 9.3 | |
| IGHV3-23*04 | IGHD2-2*03 | IGHJ6*03 | 30.9 | |
| 14 | IGHV1-69*13 | IGHD6-13*01 | IGHJ4*02 | 18.5 |
| IGHV1-69*12 | IGHD6-13*01 | IGHJ4*02 | 15.5 | |
| IGHV3-23*04 | IGHD3-9*01 | IGHJ4*02 | 22.2 | |
| 15 | IGHV1-69*09 | IGHD3-9*01 | IGHJ3*01 | 17.2 |
| IGHV3-7*02 | IGHD6-6*01 | IGHJ4*02 | 12.6 | |
| IGHV3-9*01 | IGHD6-19*01 | IGHJ6*02 | 11.8 | |
| 16 | IGHV1-3*01 | IGHD3-22*01 | IGHJ4*02 | 11.1 |
| IGHV1-69*13 | IGHD3-3*01 | IGHJ3*01 | 8.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Wu, L.; Sun, X.; Han, X.; Yan, H.; Huang, J.; Zhang, Y.; Hu, Z.; Zu, Y.; Yin, C.C.; et al. Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia. Biology 2022, 11, 161. https://doi.org/10.3390/biology11020161
Xia M, Wu L, Sun X, Han X, Yan H, Huang J, Zhang Y, Hu Z, Zu Y, Yin CC, et al. Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia. Biology. 2022; 11(2):161. https://doi.org/10.3390/biology11020161
Chicago/Turabian StyleXia, Miaoran, Lina Wu, Xiaoping Sun, Xin Han, Huige Yan, Jing Huang, Youhui Zhang, Zhihong Hu, Youli Zu, C. Cameron Yin, and et al. 2022. "Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia" Biology 11, no. 2: 161. https://doi.org/10.3390/biology11020161
APA StyleXia, M., Wu, L., Sun, X., Han, X., Yan, H., Huang, J., Zhang, Y., Hu, Z., Zu, Y., Yin, C. C., & Qiu, X. (2022). Next-Generation Sequencing Revealed a Distinct Immunoglobulin Repertoire with Specific Mutation Hotspots in Acute Myeloid Leukemia. Biology, 11(2), 161. https://doi.org/10.3390/biology11020161









