Simple Summary
Gut-associated protozoa are a heterogenous group of microbes that frequently reside within humans. The role of commensal intestinal protozoa and their interaction with the human organism is a highly complex topic, and an area of research that has been largely neglected. We argue that some protozoa species might be beneficial inhabitants of the gut that can significantly impact human health and disease. Here, we aimed to comprehensively review existing literature on the conflicting outcomes of protozoa colonization, their interaction with the bacterial microbiota, and the crosstalk between the protozoa and the host immune system. Moreover, we emphasize the importance for future studies to investigate these aspects of protozoa colonization that will undoubtedly increase our understanding of complex interactions between intestinal protozoa, other microbiota organisms, and the human host.
Abstract
The human gastrointestinal microbiota contains a diverse consortium of microbes, including bacteria, protozoa, viruses, and fungi. Through millennia of co-evolution, the host–microbiota interactions have shaped the immune system to both tolerate and maintain the symbiotic relationship with commensal microbiota, while exerting protective responses against invading pathogens. Microbiome research is dominated by studies describing the impact of prokaryotic bacteria on gut immunity with a limited understanding of their relationship with other integral microbiota constituents. However, converging evidence shows that eukaryotic organisms, such as commensal protozoa, can play an important role in modulating intestinal immune responses as well as influencing the overall health of the host. The presence of several protozoa species has recently been shown to be a common occurrence in healthy populations worldwide, suggesting that many of these are commensals rather than invading pathogens. This review aims to discuss the most recent, conflicting findings regarding the role of intestinal protozoa in gut homeostasis, interactions between intestinal protozoa and the bacterial microbiota, as well as potential immunological consequences of protozoa colonization.
1. Introduction
The mammalian gut harbors a vast number of viruses, bacteria, fungi, and protozoa (single-celled eukaryotes), collectively referred to as the microbiota [1]. Dynamic interplay between these distinct microbiota constituents and the host contributes to many essential physiological processes, including development, metabolism, and immunity [2,3]. It is becoming increasingly evident that disruption of this complex ecosystem, referred to as gut dysbiosis, contributes to the development of various gastrointestinal as well as systemic diseases, such as inflammatory bowel disease (IBD), metabolic disorder, autoimmunity, and cancer [4,5,6]. Furthermore, emerging evidence of a bidirectional crosstalk between the intestinal microbiota and the brain has linked dysbiosis to various diseases of the central nervous system, such as depression, multiple sclerosis, and Parkinson’s disease [7,8]. It is well recognized that the intestinal microbiota plays a pivotal role in the manifestations of IBD [9]. IBD is a chronic inflammatory disease of the intestinal lining that can be classified into two distinct conditions, Crohn’s disease or ulcerative colitis [10]. Although usually not fatal, IBD is associated with lowered life expectancy and significantly decreased quality of life for patients that suffer from chronic symptoms, including persistent diarrhea, abdominal pain, and rectal bleeding. Moreover, chronic inflammation in IBD has been associated with serious, often fatal comorbidities, including cancer, cardiovascular diseases, and liver diseases [11,12]. The rate of incidence of IBD has been rising dramatically since the industrial revolution, with a current global burden of more than 6 million people [13]. Although the IBD etiology remains largely unknown, it has been hypothesized that the industrial lifestyle, which includes increased use of antibiotics and a diet rich in highly processed foods, has resulted in detrimental changes to the intestinal microbiota that significantly contribute to disease risk [9,14]. Therefore, a detailed understanding of the biological roles of distinct intestinal microbial groups, their mutual interactions, as well as their impact on human disease is of paramount importance.
Since most studies have concentrated on gut-resident bacteria as the main component of the microbiota, the mechanisms and consequences of intestinal protozoa colonization have only recently begun to be clarified [3,15,16,17]. Protozoa are a diverse group of single-celled eukaryotic organisms that can be found in a variety of environments either as free-living or parasitic/symbiotic microbes. Historically, protozoa have been classified into four subgroups: amoebas, flagellates, coccidians, and ciliates, and their categorization depends on specific morphological features, such as internal structure and motility [18]. After the emergence of molecular phylogenetics, an updated classification has been proposed, integrating insights from the genomic studies with the structural and biochemical evidence. Thus, protozoa have been proposed to comprise two subkingdoms, with Choanozoa and Amoebozoa grouped together as the subkingdom Sarcomastigota, while Alveolata, Rhizaria, Excavata, and Apusozoa constitute the subkingdom Biciliata [19]. More recently, an even more updated classification has been proposed [20]. From an evolutionary point of view, eukaryotic microbes such as protozoa have co-evolved with humans and undoubtedly affected the dynamics of the gut microbiota [21]. Despite extensive strides in parasitic research, including studies of pathogenic protozoa, the role of commensal protozoa in shaping the immune landscape of the gut remains enigmatic and questioned [22,23,24]. One of the main challenges lies in the characteristic features and biological classification of commensal protozoa [25]. By definition, commensal microbes reside within the host without causing a negative health impact and are well tolerated by the immune system [26]. However, due to the highly dynamic nature of the host–microbiota interactions, a particular protozoa can be classified as commensal rather than parasitic, and vice-versa, often in a context-specific manner [21,27,28]. Furthermore, heterogeneity in experimental design, differences between protozoa species, and geographical changes in gut microbiota all result in a lack of consensus regarding the exact role of intestinal protozoa and their contribution to mucosal immune homeostasis [15,29]. It is well established that the bacterial compartment of human gut microbiota comprises a plethora of different species, ranging from beneficial to opportunistic and/or pathogenic [30]. We hypothesize that a similar paradigm also exists for intestinal protozoa that frequently inhabit the human gastrointestinal tract. From this perspective, we aim here to comprehensively review existing evidence of commensal and potentially beneficial aspects of protozoa colonization in human health and disease.
2. Intestinal Protozoa—Pathogenic, Commensal, or Beneficial?
Giardia lamblia, Entamoeba histolytica, and Cryptosporidium spp. are among the most common enteric protozoa parasites that significantly contribute to acute gastroenteritis and diarrheal disease globally [31,32]. Their virulence factors and intestinal invasion mechanisms are well characterized and have been frequently reviewed [33,34,35,36]. Besides gastrointestinal manifestations, infections with these protozoa can lead to serious health-threatening conditions, such as amoebic liver abscess and amoebic colitis [36]. However, while parasitic infections still pose a great health burden, numerous intestinal protozoa species are considered non-pathogenic (Table 1) [29,37]. Lokmer et al. applied a metagenomic approach to examine the occurrence of commensal and potentially beneficial protozoa and to study their ecological dynamics in several worldwide populations. They showed a high prevalence of Blastocystis spp., Entamoeba spp., and various other protozoa genera in healthy individuals across human populations [37]. Other epidemiological studies have shown that species such as Dientamoeba and Enteromonas are common residents of the human gut, at least in some parts of the world [37,38].

Table 1.
Human intestinal protozoa species and their characteristics.
2.1. Blastocystis spp.
Blastocystis spp. is one of the most prevalent protozoa found in humans, with an estimated 1 billion colonized individuals worldwide [22,40]. Historically, Blastocystis was predominantly characterized as a parasitic protozoa [53], but conflicting results regarding its pathogenic potential and clinical significance have emerged in several studies [40]. Blastocystis has been associated with the etiology of irritable bowel syndrome (IBS) [54,55,56,57] and IBD [58]. In contrast, other cohort studies have found no correlation between gastrointestinal symptoms and the presence of Blastocystis, either in healthy subjects or IBS patients [59,60]. Similarly, the prevalence of Blastocystis infection has been inconsistently reported to be higher in either immunocompetent or immunocompromised individuals depending on the study [61,62]. One possible explanation for this discrepancy is that Blastocystis has mainly been investigated as a causative agent in disease propagation, with limited information about its distribution in a healthy population. With recent advances in sequencing technologies and an increased number of epidemiological surveys, it has become evident that the presence of Blastocystis is a common occurrence in both healthy and symptomatic individuals, which inevitably questions its alleged pathogenicity [63,64]. To date, at least 17 Blastocystis subtypes (ST) have been identified, of which nine are found in humans (ST1-ST9), with ST1–ST4 accounting for up to 90% of all occurrences [40,65]. Recent findings suggest an association between Blastocystis subtype variation and pathogenic potential. For instance, ST2 was suggested to be a non-pathogenic type [66], while ST3 has frequently been identified as a major subtype among patients with gastrointestinal disorders [67,68]. A study by Ali et al. showed a significant association between Blastocystis subtypes and colorectal cancer (CRC) [69]. Although a similar prevalence of Blastocystis ST1, 2, and 3 were observed between CRC and non-cancer individuals, a rare ST7 was identified in CRC patients [69].
At the other end of the spectrum, beneficial roles for Blastocystis have also been proposed. Colonization with Blastocystis is associated with higher microbial diversity and richness, both of which are suggested to benefit intestinal health [22,70]. Additionally, it has been shown that body mass index is strongly negatively correlated with Blastocystis presence [71]. Several studies have reported that colonization with Blastocystis is more common in healthy subjects than in patients with active IBD, IBS, or CRC, supporting that Blastocystis might be considered a component of the healthy intestinal microbiota [58,60,71,72,73]. Tito et al. recently surveyed Blastocystis subtype prevalence and relative abundance in a Western population cohort. They found that 30% of the healthy population was Blastocystis carriers compared to only 9% in an IBD patient cohort, with a significant association between Blastocystis subtype and microbiota composition [74]. Moreover, animal studies demonstrate that colonization with Blastocystis alters the gut microenvironment in a protective manner, which may, in turn, promote faster recovery from intestinal inflammation [75,76]. The highly contradicting reports between studies describing the impact of Blastocystis colonization can be partially attributed to some unexplored variables such as the diversity of intra-subtypes within a subtype, background microbiota composition, host genetics, and diet [77].
Overall, it is becoming evident that the impact of Blastocystis colonization on human gastrointestinal health is much more complex than originally envisioned, with the outcome depending on the context and the specific subtype of the protozoa. Thus, it is essential for future studies to elucidate and characterize the contrasting consequences of colonization of different Blastocystis subtypes.
2.2. Dientamoeba fragilis
Like Blastocystis spp., colonization with Dientamoeba fragilis has been reported to exert conflicting roles in gut homeostasis. In contrast to other intestinal protozoa whose colonization prevalence is generally considered higher in the emerging nations, D. fragilis has been identified more frequently in the developed world [78,79]. However, due to differences in surveillance systems and diagnostic procedures, its prevalence might be underestimated in some regions [78]. The presence of D. fragilis has frequently been associated with disease [45], just as it has been commonly found in asymptomatic carriers [72,73,74]. Population-based case-control studies in European countries, including the Netherlands [80], Denmark [60], and Belgium [81], have uniformly reported higher D. fragilis colonization in healthy subjects compared to patients with digestive symptoms. Similar to the reported effects of Blastocystis colonization, a recent study by Rasmussen et al. showed that colonization with D. fragilis is associated with greater gut microbiota diversity compared to non-carriers [82]. Thus far, two subtypes of D. fragilis have been identified, with type 1 accounting for the majority of confirmed cases. There are minor phenotypic differences between the two subtypes and their distinction is solely based on genetic diversity. It should be noted that these subtypes have only recently been characterized and many studies have not differentiated between them. It remains unknown whether there is a difference in virulence and pathogenic potential among the two subtypes, which might explain the controversial reports regarding the consequences of D. fragilis colonization [45].
2.3. Entamoeba spp.
Other common intestinal inhabitants with a worldwide distribution are Entamoeba species, the majority of which are generally accepted as commensal organisms [83]. Currently, eight species have been identified that are able to infect humans: E. histolytica, E. bangladeshi, E. dispar, E. hartmanni, E. moshkovskii, E. coli, and E. polecki, with E. histolytica as the only one with well-established pathogenicity [84]. The worldwide frequency of Entamoeba occurrence in humans is estimated at 3.5%. However, the prevalence of commensal Entamoeba spp. has largely been underestimated due to high morphological and genetic similarity with the invasive E. histolytica [83,84]. Microscopy, the most widely used method for the detection of Entamoeba organisms, is not always sufficient for differentiating between the invasive E. histolytica and non-pathogenic strains of Entamoeba [85]. Increased use of molecular diagnostic methods has recently revealed that colonization with commensal Entamoeba spp. is overall more common than infections with E. histolytica [85]. A recent cross-sectional study from North India found a higher prevalence of Entamoeba spp. in asymptomatic subjects compared to cases with intestinal symptoms, with E. dispar identified as the predominant species [51]. In agreement with observations made for other commensal protozoa species, individuals colonized with non-pathogenic Entamoeba spp. display higher gut microbiota diversity [86]. Additionally, Entamoeba-colonized individuals show a shift in the microbiota composition towards a more eubiotic composition, characterized by a decrease of genera associated with autoimmune and inflammatory disease and a corresponding increase of species linked to beneficial effects on intestinal health [87].
Overall, colonization with the above-mentioned protozoa species seems to be of commensal or beneficial nature, especially considering that their colonization is linked to an increase of microbiota diversity, as opposed to infections with pathogenic protozoa species, which often lead to decreased microbiota diversity [88]. However, the contradictory findings between studies support that intestinal protozoa, similar to intestinal bacteria, are a heterogeneous group of organisms that comprises both commensal as well as pathogenic or opportunistic species, with significant variability existing within one species as well, as is the case for Blastocystis. Therefore, their importance in human health and disease appears to be largely dependent on the characteristics of a particular species. Furthermore, assessment of the clinical significance of commensal protozoa species in humans relies on appropriate detection methods and accurate species differentiation [37]. DNA-based methods have generally been considered more sensitive for protozoa detection than microscopy, which, until recently, has been the gold standard [89]. On the other hand, DNA-based methods, such as quantitative PCR, can accurately identify the presence of different protozoa species, but fall short in investigating inter-taxa relationships between different microbiota members as well as in determining the relative abundance of a particular species [90]. With recent advances in next generation sequencing strategies, such as e.g., metagenomic approaches, there is an opportunity for greater detection accuracy, detailed taxonomical information, as well as characterization of complex interactions between various microbial populations [37].
3. Protozoa–Microbiota Interactions
The various communities of intestinal bacteria play a fundamental role in determining human health. It is generally suggested that high intestinal microbial diversity is a hallmark of a healthy and resilient gut microbiota [91]. Emerging studies consistently report increased bacterial diversity as well as community compositional changes evident in protozoa-colonized individuals (Table 2) [21,22,70,87]. Among the characteristic features of Blastocystis colonization is a higher abundance of specific taxa within Firmicutes, especially those from the Clostridia class, such as Ruminococcaceae and Prevotellaceae families, and a general decrease of Bacteroides abundance [71,74,92]. Furthermore, Blastocystis carriers show a significant decrease of Enterobacteriaceae and Proteobacteria when compared to Blastocystis-free subjects [70,71]. Interestingly, Proteobacteria and several species within the Enterobacteriaceae family can be considered “pathogenic” and linked to microbial dysbiosis associated with the development and pathogenesis of IBD [93,94,95]. Moreover, the presence of Blastocystis is strongly associated with the abundance of archaeal organisms, primarily Methanobrevibacter smithii [70,71,74]. M. smithii has been shown to play an important role in human health, supporting the digestion of glycans through the removal of bacterial fermentation end products [96]. M. smithii, together with members of the Faecalibacterium and Roseburia genera that are also enriched in Blastocystis-colonized individuals [22,70], increase the production of the short-chain fatty acid butyrate [97]. Butyrate has well-established beneficial effects on gut health, serving as an important energy source for colonic epithelial cells and acting as an inhibitor of gut inflammation [97,98]. Butyrate-producing bacteria, specifically Faecalibacterium prausnitzii and Roseburia spp., appear to be significantly reduced in patients with Crohn’s disease and have emerged as potential therapeutics for IBD [97,99,100]. Furthermore, a higher ratio of Faecalibacterium prausnitzii to Escherichia coli, as observed in Blastocystis- and Entamoeba- colonized individuals, has been associated with a healthy and balanced microbial ecosystem [87]. This strongly suggests that colonization with Blastocystis selectively reduces the abundance of pathogenic bacteria and induces compositional changes to the microbiota that might be beneficial for the maintenance of intestinal homeostasis.
On the other hand, adverse associations between Blastocystis colonization and eubiotic microbial profile have also been described. Several studies have reported a decrease of Bifidobacterium in individuals colonized with Blastocystis [70,101]. Bifidobacterium spp. have been associated with homeostatic functions within the gut, including protection of the epithelial barrier and regulation of inflammation [102]. Accordingly, a study by Alzate et al. showed that children colonized with Blastocystis exhibited markedly reduced abundance of the highly beneficial Akkermansia spp. compared to children that were Blastocystis-free [48]. These results are in line with those recently reported by Caudet et al., who observed decreased abundance of Akkermansia spp. and Bifidobacterium spp. in obese Blastocystis-colonized adults [101]. Akkermansia spp. has emerged as one of the important health-promoting microbes, with functions spanning from maintenance of the gut lining to protection against inflammation [103,104]. In contrast to earlier findings, where Blastocystis colonization has been associated with a decline of Lactobacillus spp. [22], Caudet et al. reported a marked increase of Lactobacillus spp. in Blastocystis-colonized individuals [101]. A possible explanation for these contradictory findings might be differences in the Blastocystis subtype. Of note, the importance of Blastocystis subtype characterization when investigating the relationship between Blastocystis colonization and gut microbiota composition has been recently emphasized in a study demonstrating an inverse association between Blastocystis subtypes and Akkermansia spp. relative abundance [74]. Blastocystis ST3 showed a negative correlation, whereas Blastocystis ST4 was strongly positively correlated to Akkermansia and M. smithii abundance [74]. Accordingly, Blastocystis ST4 was shown in a separate study to ameliorate colonic inflammation in murine colonization models via compositional changes to the gut microbiota that included expansion of Akkermansia, as well as modulation of host-immune responses [76]. Furthermore, the presence of Blastocystis ST7 in diarrhea patients has been recently shown to be associated with decreased microbiota diversity and bacterial composition changes, characterized by significant enrichment of bacteria belonging to the Enterobacteriaceae family as well as Escherichia coli, compared to non-infected patients [105]. These results indicate that the impact of Blastocystis colonization on gut microbiota composition and structure is subtype-specific, highlighting the importance of Blastocystis subtype characterization in future studies.
Research on microbiota composition associated with D. fragilis colonization is limited. However, a study conducted in Denmark investigating the microbial profile in D. fragilis-positive children revealed 16 bacterial genera that were significantly more abundant in colonized children [82]. Some of the most enriched bacterial genera in D. fragilis carriers were Victivallis, Oscillibacter and Coprococcus, whereas Flavonifractor was enriched in non-colonized children. After the removal of D. fragilis by metronidazole treatment, the abundance of Flavonifractor increased while other bacteria, such as Coproccocus, were reduced in previously colonized children that were cleared of D. fragilis. Evaluating microbiota composition after metronidazole treatment should be done cautiously since this drug is effective against most anaerobic bacteria [106]. However, the microbial heterogeneity caused by metronidazole treatment was transient, with relative abundance reverting to pre-treatment baseline levels within 8 weeks for all genera except for Flavonifractor, which kept increasing in children that lost D. fragilis [82]. Flavonifractor has frequently been associated with disease due to its ability to degrade beneficial anti-carcinogenic flavonoids [107]. Recently, Flavonifractor plautii in the intestinal microbiota has been identified as the key bacterium associated with sporadic young-onset CRC [107,108]. Therefore, colonization with D. fragilis appears to be linked to beneficial changes in the composition of gut bacteria, thus potentially exerting protective effects against dysbiosis-related diseases.
Colonization with Entamoeba spp. results in increased microbiota diversity and compositional changes, characterized by an increase of Firmicutes taxa, such as Ruminococcaceae, coupled with a significant decrease of Bacteroides [21,86]. Interestingly, a reduced ratio of Firmicutes to Bacteroides causes loss of microbial diversity as well as dysbiosis linked to the progression of IBD, CRC, and type 2 diabetes [109,110]. Recently, it was shown that healthy Colombian children colonized with Entamoeba coli exhibit a significant enrichment of Akkermansia in their gut microbiota compared to non-colonized children [48]. Moreover, besides a general increase of bacterial richness, a significant enhancement of Coprococcus and Alistipes was observed in colonized children [48]. Coprococcus is an important anaerobic and butyrate-producing bacterium [111], and decreased abundance of Coprococcus has been implicated in the pathogenesis of CRC [112]. Furthermore, it was recently demonstrated that Coproccocus, as well as other butyrate-producing bacteria, play an important role in language development and cognitive functions in preadolescent children [111].
Together, these studies indicate that commensal gut protozoa significantly remodel the intestinal bacterial niche, potentially creating a favorable microenvironment beneficial for the host. A common observation across the different protozoa species seems to be an enrichment of SCFA-producing bacteria. Importantly, this is in contrast to what has been demonstrated for pathogenic protozoa, e.g., Cryptosporidium, where increased infection severity corresponded with a decreased level of fecal SCFA content [113].
However, it is mechanistically unclear how protozoa influence bacterial composition in the gut microbiota. The possibilities include direct modulation e.g., via preferential feeding on specific bacterial species or secretion of metabolites that modulate the fitness of specific bacteria [114,115]. On the other hand, it has been demonstrated that microbiota composition can influence the severity of protozoa infection, as was recently shown for Cryptosporidium [113]. This suggests a bidirectional interkingdom crosstalk between different microbiota constituents and supports that background state and composition of intestinal microbiota might play a role in determining the outcomes of commensal protozoa colonization. Additionally, as discussed below in Chapter 4, it is also plausible that changes in bacterial composition are a consequence of direct interactions between the protozoa and the host. Future studies will surely shed more light on the precise mechanisms by which protozoa remodel and are, themselves, influenced by the intestinal bacterial microbiome.

Table 2.
Colonization with different protozoa species and the associated bacterial changes in healthy individuals.
Table 2.
Colonization with different protozoa species and the associated bacterial changes in healthy individuals.
| Protozoa spp. | Detection Method | Alterations to the Gut Bacterial Microbiota in Colonized Individuals | Ref. | Characteristics of the Enriched Bacterial Species |
|---|---|---|---|---|
| Blastocystis | Real-time PCR | Increase of bacterial genera: Acetanaerobacterium, Acetivibrio, Coprococcus, Hespellia, Oscillibacter, Papillibacter, Sporobacter, Ruminococcus, Prevotella, Roseburia, Faecalibacterium Decrease of bacterial families: Enterococcaceae, Streptococcaceae, Lactobacillaceae Enterobacteriaceae | [22] | Acetanaerobacterium: anaerobic, fermentation of acetate ethanol [116] Coprococcus: anaerobic, vitamin B, butyrate- and acetate production [111,117] Hespellia: anaerobic, butyrate production [118] Oscillibacter: anaerobic, glucose oxidation [119] Papillibacter: anaerobic, butyrate production [120] Ruminococcus: anaerobic, metabolism of complex polysaccharides [121] Prevotella: anaerobic, metabolism of polysaccharides [122] Roseburia: anaerobic, butyrate-production [120] Feacalibacterium: anaerobic, butyrate and other SCFA production [120] |
| Blastocystis (ST1-6) | Metagenomics | General increase of Firmicutes phyla and Clostridia order. Decrease of Bacteroides genus Increase of bacterial species: Methanobrevibacter smithii, Akkermansia muciniphila, Butyrivibrio crossotus, Eubacterium siraeum, Coprococcus catus, Prevotella copri, Eubacterium rectale, Bifidobacterium adolescentis, Faecalibacterium prausnitzii, Treponema succinifaciens Decrease of bacterial species: Ruminococcus gnavus, Dialister invisus, Escherichia coli, Bacteroides thetaiotamicron, Bacteroides fragilis, Bacteroides vulgatus, Bacteroides uniformis, Bacteroidesovatus, Bacteroides stercoris | [71] | Methanobrevibacter: methanogen, anaerobic, SCFA-production [96] Akkermansia municiphila: anaerobic, mucin degrading, SCFA-production [104] Butyrivibrio crossotus: anaerobic, butyrate-production [120] Eubacterium siraeum: anaerobic, degradation of xylans and ferulic acid production [123] Coprococcus catus: anaerobic, SCFA-production [117] Eubacterium rectale: anaerobic, SCFA-production [120] Bifidobacterium adolescentis: anaerobic, SCFA- and folate production [124] Treponema succinifaciens: anaerobic, SCFA-production [125] |
| Blastocystis (ST3) | Microscopic evaluation and real-time PCR | General increase of Prevotellaceae, Methanobacteriaceae, Clostridiaceae Lachnospiraceae, Erysipelotrichaceae and Pasteurellaceae family. Decrease of Bacteroidaceae and Veillonecellaceae family. Increase of bacterial genera: Prevotella, Methanobrevibacter, Ruminococcus Decrease of bacterial genera: Bacteroides | [126] | Prevotella: anaerobic, metabolism of polysaccharides [122] Methanobrevibacter: methanogen, anaerobic, SCFA-production [96] Ruminococcus: anaerobic, metabolism of complex polysaccharides [121] |
| D. fragilis | Microscopic evaluation, multiplex qPCR and real-time PCR | General Decrease of Bacteroides. Increase of bacterial genera: Akkermansia muciniphila, Methanobrevibacter smithii, Butyrivibrio crossotus, Alistipes, Victivallis, Oscillibacter, Eubacterium, Coproccus, Bifidobacterium adolescentis, Bifidobacterium longum, Ruminococcus bromii, Prevotella copri, Decrease of bacterial genera: Flavonifractor, Parabacteroides distasonis, Bacteroides fragilis,Clostridium leptum, | [73,82] | Methanobrevibacter: methanogen, anaerobic, SCFA-production [96] Akkermansia municiphila: anaerobic, mucin degrading, SCFA-production [104] Victivallis: anaerobic and sugar fermenting [127] Oscillibacter: anaerobic, glucose oxidation [119] Coprococcus: anaerobic, vitamin B, butyrate- and acetate production [111,117] Bifidobacterium adolescentis: anaerobic, SCFA- and folate production [124] Eubacterium siraeum: anaerobic, degradation of xylans and ferulic acid production [123] |
| Entamoeba spp. | Microscopic evaluation and metagenomics | General increase of taxa Clostridiales, Ruminococcaceae. Decrease of Bacteroides, Prevotella and Fusobacteria Increase of bacterial genera Akkarmensia municiphila, Coprococcus, Alistepes Decrease of bacterial genera: Blautia, Streptococcus | [21,48,86] | Alistepes: anaerobic, hydrolysis of tryptophan to indole [128] Coprococcus: anaerobic, vitamin B, butyrate- and acetate production [111,117] Akkermansia municiphila: anaerobic, mucin degrading, SCFA-production [104] |
| Entamoeba and Blastocystis | Nested-PCR | Increase of Faecalibacterium prausnitziim. Decrease of Escherichia coli | [87] | Feacalibacterium prausnitziim: anaerobic, butyrate and other SCFA production [120] |
4. The Impact of Commensal Gut Protozoa on the Host Immune System
Research studies describing the interaction between commensal protozoa species and the mammalian immune system are scarce. Most of the available reports are based on in vitro studies as well as animal models. Although these likely do not fully reflect human (patho)physiology, they still provide valuable insights into potential immunological consequences of intestinal protozoa colonization.
Recently, it was shown that colonization with Blastocystis ST4 attenuates colonic inflammation in a dextran sulfate sodium (DSS)-induced colitis mouse model via induction of T helper (Th) 2 cells and T regulatory (Treg) cells [76]. Mice colonized with Blastocystis ST4 showed a decrease of tumor necrosis factor-α expressing (TNF) CD4+ T-cells and an upregulation of signature Th2 cytokines interleukin (IL)-4, IL-5, and IL-13, as well as the anti-inflammatory cytokine IL-10 [76]. Additionally, a marked increase of abundance of SCFA-producing bacteria, such as Ruminococcaceae and Roseburia, was observed following Blastocystis ST4 colonization. Analysis of the SCFA content in feces from colitic mice that had received fecal matter transplant from Blastocystis ST4-colonized mice revealed enrichment of 6 SCFAs (butyric, isobutyric, valeric, isovaleric, 2-methylbutyric, and caproic acid) compared to mice that received fecal matter transplant from Blastocystis-free mice [76]. Importantly, recent reports have repeatedly suggested a highly beneficial role of SCFAs on gut homeostasis and immune modulation [129]. SCFAs in the intestinal lumen are absorbed by colonocytes where they enter the citric acid cycle and are used for energy production. Unmetabolized SCFAs enter the systemic circulation and travel to different organs, serving as substrates or signaling molecules for various cellular processes such as chemotaxis, proliferation, and differentiation [130,131]. SCFAs achieve this by acting as histone deacetylase (HDAC) inhibitors as well as activators of cell surface receptors [130]. It has been demonstrated that butyrate, created by SCFA-producing microorganisms in the intestines, can facilitate generation of extrathymic Tregs via enhanced acetylation of Foxp3 locus in CD4+ T cells. Moreover, butyrate was shown to induce gene expression changes in dendritic cells, characterized by decreased expression of pro-inflammatory cytokines and, consequently, prompting Treg differentiation [132]. Thus, one potential beneficial mechanism of protozoa colonization might be by indirect modulation of the immune system and its skewing towards a Th2/Treg-dominating response via enrichment of SCFA-producing bacteria in the gut microbiota.
A recent in vitro study showed that Blastocystis ST4 could decrease the growth of the common pathogen Bacteroides vulgatus by inducing reactive oxygen species and the expression of genes associated with oxidative stress. Moreover, a significant reduction in intestinal epithelial permeability was observed in co-cultures with Blastocystis ST4, suggesting that this Blastocystis subtype protects the intestinal barrier from opportunistic bacterial species, at least in vitro [133]. In agreement with that, it was previously demonstrated that Tritrichomonas musculis (T. musculis), a commensal of the rodent microbiota and the closest ortholog to D. fragilis, can protect against the mucosal bacterial infection with Salmonella typhimurium via induction of intestinal epithelium-derived IL-18 and inflammasome activation. However, due to sustained inflammation characterized by increased numbers of IFN-γ-producing CD4+ Th1 and IL-17-producing Th17 cells within the colonic tissue, mice colonized with T. musculis were more susceptible to experimental gut inflammation and cancer [16].
On the other hand, Blastocystis ST7 has been suggested to have immunocompromising functions, and several potential virulence factors have been identified that could support the notion of pathogenicity. Antigens from Blastocystis ST7 have been reported to induce the mitogen-activated protein kinase-dependent expression of pro-inflammatory cytokines such as IL-1β, IL-6, and tumor necrosis factor, in macrophages, mouse intestinal explants, and colonic tissue [134]. Furthermore, Blastocystis ST7 has a significantly higher activity of cysteine proteases compared to other Blastocystis subtypes. Cysteine proteases are a characteristic feature of parasitic protozoa (e.g., Entamoeba histolytica and Cryptosporidium spp.) that have been shown to facilitate invasion of host tissue, as well as immune evasion [135]. For instance, Entamoeba histolytica utilizes cysteine proteases for degradation of colonic mucins and extracellular matrix components that lead to separation of epithelial cells, consequently breaching the epithelial barrier and enabling the protozoa to invade the host tissue [136,137]. Additionally, cysteine proteases are potent modulators of the host immune defense via direct degradation of immunoglobulin A (IgA), IgG, and IL-18, as well as attenuation of protective Th1-type responses (Figure 1) [135]. Besides Blastocystis ST7, no evidence of a similar mechanism has been found in other protozoa species thus far. While Blastocystis ST4 did not change the epithelial permeability, Blastocystis ST7 induced significant epithelial barrier disruption in epithelial cell lines as well as degradation of IgA [138,139]. Taken together, these data again point toward subtype-specific effects of Blastocystis on immune modulation, with ST7 emerging as a major immune-compromising subspecies.
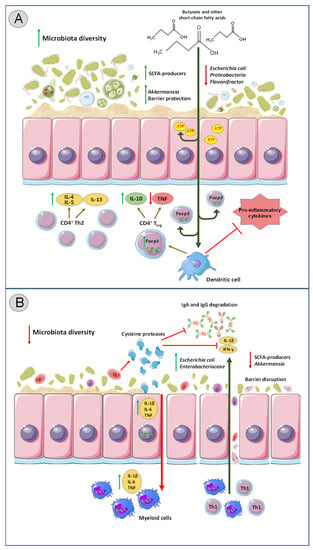
Figure 1.
The impact of intestinal protozoa colonization on gut immunity and bacterial composition. (A) Colonization with commensal protozoa species (e.g., Entamoeba spp. besides Entamoeba histolytica) leads to increased microbial diversity and remodeling of bacterial communities in the gut. SCFA (short-chain fatty acids) producers are enriched, and pathogenic bacterial species such as Escherichia coli and Proteobacteria are reduced. SCFAs in the intestinal lumen are absorbed by epithelial cells for energy production, while unmetabolized SCFAs enter the systemic circulation. Outside the gut, SCFAs facilitate generation of extrathymic Tregs via enhanced acetylation of Foxp3 locus in CD4+ T cells as well as directly affect gene expression in dendritic cells, thus prompting Treg differentiation and consequently IL-10 production. Colonization with commensal protozoa strains leads to polarization of T cell responses towards Th2-dominated profile characterized by IL-4, IL-5, and IL-13 secretion as well as downregulation of TNF. Overall, this results in decreased pro-inflammatory response within the intestines. (B) Colonization with pathogenic protozoa species (such as Cryptosporidium and Entamoeba histolytica) can lead to decreased microbiota diversity, enrichment of pathogenic bacterial species, and a decline in abundance of beneficial bacteria. Cysteine proteases produced by pathogenic protozoa compromise the intestinal epithelial barrier by depleting colonic mucin and create gaps between colonocytes, enabling the parasites to breach the epithelial barrier and invade the tissue. Cysteine proteases also degrade IgA, IgG and IL-18, as well as inhibit Th1-type responses. Pathogenic protozoa also upregulate IL-1B, IL-6, and TNF in the intestines. Overall, these triggers aggravated inflammatory response within the gut. SCFA: Short chain fatty acid, IL: Interleukin, Tregs: T regulatory Th: T helper, TNF: tumor necrosis factor.
Another important factor that has been often overlooked in experimental studies of protozoa colonization models is the dynamic temporal change that occurs to the microbiota and gut immunity. For example, it has been recently shown that Blastocystis ST3 can protect from intestinal inflammation, but only after prolonged colonization time. Short-term exposure experiments, where rat colitis was induced 3 weeks post-colonization with Blastocystis ST3, revealed no difference on disease activity; in contrast, long-term exposure (13 weeks post-colonization) resulted in faster recovery and protection from colitis [75]. This might suggest that despite being initially deleterious or neutral, over time, Blastocystis ST3 supports a more balanced intestinal microbial ecosystem that is more suitable to control disturbances.
5. Conclusions and Future Perspectives
Intestinal protozoa have co-evolved with humans, and their interactions with the human host seem to be highly dynamic and variable, with some species and subtypes exhibiting beneficial properties, while others manifest adverse immunomodulatory effects. The fact that many of these protozoa species cause dormant persistent colonization that often leads to life-long affiliation with their host, points towards commensalism or even symbiosis rather than parasitism. In line with that, colonization with intestinal protozoa appears to significantly increase the diversity of the gut microbiota and selectively modulate the composition of different bacterial communities. Some outstanding questions remain as to whether a therapeutic impact might be achieved by diversification of the human gut via controlled colonization with commensal protozoa strains or by FMT from protozoa-colonized healthy donors to patients with IBD or other gastrointestinal diseases. FMT is an emerging therapy with a successful track record against severe intestinal bacterial infections and a potential therapeutic candidate against diseases associated with microbial dysbiosis [140]. Currently, the presence of protozoa species in human fecal matter donors is an exclusion criterion due to the ongoing debate regarding their pathogenicity. Some preliminary studies in patients subjected to Blastocystis transmission have shown no adverse effects in recipients after colonization [140,141]. Investigating whether such modulation of the gut microbiota would be beneficial may be of utmost importance for the development of novel treatment strategies. On the same note, modulation of the host immunity towards a dominant anti-inflammatory response (directly or indirectly through alterations of the bacterial compartment) might constitute an attractive target, especially in relation to autoimmunity. However, more mechanistic studies are needed to decipher the highly complex relationship between commensal protozoa, microbiota, and the host immune responses. While the consensus concerning the role of intestinal protozoa species in health and disease has not been reached, it is becoming increasingly clear that their presence within the gut is not inconsequential to the human host. In-depth characterization of the significance of commensal protozoa colonization might potentially unravel important changes in immunological mechanisms and microbiota dynamics that can greatly benefit our understanding of human intestinal homeostasis.
Author Contributions
Conceptualization, M.D. and J.B.M.; writing—original draft preparation, M.D.; figures and tables: M.D.; writing—review and editing, B.P., J.B.M. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from Novo Nordisk Foundation (No. 0058349), Brødrene Hartmanns Fond (No. A38338), A.P. Møller Lægevidenskabens Fremme (No. 20-L-0310), and Torben og Alice Frimodts Fond (No. 10204).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The Figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef]
- Xiong, J.; Hu, H.; Xu, C.; Yin, J.; Liu, M.; Zhang, L.; Duan, Y.; Huang, Y. Development of gut microbiota along with its metabolites of preschool children. BMC Pediatr. 2022, 22, 25. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Maeda, Y.; Takeda, K. Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, K.; Wu, Y.; Yang, Y.; Tso, P.; Wu, Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 2017, 8, 942. [Google Scholar] [CrossRef]
- Rogers, G.B.; Keating, D.J.; Young, R.L.; Wong, M.L.; Licinio, J.; Wesselingh, S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry 2016, 21, 738–748. [Google Scholar] [CrossRef]
- Maiuolo, J.; Gliozzi, M.; Musolino, V.; Carresi, C.; Scarano, F.; Nucera, S.; Scicchitano, M.; Oppedisano, F.; Bosco, F.; Ruga, S.; et al. The Contribution of Gut Microbiota-Brain Axis in the Development of Brain Disorders. Front. Neurosci. 2021, 15, 616883. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- García, M.J.; Pascual, M.; Del Pozo, C.; Díaz-González, A.; Castro, B.; Rasines, L.; Crespo, J.; Rivero, M. Impact of immune-mediated diseases in inflammatory bowel disease and implications in therapeutic approach. Sci. Rep. 2020, 10, 10731. [Google Scholar] [CrossRef]
- Argollo, M.; Gilardi, D.; Peyrin-Biroulet, C.; Chabot, J.F.; Peyrin-Biroulet, L.; Danese, S. Comorbidities in inflammatory bowel disease: A call for action. Lancet Gastroenterol. Hepatol. 2019, 4, 643–654. [Google Scholar] [CrossRef]
- Matos, R.; Lencastre, L.; Rocha, V.; Torres, S.; Vieira, F.; Barbosa, M.R.; Ascenção, J.; Guerra, M.P. Quality of life in patients with inflammatory bowel disease: The role of positive psychological factors. Health Psychol. Behav. Med. 2021, 9, 989–1005. [Google Scholar] [CrossRef]
- Silangcruz, K.; Nishimura, Y.; Czech, T.; Kimura, N.; Hagiya, H.; Koyama, T.; Otsuka, F. Impact of the World Inflammatory Bowel Disease Day and Crohn’s and Colitis Awareness Week on Population Interest Between 2016 and 2020: Google Trends Analysis. JMIR Infodemiol. 2021, 1, e32856. [Google Scholar] [CrossRef]
- Cui, G.; Liu, H.; Xu, G.; Laugsand, J.B.; Pang, Z. Exploring Links Between Industrialization, Urbanization, and Chinese Inflammatory Bowel Disease. Front. Med. 2021, 8, 757025. [Google Scholar] [CrossRef]
- Lukeš, J.; Stensvold, C.R.; Jirků-Pomajbíková, K.; Wegener Parfrey, L. Are Human Intestinal Eukaryotes Beneficial or Commensals? PLoS Pathog. 2015, 11, e1005039. [Google Scholar] [CrossRef]
- Chudnovskiy, A.; Mortha, A.; Kana, V.; Kennard, A.; Ramirez, J.D.; Rahman, A.; Remark, R.; Mogno, I.; Ng, R.; Gnjatic, S.; et al. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 2016, 167, 444–456.e414. [Google Scholar] [CrossRef]
- Wei, Y.; Gao, J.; Kou, Y.; Meng, L.; Zheng, X.; Liang, M.; Sun, H.; Liu, Z.; Wang, Y. Commensal Bacteria Impact a Protozoan’s Integration into the Murine Gut Microbiota in a Dietary Nutrient-Dependent Manner. Appl. Environ. Microbiol. 2020, 86, e00303-20. [Google Scholar] [CrossRef]
- Issa, R. Non-pathogenic protozoa. Int. J. Pharm. Pharm. Sci. 2014, 6, 30–40. [Google Scholar]
- Cavalier-Smith, T. Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol. 2003, 39, 338–348. [Google Scholar] [CrossRef]
- Ocaña, K.A.; Dávila, A.M. Phylogenomics-based reconstruction of protozoan species tree. Evol. Bioinform. Online 2011, 7, 107–121. [Google Scholar] [CrossRef]
- Even, G.; Lokmer, A.; Rodrigues, J.; Audebert, C.; Viscogliosi, E.; Ségurel, L.; Chabé, M. Changes in the Human Gut Microbiota Associated With Colonization by Blastocystis sp. and Entamoeba spp. in Non-Industrialized Populations. Front. Cell. Infect. Microbiol. 2021, 11, 533528. [Google Scholar] [CrossRef]
- Audebert, C.; Even, G.; Cian, A.; Safadi, D.E.; Certad, G.; Delhaes, L.; Pereira, B.; Nourrisson, C.; Poirier, P.; Wawrzyniak, I.; et al. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 2016, 6, 25255. [Google Scholar] [CrossRef] [PubMed]
- von Huth, S.; Thingholm, L.B.; Kofoed, P.-E.; Bang, C.; Rühlemann, M.C.; Franke, A.; Holmskov, U. Intestinal protozoan infections shape fecal bacterial microbiota in children from Guinea-Bissau. PLoS Negl. Trop. Dis. 2021, 15, e0009232. [Google Scholar] [CrossRef]
- Chabé, M.; Lokmer, A.; Ségurel, L. Gut Protozoa: Friends or Foes of the Human Gut Microbiota? Trends Parasitol. 2017, 33, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Maritz, J.M.; Land, K.M.; Carlton, J.M.; Hirt, R.P. What is the importance of zoonotic trichomonads for human health? Trends Parasitol. 2014, 30, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Deng, L.; Wojciech, L.; Gascoigne, N.R.J.; Peng, G.; Tan, K.S.W. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 2021, 17, e1009253. [Google Scholar] [CrossRef]
- Wawrzyniak, I.; Poirier, P.; Viscogliosi, E.; Dionigia, M.; Texier, C.; Delbac, F.; Alaoui, H.E. Blastocystis, an unrecognized parasite: An overview of pathogenesis and diagnosis. Ther. Adv. Infect. Dis. 2013, 1, 167–178. [Google Scholar] [CrossRef]
- Leung, J.M.; Graham, A.L.; Knowles, S.C.L. Parasite-Microbiota Interactions With the Vertebrate Gut: Synthesis Through an Ecological Lens. Front. Microbiol. 2018, 9, 843. [Google Scholar] [CrossRef]
- Panthee, B.; Gyawali, S.; Panthee, P.; Techato, K. Environmental and Human Microbiome for Health. Life 2022, 12, 456. [Google Scholar] [CrossRef]
- Thompson, R.C.A.; Ash, A. Molecular epidemiology of Giardia and Cryptosporidium infections. Infect. Genet. Evol. 2016, 40, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kurenzvi, L.; Sebunya, T.K.; Coetzee, T.; Paganotti, G.M.; Teye, M.V. Prevalence of Cryptosporidium parvum, Giardia intestinalis and molecular characterization of group A rotavirus associated with diarrhea in children below five years old in Gaborone, Botswana. Pan. Afr. Med. J. 2020, 37, 159. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Padalia, J.; Moonah, S. Tissue Destruction Caused by Entamoeba histolytica Parasite: Cell Death, Inflammation, Invasion, and the Gut Microbiome. Curr. Clin. Microbiol. Rep. 2019, 6, 51–57. [Google Scholar] [CrossRef]
- Guérin, A.; Striepen, B. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host. Microbe. 2020, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.D. Giardia duodenalis: Biology and Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00024-19. [Google Scholar] [CrossRef]
- Marie, C.; Petri, W.A., Jr. Regulation of virulence of Entamoeba histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef]
- Lokmer, A.; Cian, A.; Froment, A.; Gantois, N.; Viscogliosi, E.; Chabé, M.; Ségurel, L. Use of shotgun metagenomics for the identification of protozoa in the gut microbiota of healthy individuals from worldwide populations with various industrialization levels. PLoS ONE 2019, 14, e0211139. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Walters, W.A.; Lauber, C.L.; Clemente, J.C.; Berg-Lyons, D.; Teiling, C.; Kodira, C.; Mohiuddin, M.; Brunelle, J.; Driscoll, M.; et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 2014, 5, 298. [Google Scholar] [CrossRef]
- Schuster, F.L.; Ramirez-Avila, L. Current world status of Balantidium coli. Clin. Microbiol. Rev. 2008, 21, 626–638. [Google Scholar] [CrossRef]
- Andersen, L.O.B.; Stensvold, C.R. Blastocystis in Health and Disease: Are We Moving from a Clinical to a Public Health Perspective? J. Clin. Microbiol. 2016, 54, 524–528. [Google Scholar] [CrossRef]
- Mathison, B.A.; Pritt, B.S. Parasites of the Gastrointestinal Tract. In Encyclopedia of Infection and Immunity; Rezaei, N., Ed.; Elsevier: Oxford, UK, 2022; pp. 136–203. [Google Scholar] [CrossRef]
- Dubey, J.P.; Almeria, S. Cystoisospora belli infections in humans: The past 100 years. Parasitology 2019, 146, 1490–1527. [Google Scholar] [CrossRef]
- Ryan, U.; Fayer, R.; Xiao, L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology 2014, 141, 1667–1685. [Google Scholar] [CrossRef]
- Almeria, S.; Cinar, H.N.; Dubey, J.P. Cyclospora cayetanensis and Cyclosporiasis: An Update. Microorganisms 2019, 7, 317. [Google Scholar] [CrossRef]
- Stark, D.; Barratt, J.; Chan, D.; Ellis, J.T. Dientamoeba fragilis, the Neglected Trichomonad of the Human Bowel. Clin. Microbiol. Rev. 2016, 29, 553–580. [Google Scholar] [CrossRef]
- Poulsen, C.S.; Stensvold, C.R. Systematic review on Endolimax nana: A less well studied intestinal ameba. Trop. Parasitol. 2016, 6, 8–29. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.A. Entamoeba bangladeshi: An insight. Trop. Parasitol. 2014, 4, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Alzate, J.F.; Toro-Londoño, M.; Cabarcas, F.; Garcia-Montoya, G.; Galvan-Diaz, A. Contrasting microbiota profiles observed in children carrying either Blastocystis spp. or the commensal amoebas Entamoeba coli or Endolimax nana. Sci. Rep. 2020, 10, 15354. [Google Scholar] [CrossRef]
- Pysova, I.; Tumova, P.; Tolarova, V.; Nohynkova, E. Nonpathogenic Entamoeba dispar quickly outgrows pathogenic Entamoeba histolytica in mixed xenic cultures. Lett. Appl. Microbiol. 2009, 48, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Winiecka-Krusnell, J.; Lier, T.; Lebbad, M. Evaluation of a PCR Method for Detection of Entamoeba polecki, with an Overview of Its Molecular Epidemiology. J. Clin. Microbiol. 2018, 56, e00154-18. [Google Scholar] [CrossRef]
- Singh, A.; Banerjee, T.; Khan, U.; Shukla, S.K. Epidemiology of clinically relevant Entamoeba spp. (E. histolytica/dispar/moshkovskii/bangladeshi): A cross sectional study from North India. PLoS Negl. Trop. Dis. 2021, 15, e0009762. [Google Scholar] [CrossRef]
- Li, W.C.; Ying, M.; Gong, P.T.; Li, J.H.; Yang, J.; Li, H.; Zhang, X.C. Pentatrichomonas hominis: Prevalence and molecular characterization in humans, dogs, and monkeys in Northern China. Parasitol. Res. 2016, 115, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; van der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Nourrisson, C.; Scanzi, J.; Pereira, B.; NkoudMongo, C.; Wawrzyniak, I.; Cian, A.; Viscogliosi, E.; Livrelli, V.; Delbac, F.; Dapoigny, M.; et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: Comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE 2014, 9, e111868. [Google Scholar] [CrossRef]
- Poirier, P.; Wawrzyniak, I.; Vivarès, C.P.; Delbac, F.; El Alaoui, H. New insights into Blastocystis spp.: A potential link with irritable bowel syndrome. PLoS Pathog. 2012, 8, e1002545. [Google Scholar] [CrossRef]
- Shafiei, Z.; Esfandiari, F.; Sarkari, B.; Rezaei, Z.; Fatahi, M.R.; Hosseini Asl, S.M.K. Parasitic infections in irritable bowel syndrome patients: Evidence to propose a possible link, based on a case-control study in the south of Iran. BMC Res. Notes 2020, 13, 264. [Google Scholar] [CrossRef]
- Kosik-Bogacka, D.; Lepczyńska, M.; Kot, K.; Szkup, M.; Łanocha-Arendarczyk, N.; Dzika, E.; Grochans, E. Prevalence, subtypes and risk factors of Blastocystis spp. infection among pre- and perimenopausal women. BMC Infect. Dis. 2021, 21, 1125. [Google Scholar] [CrossRef]
- Petersen, A.M.; Stensvold, C.R.; Mirsepasi, H.; Engberg, J.; Friis-Møller, A.; Porsbo, L.J.; Hammerum, A.M.; Nordgaard-Lassen, I.; Nielsen, H.V.; Krogfelt, K.A. Active ulcerative colitis associated with low prevalence of Blastocystis and Dientamoeba fragilis infection. Scand. J. Gastroenterol. 2013, 48, 638–639. [Google Scholar] [CrossRef] [PubMed]
- Leder, K.; Hellard, M.E.; Sinclair, M.I.; Fairley, C.K.; Wolfe, R. No correlation between clinical symptoms and Blastocystis hominis in immunocompetent individuals. J. Gastroenterol. Hepatol. 2005, 20, 1390–1394. [Google Scholar] [CrossRef]
- Krogsgaard, L.R.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: A population-based case-control study. Clin. Gastroenterol. Hepatol. 2015, 13, 507–513.e502. [Google Scholar] [CrossRef]
- Gassama, A.; Sow, P.S.; Fall, F.; Camara, P.; Guèye-N’diaye, A.; Seng, R.; Samb, B.; M’Boup, S.; Aïdara-Kane, A. Ordinary and opportunistic enteropathogens associated with diarrhea in Senegalese adults in relation to human immunodeficiency virus serostatus. Int. J. Infect. Dis. 2001, 5, 192–198. [Google Scholar] [CrossRef]
- Sarzhanov, F.; Dogruman-Al, F.; Santin, M.; Maloney, J.G.; Gureser, A.S.; Karasartova, D.; Taylan-Ozkan, A. Investigation of neglected protists Blastocystis sp. and Dientamoeba fragilis in immunocompetent and immunodeficient diarrheal patients using both conventional and molecular methods. PLoS Negl. Trop. Dis. 2021, 15, e0009779. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.D.; Sánchez, L.V.; Bautista, D.C.; Corredor, A.F.; Flórez, A.C.; Stensvold, C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014, 22, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, P.D.; Stensvold, C.R.; Rajilić-Stojanović, M.; Heilig, H.G.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Sulżyc-Bielicka, V.; Kołodziejczyk, L.; Adamska, M.; Skotarczak, B.; Jaczewska, S.; Safranow, K.; Bielicki, P.; Kładny, J.; Bielicki, D. Colorectal cancer and Blastocystis sp. infection. Parasit. Vectors 2021, 14, 200. [Google Scholar] [CrossRef]
- Dogruman-Al, F.; Dagci, H.; Yoshikawa, H.; Kurt, O.; Demirel, M. A possible link between subtype 2 and asymptomatic infections of Blastocystis hominis. Parasitol. Res. 2008, 103, 685–689. [Google Scholar] [CrossRef]
- Khademvatan, S.; Masjedizadeh, R.; Yousefi-Razin, E.; Mahbodfar, H.; Rahim, F.; Yousefi, E.; Foroutan, M. PCR-based molecular characterization of Blastocystis hominis subtypes in southwest of Iran. J. Infect. Public Health 2018, 11, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.S.; Whipps, C.M.; Ganac, R.D.; Hudson, N.R.; Boorom, K. Association of Blastocystis subtype 3 and 1 with patients from an Oregon community presenting with chronic gastrointestinal illness. Parasitol. Res. 2009, 104, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.H.; Ismail, M.A.M.; El-Badry, A.A.; Abu-Sarea, E.Y.; Dewidar, A.M.; Hamdy, D.A. An Association Between Blastocystis Subtypes and Colorectal Cancer Patients: A Significant Different Profile from Non-cancer Individuals. Acta Parasitol. 2022, 67, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Kodio, A.; Coulibaly, D.; Koné, A.K.; Konaté, S.; Doumbo, S.; Guindo, A.; Bittar, F.; Gouriet, F.; Raoult, D.; Thera, M.A.; et al. Blastocystis Colonization Is Associated with Increased Diversity and Altered Gut Bacterial Communities in Healthy Malian Children. Microorganisms 2019, 7, 649. [Google Scholar] [CrossRef]
- Beghini, F.; Pasolli, E.; Truong, T.D.; Putignani, L.; Cacciò, S.M.; Segata, N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017, 11, 2848–2863. [Google Scholar] [CrossRef]
- Rossen, N.G.; Bart, A.; Verhaar, N.; van Nood, E.; Kootte, R.; de Groot, P.F.; D’Haens, G.R.; Ponsioen, C.Y.; van Gool, T. Low prevalence of Blastocystis sp. in active ulcerative colitis patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Krogsgaard, L.R.; Andersen, L.O.B.; Johannesen, T.B.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin. Gastroenterol. Hepatol. 2018, 9, e161. [Google Scholar] [CrossRef] [PubMed]
- Tito, R.Y.; Chaffron, S.; Caenepeel, C.; Lima-Mendez, G.; Wang, J.; Vieira-Silva, S.; Falony, G.; Hildebrand, F.; Darzi, Y.; Rymenans, L.; et al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 2019, 68, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Billy, V.; Lhotská, Z.; Jirků, M.; Kadlecová, O.; Frgelecová, L.; Parfrey, L.W.; Pomajbíková, K.J. Blastocystis Colonization Alters the Gut Microbiome and, in Some Cases, Promotes Faster Recovery From Induced Colitis. Front. Microbiol. 2021, 12, 641483. [Google Scholar] [CrossRef]
- Deng, L.; Wojciech, L.; Png, C.W.; Koh, E.Y.; Aung, T.T.; Kioh, D.Y.Q.; Chan, E.C.Y.; Malleret, B.; Zhang, Y.; Peng, G.; et al. Experimental colonization with Blastocystis ST4 is associated with protective immune responses and modulation of gut microbiome in a DSS-induced colitis mouse model. Cell Mol. Life Sci. 2022, 79, 245. [Google Scholar] [CrossRef]
- Lepczyńska, M.; Białkowska, J.; Dzika, E.; Piskorz-Ogórek, K.; Korycińska, J. Blastocystis: How do specific diets and human gut microbiota affect its development and pathogenicity? Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M. Molecular epidemiology of Dientamoeba fragilis. Acta Trop. 2018, 184, 73–77. [Google Scholar] [CrossRef]
- Röser, D.; Simonsen, J.; Nielsen, H.V.; Stensvold, C.R.; Mølbak, K. Dientamoeba fragilis in Denmark: Epidemiological experience derived from four years of routine real-time PCR. Eur. J. Clin. Microbiol. 2013, 32, 1303–1310. [Google Scholar] [CrossRef]
- Bruijnesteijn van Coppenraet, L.E.; Dullaert-de Boer, M.; Ruijs, G.J.; van der Reijden, W.A.; van der Zanden, A.G.; Weel, J.F.; Schuurs, T.A. Case-control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: Application of molecular detection. Clin. Microbiol. Infect. 2015, 21, 592.e9–592.e19. [Google Scholar] [CrossRef]
- Brands, M.R.; Van de Vijver, E.; Haisma, S.M.; Heida, A.; van Rheenen, P.F. No association between abdominal pain and Dientamoeba in Dutch and Belgian children. Arch. Dis. Child. 2019, 104, 686–689. [Google Scholar] [CrossRef]
- Gotfred-Rasmussen, H.; Stensvold, C.R.; Ingham, A.C.; Johannesen, T.B.; Andersen, L.O.B.; Röser, D.; Nielsen, H.V. Impact of Metronidazole Treatment and Dientamoeba Fragilis Colonization on Gut Microbiota Diversity. J. Pediatr. Gastroenterol. Nutr. 2021, 73, 23–29. [Google Scholar] [CrossRef] [PubMed]
- López, M.C.; León, C.M.; Fonseca, J.; Reyes, P.; Moncada, L.; Olivera, M.J.; Ramírez, J.D. Molecular Epidemiology of Entamoeba: First Description of Entamoeba moshkovskii in a Rural Area from Central Colombia. PLoS ONE 2015, 10, e0140302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El-Dib, N.A.; Khater, M.M. Entamoeba. In Encyclopedia of Infection and Immunity; Rezaei, N., Ed.; Elsevier: Oxford, UK, 2022; pp. 492–512. [Google Scholar] [CrossRef]
- Cui, Z.; Li, J.; Chen, Y.; Zhang, L. Molecular epidemiology, evolution, and phylogeny of Entamoeba spp. Infect. Genet. Evol. 2019, 75, 104018. [Google Scholar] [CrossRef] [PubMed]
- Morton, E.R.; Lynch, J.; Froment, A.; Lafosse, S.; Heyer, E.; Przeworski, M.; Blekhman, R.; Ségurel, L. Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by Entamoeba and Subsistence. PLoS Genet. 2015, 11, e1005658. [Google Scholar] [CrossRef] [PubMed]
- Valerio, I.; Floriana, S.; Valentina, T.; Fabrizio, P.; Anatole, M.; Veronica Di, C.; David Di, C.; Serena, S.; Federica, B.; Rossella, D.A. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte d’Ivoire. J. Infect. Dev. Ctries. 2016, 10, 8179. [Google Scholar] [CrossRef]
- Mejia, R.; Damania, A.; Jeun, R.; Bryan, P.E.; Vargas, P.; Juarez, M.; Cajal, P.S.; Nasser, J.; Krolewiecki, A.; Lefoulon, E.; et al. Impact of intestinal parasites on microbiota and cobalamin gene sequences: A pilot study. Parasit. Vectors 2020, 13, 200. [Google Scholar] [CrossRef]
- Chihi, A.; O’Brien Andersen, L.; Aoun, K.; Bouratbine, A.; Stensvold, C.R. Amplicon-based next-generation sequencing of eukaryotic nuclear ribosomal genes (metabarcoding) for the detection of single-celled parasites in human faecal samples. Parasit. Epidemiol. Control. 2022, 17, e00242. [Google Scholar] [CrossRef]
- Weber, A.A.T.; Pawlowski, J. Can Abundance of Protists Be Inferred from Sequence Data: A Case Study of Foraminifera. PLoS ONE 2013, 8, e56739. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Garcia Yunta, R.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016, 1, 16088. [Google Scholar] [CrossRef]
- O’Brien Andersen, L.; Karim, A.B.; Roager, H.M.; Vigsnæs, L.K.; Krogfelt, K.A.; Licht, T.R.; Stensvold, C.R. Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur. J. Clin. Microbiol. 2016, 35, 1427–1431. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Vester-Andersen, M.K.; Mirsepasi-Lauridsen, H.C.; Prosberg, M.V.; Mortensen, C.O.; Träger, C.; Skovsen, K.; Thorkilgaard, T.; Nøjgaard, C.; Vind, I.; Krogfelt, K.A.; et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci. Rep. 2019, 9, 13473. [Google Scholar] [CrossRef] [PubMed]
- Camara, A.; Konate, S.; Tidjani Alou, M.; Kodio, A.; Togo, A.H.; Cortaredona, S.; Henrissat, B.; Thera, M.A.; Doumbo, O.K.; Raoult, D.; et al. Clinical evidence of the role of Methanobrevibacter smithii in severe acute malnutrition. Sci. Rep. 2021, 11, 5426. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Tong, M.; Li, X.; Wegener Parfrey, L.; Roth, B.; Ippoliti, A.; Wei, B.; Borneman, J.; McGovern, D.P.B.; Frank, D.N.; Li, E.; et al. A Modular Organization of the Human Intestinal Mucosal Microbiota and Its Association with Inflammatory Bowel Disease. PLoS ONE 2013, 8, e80702. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low Counts of Faecalibacterium prausnitzii in Colitis Microbiota. Inflamm. Bowel. Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Caudet, J.; Trelis, M.; Cifre, S.; Soriano, J.M.; Rico, H.; Merino-Torres, J.F. Interplay between Intestinal Bacterial Communities and Unicellular Parasites in a Morbidly Obese Population: A Neglected Trinomial. Nutrients 2022, 14, 3211. [Google Scholar] [CrossRef]
- Yao, S.; Zhao, Z.; Wang, W.; Liu, X. Bifidobacterium Longum: Protection against Inflammatory Bowel Disease. J. Immunol. Res. 2021, 2021, 8030297. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; Chia, L.W.; Aalvink, S.; Chamlagain, B.; Piironen, V.; Knol, J.; de Vos, W.M. Microbial Metabolic Networks at the Mucus Layer Lead to Diet-Independent Butyrate and Vitamin B(12) Production by Intestinal Symbionts. mBio 2017, 8, e00770-17. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Lee, J.W.J.; Tan, K.S.W. Infection with pathogenic Blastocystis ST7 is associated with decreased bacterial diversity and altered gut microbiome profiles in diarrheal patients. Parasit. Vectors 2022, 15, 312. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.D.; Klutman, N.E.; Lamp, K.C. Metronidazole. A therapeutic review and update. Drugs 1997, 54, 679–708. [Google Scholar] [CrossRef]
- Gupta, A.; Dhakan, D.B.; Maji, A.; Saxena, R.; P K, V.P.; Mahajan, S.; Pulikkan, J.; Kurian, J.; Gomez, A.M.; Scaria, J.; et al. Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India. mSystems 2019, 4, e00438-19. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L.; Shi, D.; Kong, C.; Liu, J.; Liu, G.; Li, X.; Ma, Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat. Commun. 2021, 12, 6757. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Y.; Gu, J. Clinical Implications of the Associations Between Intestinal Microbiome and Colorectal Cancer Progression. Cancer Manag. Res. 2020, 12, 4117–4128. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut Microbiome Profiles Are Associated With Type 2 Diabetes in Urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef]
- Kort, R.; Schlösser, J.; Vazquez, A.R.; Atukunda, P.; Muhoozi, G.K.M.; Wacoo, A.P.; Sybesma, W.F.H.; Westerberg, A.C.; Iversen, P.O.; Schoen, E.D. Model Selection Reveals the Butyrate-Producing Gut Bacterium Coprococcus eutactus as Predictor for Language Development in 3-Year-Old Rural Ugandan Children. Front. Microbiol. 2021, 12, 681485. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; de’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Charania, R.; Wade, B.E.; McNair, N.N.; Mead, J.R. Changes in the Microbiome of Cryptosporidium-Infected Mice Correlate to Differences in Susceptibility and Infection Levels. Microorganisms 2020, 8, 879. [Google Scholar] [CrossRef] [PubMed]
- Olajide, J.S.; Cai, J. Perils and Promises of Pathogenic Protozoan Extracellular Vesicles. Front. Cell. Infect. Microbiol. 2020, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Vergara, G.; Hoque, M.M.; McDougald, D.; Noorian, P. The Impact of Protozoan Predation on the Pathogenicity of Vibrio cholerae. Front. Microbiol. 2020, 11, 17. [Google Scholar] [CrossRef]
- Chen, S.Y.; Niu, L.L.; Dong, X.Z. Hydrogen production from glucose by Acetanaerobacterium elongatum. Wei Sheng Wu Xue Bao 2006, 46, 233–237. [Google Scholar] [PubMed]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Ghosh, S.; Pramanik, S. Structural diversity, functional aspects and future therapeutic applications of human gut microbiome. Arch. Microbiol. 2021, 203, 5281–5308. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Kim, H.-E.; Park, J.I.; Cho, H.; Kwak, M.-J.; Kim, B.-Y.; Yang, S.H.; Lee, J.P.; Kim, D.K.; Joo, K.W.; et al. The Association between Gut Microbiota and Uremia of Chronic Kidney Disease. Microorganisms 2020, 8, 907. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [PubMed]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Precup, G.; Vodnar, D.C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef]
- Duncan, S.H.; Russell, W.R.; Quartieri, A.; Rossi, M.; Parkhill, J.; Walker, A.W.; Flint, H.J. Wheat bran promotes enrichment within the human colonic microbiota of butyrate-producing bacteria that release ferulic acid. Environ. Microbiol. 2016, 18, 2214–2225. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Tanes, C.; Walker, E.M.; Slisarenko, N.; Gerrets, G.L.; Grasperge, B.F.; Qin, X.; Jazwinski, S.M.; Bushman, F.D.; Bittinger, K.; Rout, N. Gut Microbiome Changes Associated with Epithelial Barrier Damage and Systemic Inflammation during Antiretroviral Therapy of Chronic SIV Infection. Viruses 2021, 13, 1567. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, S.; Furzi, F.; Fontanelli Sulekova, L.; Taliani, G.; Mattiucci, S. Occurrence of Blastocystis-subtypes in patients from Italy revealed association of ST3 with a healthy gut microbiota. Parasite Epidemiol. Control 2020, 9, e00134. [Google Scholar] [CrossRef]
- van Passel, M.W.; Kant, R.; Palva, A.; Lucas, S.; Copeland, A.; Lapidus, A.; Glavina del Rio, T.; Dalin, E.; Tice, H.; Bruce, D.; et al. Genome sequence of Victivallis vadensis ATCC BAA-548, an anaerobic bacterium from the phylum Lentisphaerae, isolated from the human gastrointestinal tract. J. Bacteriol. 2011, 193, 2373–2374. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Carretta, M.D.; Quiroga, J.; López, R.; Hidalgo, M.A.; Burgos, R.A. Participation of Short-Chain Fatty Acids and Their Receptors in Gut Inflammation and Colon Cancer. Front. Physiol. 2021, 12, 662739. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Deng, L.; Tan, K.S.W. Interactions between Blastocystis subtype ST4 and gut microbiota in vitro. Parasit. Vectors 2022, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.X.; Png, C.W.; Tay, C.Y.; Teo, J.D.; Jiao, H.; Lehming, N.; Tan, K.S.; Zhang, Y. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect. Immun. 2014, 82, 4789–4801. [Google Scholar] [CrossRef] [PubMed]
- Siqueira-Neto, J.L.; Debnath, A.; McCall, L.I.; Bernatchez, J.A.; Ndao, M.; Reed, S.L.; Rosenthal, P.J. Cysteine proteases in protozoan parasites. PLoS Negl. Trop. Dis. 2018, 12, e0006512. [Google Scholar] [CrossRef] [PubMed]
- Meléndez-López, S.G.; Herdman, S.; Hirata, K.; Choi, M.H.; Choe, Y.; Craik, C.; Caffrey, C.R.; Hansell, E.; Chávez-Munguía, B.; Chen, Y.T.; et al. Use of recombinant Entamoeba histolytica cysteine proteinase 1 to identify a potent inhibitor of amebic invasion in a human colonic model. Eukaryot. Cell 2007, 6, 1130–1136. [Google Scholar] [CrossRef]
- Takeuchi, A.; Phillips, B.P. Electron Microscope Studies of Experimental Entamoeba Histolytica Infection in the Guinea Pig: I. Penetration of the Intestinal Epithelium by Trophozoites. Am. J. Trop. Med. 1975, 24, 34–48. [Google Scholar] [CrossRef]
- Wu, Z.; Mirza, H.; Tan, K.S.W. Intra-Subtype Variation in Enteroadhesion Accounts for Differences in Epithelial Barrier Disruption and Is Associated with Metronidazole Resistance in Blastocystis Subtype-7. PLoS Negl. Trop. Dis. 2014, 8, e2885. [Google Scholar] [CrossRef]
- Puthia, M.K.; Vaithilingam, A.; Lu, J.; Tan, K.S. Degradation of human secretory immunoglobulin A by Blastocystis. Parasitol. Res. 2005, 97, 386–389. [Google Scholar] [CrossRef]
- Tan, P.; Li, X.; Shen, J.; Feng, Q. Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease: An Update. Front. Pharmacol. 2020, 11, 574533. [Google Scholar] [CrossRef]
- Terveer, E.M.; van Gool, T.; Ooijevaar, R.E.; Sanders, I.; Boeije-Koppenol, E.; Keller, J.J.; Bart, A.; Kuijper, E.J. Human Transmission of Blastocystis by Fecal Microbiota Transplantation Without Development of Gastrointestinal Symptoms in Recipients. Clin. Infect. Dis. 2020, 71, 2630–2636. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).