Integrating Genome-Wide Association Study with RNA-Sequencing Reveals HDAC9 as a Candidate Gene Influencing Loin Muscle Area in Beijing Black Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and LMA Measurement
2.2. Genotyping and Quality Control
2.3. Genome-Wide Association Study
2.4. Total RNA Isolation, Purification, and Quantification
2.5. cDNA Library Construction and Sequencing
2.6. Data Quality Control and Differential Gene Expression Data Analysis
2.7. Gene Ontology (GO) Annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of DEGs
2.8. Quantitative Real-Time PCR (qRT-PCR) Validation
3. Results
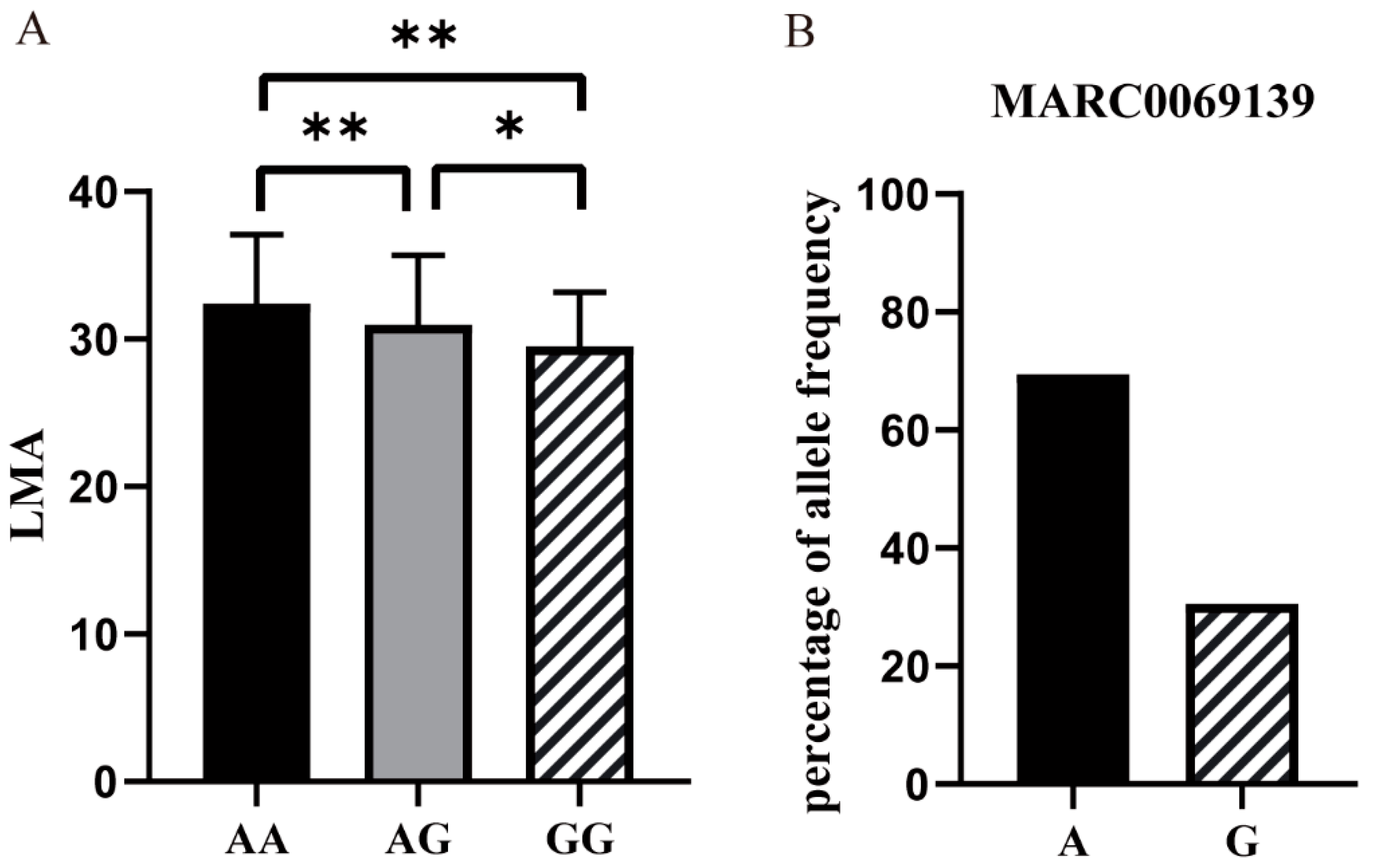
3.1. Genome-Wide Association SNPs for LMA
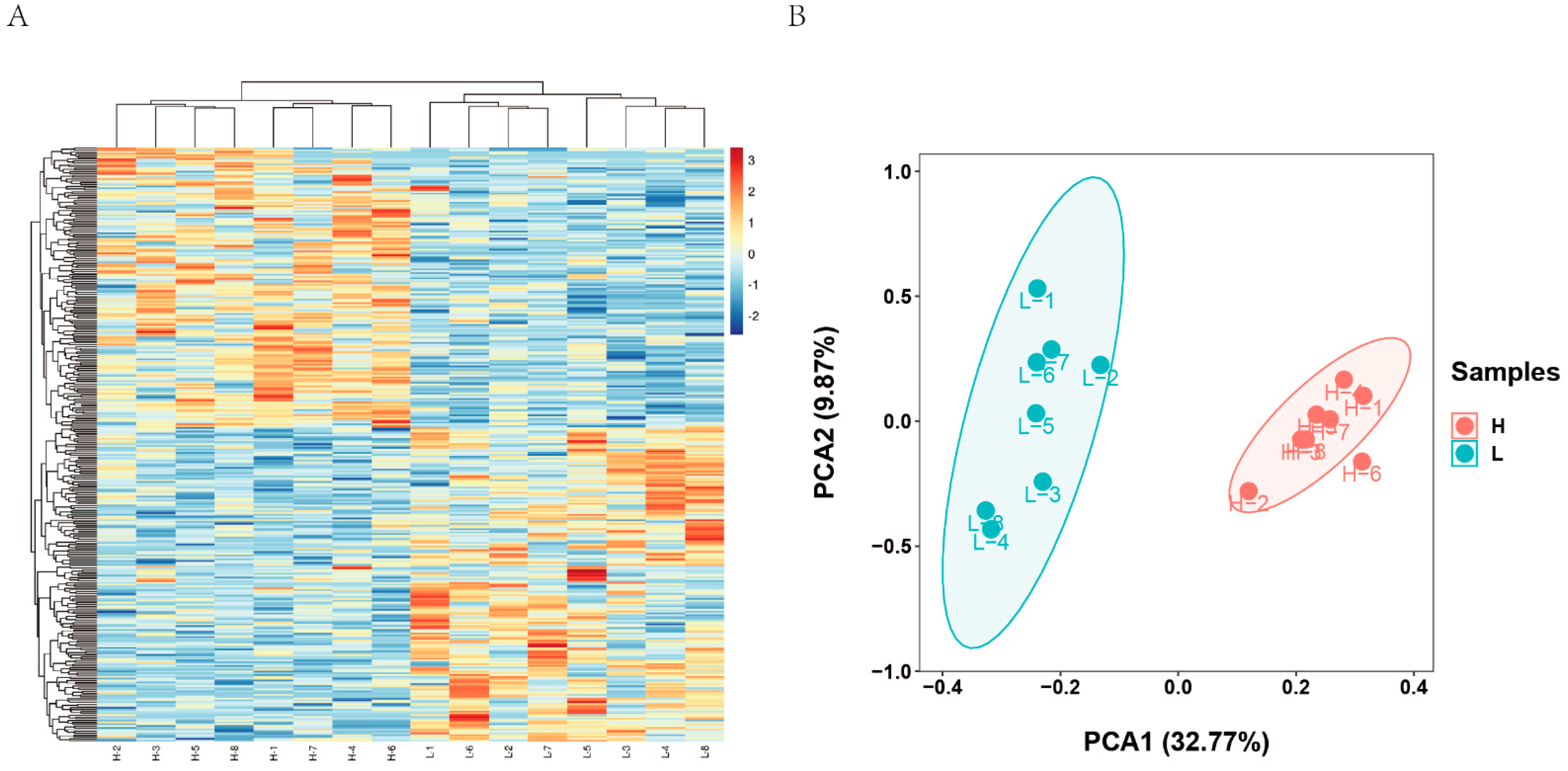
3.2. RNA-Seq Data
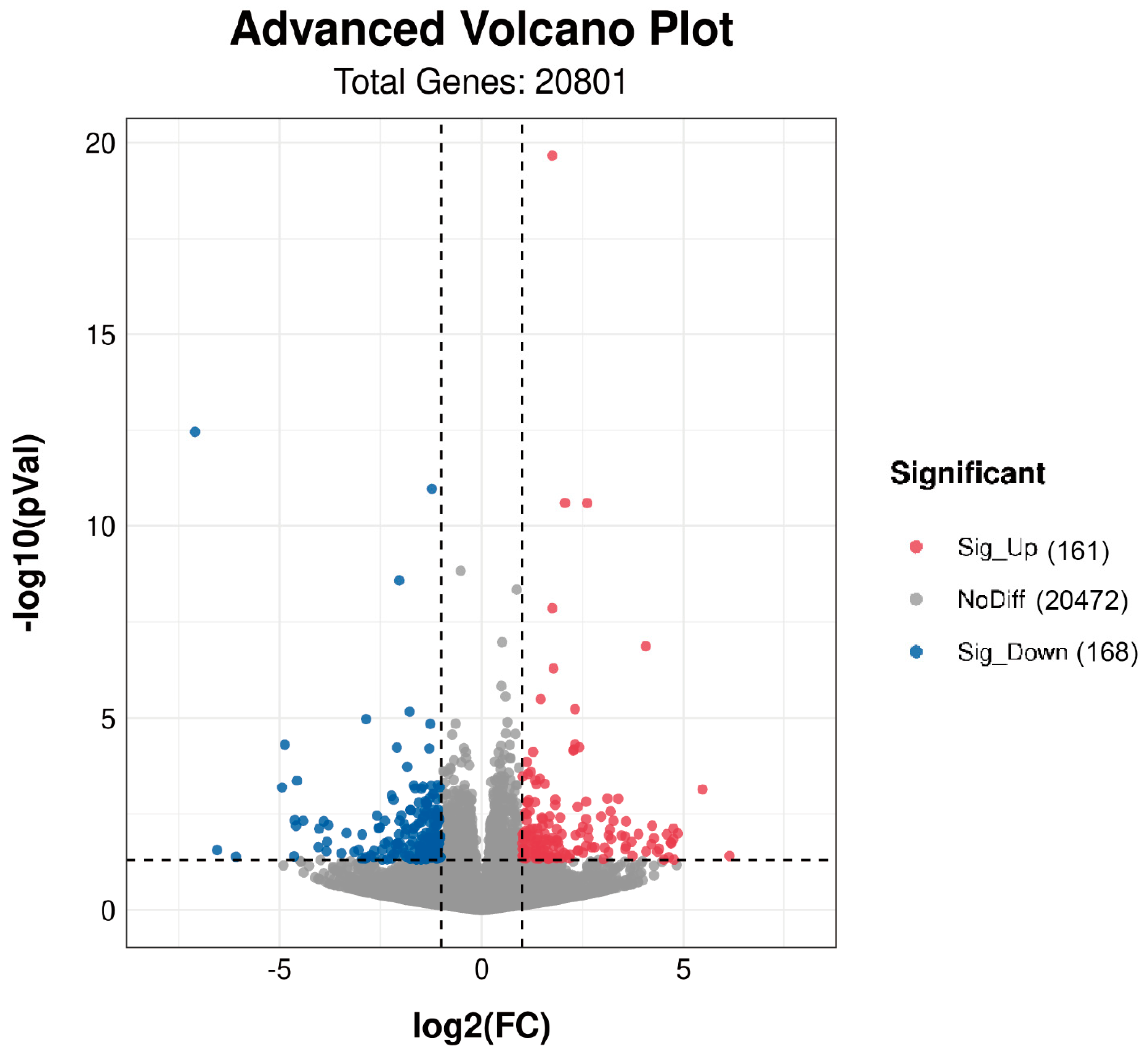
3.3. DEGs between Higher and Lower LMA Groups
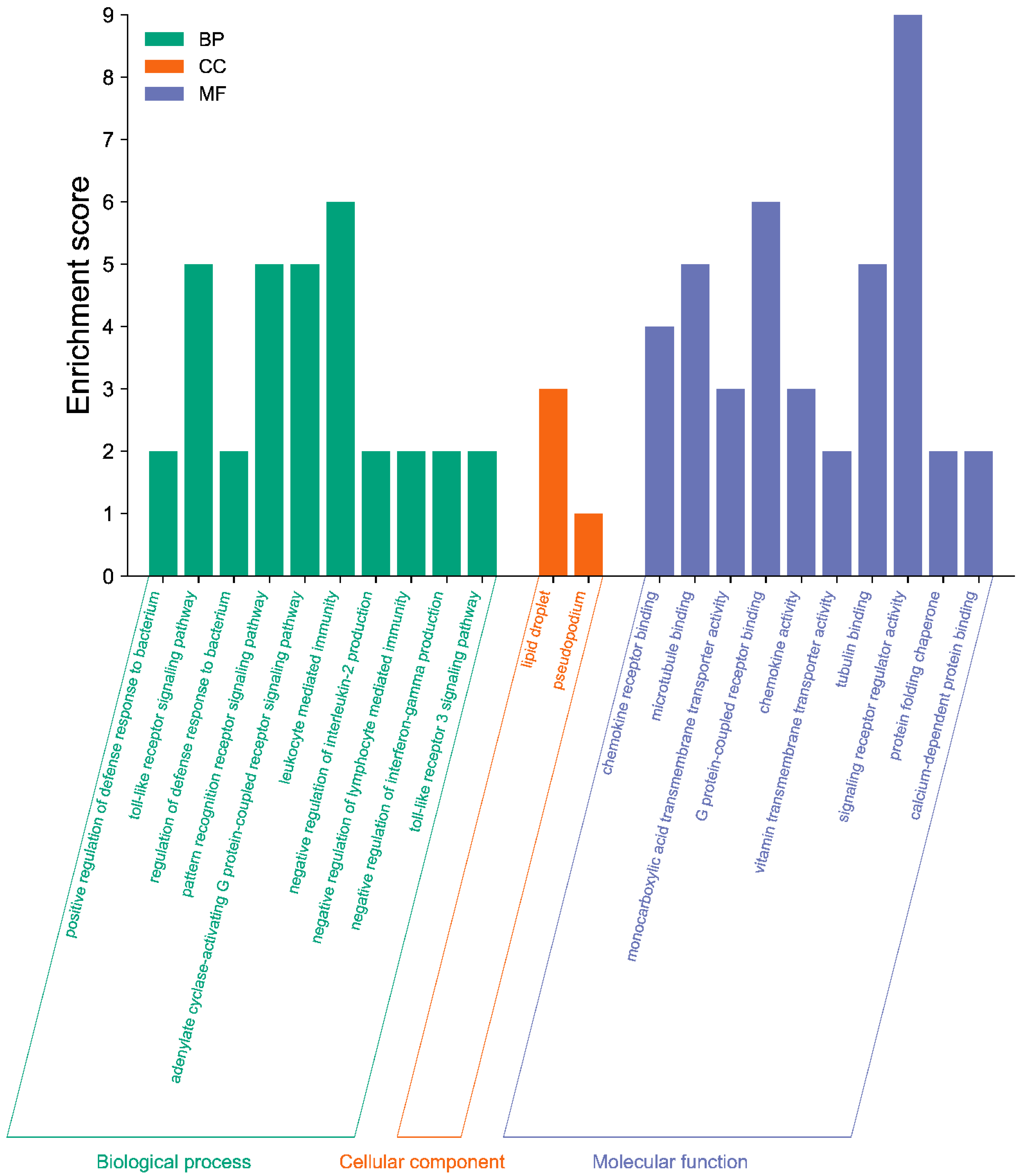
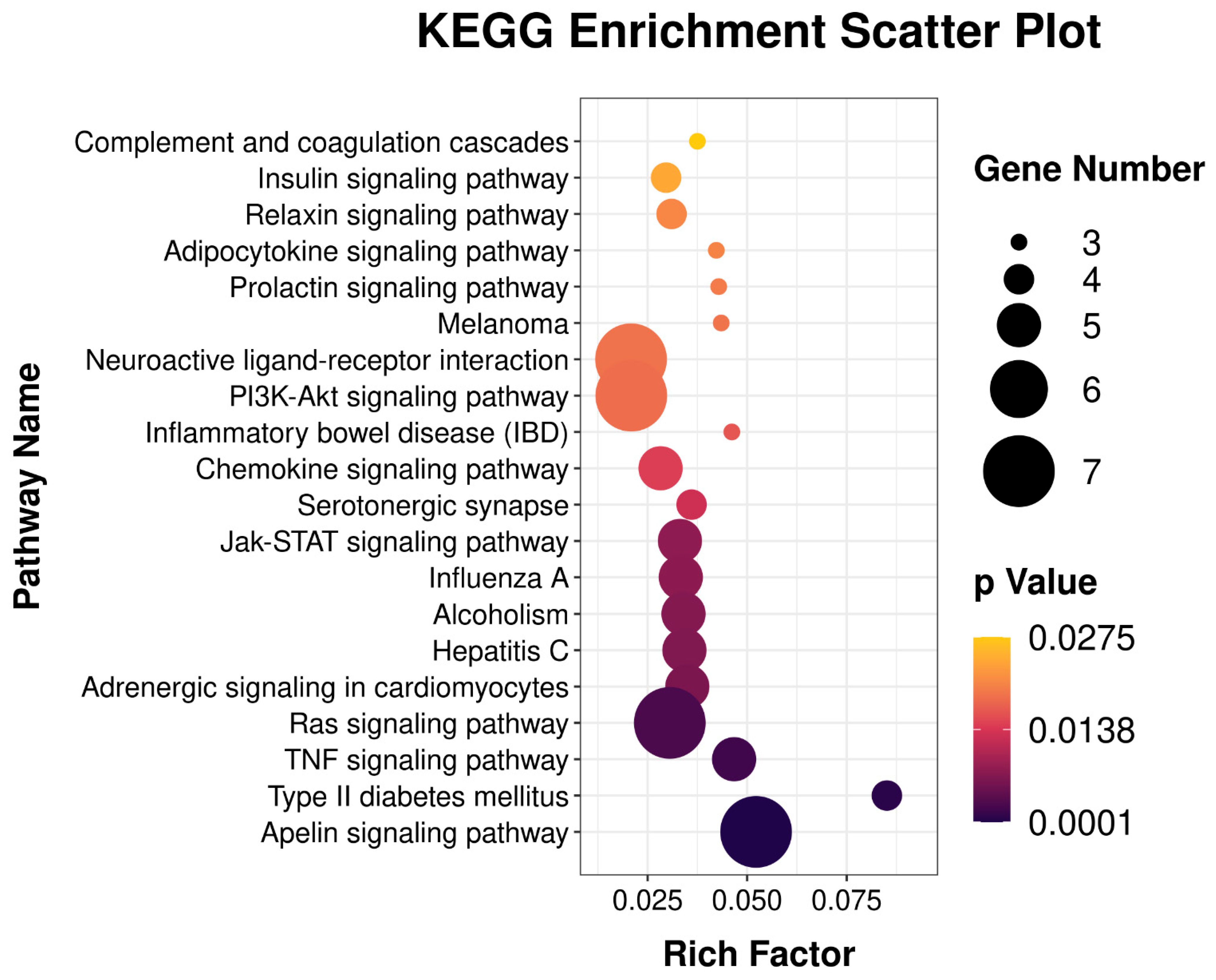
3.4. GO Enrichment and KEGG Pathway Analysis
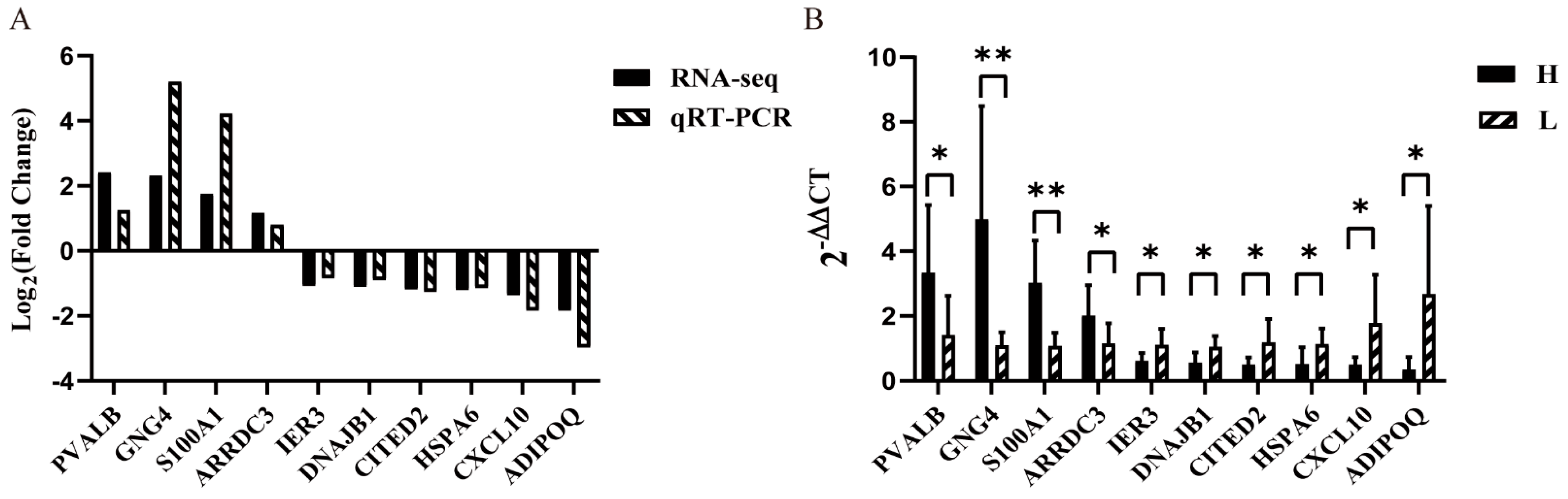
3.5. Validation of Gene Expression Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friesen, K.G.; Nelssen, J.L.; Goodband, R.D.; Tokach, M.D.; Unruh, J.A.; Kropf, D.H.; Kerr, B.J. The effect of dietary lysine on growth, carcass composition, and lipid metabolism in high-lean growth gilts fed from 72 to 136 kilograms. J. Anim. Sci. 1995, 73, 3392–3401. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, D.; Hernandez-Sanchez, J.; Chen, J.; Liu, C.; Wu, Z.; Fang, M.; Li, N. Genome Wide Association Analysis Reveals New Production Trait Genes in a Male Duroc Population. PLoS ONE 2015, 10, e0139207. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Kadowaki, H.; Shibata, T.; Uchida, H.; Nishida, A. Selection for daily gain, loin-eye area, backfat thickness and intramuscular fat based on desired gains over seven generations of Duroc pigs. Livest. Prod. Sci. 2005, 97, 193–202. [Google Scholar] [CrossRef]
- Kuhlers, D.L.; Nadarajah, K.; Jungst, S.B.; Anderson, B.L. Genetic selection for real-time ultrasound loin eye area in a closed line of Landrace pigs. Livest. Prod. Sci. 2001, 72, 225–231. [Google Scholar] [CrossRef]
- Zhuang, Z.; Li, S.; Ding, R.; Yang, M.; Zheng, E.; Yang, H.; Gu, T.; Xu, Z.; Cai, G.; Wu, Z.; et al. Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations. PLoS ONE 2019, 14, e0218263. [Google Scholar] [CrossRef]
- He, Y.; Ma, J.; Zhang, F.; Hou, L.; Chen, H.; Guo, Y.; Zhang, Z. Multi-breed genome-wide association study reveals heterogeneous loci associated with loin eye area in pigs. J. Appl. Genet. 2016, 57, 511–518. [Google Scholar] [CrossRef]
- Jung, E.J.; Park, H.B.; Lee, J.B.; Yoo, C.K.; Kim, B.M.; Kim, H.I.; Kim, B.W.; Lim, H.T. Genome-wide association analysis identifies quantitative trait loci for growth in a Landrace purebred population. Anim. Genet. 2014, 45, 442–444. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, L.; Li, Y.; Li, W.; Yan, H.; Liu, X.; Zhao, K.; Wang, L. Substitution within erythropoietin receptor gene D1 domain associated with litter size in Beijing Black pig, Sus scrofa. Anim. Sci. J. 2011, 82, 627–632. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Shiroguchi, K.; Jia, T.Z.; Sims, P.A.; Xie, X.S. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl. Acad. Sci. USA 2012, 109, 1347–1352. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Edwards, D.B.; Ernst, C.W.; Raney, N.E.; Doumit, M.E.; Hoge, M.D.; Bates, R.O. Quantitative trait locus mapping in an F2 Duroc x Pietrain resource population: II. Carcass and meat quality traits. J. Anim. Sci. 2008, 86, 254–266. [Google Scholar] [CrossRef]
- Cho, I.C.; Yoo, C.K.; Lee, J.B.; Jung, E.J.; Han, S.H.; Lee, S.S.; Ko, M.S.; Lim, H.T.; Park, H.B. Genome-wide QTL analysis of meat quality-related traits in a large F2 intercross between Landrace and Korean native pigs. Genet. Sel. Evol. 2015, 47, 7. [Google Scholar] [CrossRef] [PubMed]
- Paszek, A.A.; Wilkie, P.J.; Flickinger, G.H.; Miller, L.M.; Louis, C.F.; Rohrer, G.A.; Alexander, L.J.; Beattie, C.W.; Schook, L.B. Interval mapping of carcass and meat quality traits in a divergent swine cross. Anim. Biotechnol. 2001, 12, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Ponsuksili, S.; Chomdej, S.; Murani, E.; Bläser, U.; Schreinemachers, H.J.; Schellander, K.; Wimmers, K. SNP detection and genetic mapping of porcine genes encoding enzymes in hepatic metabolic pathways and evaluation of linkage with carcass traits. Anim. Genet. 2005, 36, 477–483. [Google Scholar] [CrossRef]
- Mankoo, B.S.; Collins, N.S.; Ashby, P.; Grigorieva, E.; Pevny, L.H.; Candia, A.; Wright, C.V.; Rigby, P.W.; Pachnis, V. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature 1999, 400, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Macharia, R.; Matsakas, A.; Valasek, P.; Mankoo, B.S.; Patel, K. A hypoplastic model of skeletal muscle development displaying reduced foetal myoblast cell numbers, increased oxidative myofibres and improved specific tension capacity. Dev. Biol. 2010, 343, 51–62. [Google Scholar] [CrossRef]
- Barton, E.R.; Pacak, C.A.; Stoppel, W.L.; Kang, P.B. The ties that bind: Functional clusters in limb-girdle muscular dystrophy. Skelet. Muscle 2020, 10, 22. [Google Scholar] [CrossRef]
- Gençpınar, P.; Uyanık, G.; Haspolat, Ş.; Oygür, N.; Duman, Ö. Clinical and Molecular Manifestations of Congenital Muscular Alpha-Dystroglycanopathy due to an ISPD Gene Mutation. Neurophysiology 2019, 51, 373–378. [Google Scholar] [CrossRef]
- Song, D.; Fu, X.; Ge, L.; Chang, X.; Wei, C.; Liu, J.; Yang, H.; Qu, S.; Bao, X.; Toda, T.; et al. A splice site mutation c.1251G>A of ISPD gene is a common cause of congenital muscular dystrophy in Chinese patients. Clin. Genet. 2020, 97, 789–790. [Google Scholar] [CrossRef]
- Badshah, I.I.; Brown, S.; Weibel, L.; Rose, A.; Way, B.; Sebire, N.; Inman, G.; Harper, J.; O’Shaughnessy, R.F.L. Differential expression of secreted factors SOSTDC1 and ADAMTS8 cause profibrotic changes in linear morphoea fibroblasts. Br. J. Dermatol. 2019, 180, 1135–1149. [Google Scholar] [CrossRef]
- Saita, S.; Shirane, M.; Ishitani, T.; Shimizu, N.; Nakayama, K.I. Role of the ANKMY2-FKBP38 axis in regulation of the Sonic hedgehog (Shh) signaling pathway. J. Biol. Chem. 2014, 289, 25639–25654. [Google Scholar] [CrossRef]
- Kozel, C.; Thompson, B.; Hustak, S.; Moore, C.; Nakashima, A.; Singh, C.R.; Reid, M.; Cox, C.; Papadopoulos, E.; Luna, R.E.; et al. Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation. Nucleic Acids Res. 2016, 44, 8704–8713. [Google Scholar] [CrossRef]
- Li, G.; Lu, A.; Chen, A.; Geng, S.; Xu, Y.; Chen, X.; Yang, J. BZW2/5MP1 acts as a promising target in hepatocellular carcinoma. J. Cancer 2021, 12, 5125–5135. [Google Scholar] [CrossRef]
- Wang, Y.W.; Zhao, S.; Yuan, X.Y.; Liu, Y.; Zhang, K.; Wang, J.; Zhu, J.; Ma, R. miR-4732-5p promotes breast cancer progression by targeting TSPAN13. J. Cell. Mol. Med. 2019, 23, 2549–2557. [Google Scholar] [CrossRef]
- Barraclough, D.L.; Platt-Higgins, A.; de Silva Rudland, S.; Barraclough, R.; Winstanley, J.; West, C.R.; Rudland, P.S. The metastasis-associated anterior gradient 2 protein is correlated with poor survival of breast cancer patients. Am. J. Pathol. 2009, 175, 1848–1857. [Google Scholar] [CrossRef]
- Liu, D.; Rudland, P.S.; Sibson, D.R.; Platt-Higgins, A.; Barraclough, R. Human homologue of cement gland protein, a novel metastasis inducer associated with breast carcinomas. Cancer Res. 2005, 65, 3796–3805. [Google Scholar] [CrossRef]
- Park, K.; Chung, Y.J.; So, H.; Kim, K.; Park, J.; Oh, M.; Jo, M.; Choi, K.; Lee, E.J.; Choi, Y.L.; et al. AGR2, a mucinous ovarian cancer marker, promotes cell proliferation and migration. Exp. Mol. Med. 2011, 43, 91–100. [Google Scholar] [CrossRef]
- Zhang, J.S.; Gong, A.; Cheville, J.C.; Smith, D.I.; Young, C.Y. AGR2, an androgen-inducible secretory protein overexpressed in prostate cancer. Genes Chromosomes Cancer 2005, 43, 249–259. [Google Scholar] [CrossRef]
- Dumartin, L.; Whiteman, H.J.; Weeks, M.E.; Hariharan, D.; Dmitrovic, B.; Iacobuzio-Donahue, C.A.; Brentnall, T.A.; Bronner, M.P.; Feakins, R.M.; Timms, J.F.; et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011, 71, 7091–7102. [Google Scholar] [CrossRef]
- Gomez, A.; Bindesbøll, C.; Satheesh, S.V.; Grimaldi, G.; Hutin, D.; MacPherson, L.; Ahmed, S.; Tamblyn, L.; Cho, T.; Nebb, H.I.; et al. Characterization of TCDD-inducible poly-ADP-ribose polymerase (TIPARP/ARTD14) catalytic activity. Biochem. J. 2018, 475, 3827–3846. [Google Scholar] [CrossRef]
- MacPherson, L.; Tamblyn, L.; Rajendra, S.; Bralha, F.; McPherson, J.P.; Matthews, J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013, 41, 1604–1621. [Google Scholar] [CrossRef]
- Ema, M.; Ohe, N.; Suzuki, M.; Mimura, J.; Sogawa, K.; Ikawa, S.; Fujii-Kuriyama, Y. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J. Biol. Chem. 1994, 269, 27337–27343. [Google Scholar] [CrossRef]
- Sadik, A.; Somarribas Patterson, L.F.; Öztürk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfänder, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270.e34. [Google Scholar] [CrossRef] [PubMed]
- Rincón, G.; Islas-Trejo, A.; Casellas, J.; Ronin, Y.; Soller, M.; Lipkin, E.; Medrano, J.F. Fine mapping and association analysis of a quantitative trait locus for milk production traits on Bos taurus autosome 4. J. Dairy Sci. 2009, 92, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Hara, K.; Kubota, N.; Terauchi, Y.; Tobe, K.; Froguel, P.; Nagai, R.; Kadowaki, T. Dual roles of adiponectin/Acrp30 in vivo as an anti-diabetic and anti-atherogenic adipokine. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2003, 3, 243–254. [Google Scholar] [CrossRef]
- Krause, M.P.; Liu, Y.; Vu, V.; Chan, L.; Xu, A.; Riddell, M.C.; Sweeney, G.; Hawke, T.J. Adiponectin is expressed by skeletal muscle fibers and influences muscle phenotype and function. Am. J. Physiol. Cell Physiol. 2008, 295, C203–C212. [Google Scholar] [CrossRef]
- Kayan, A.; Uddin, M.J.; Cinar, M.U.; Grosse-Brinkhaus, C.; Phatsara, C.; Wimmers, K.; Ponsuksili, S.; Tesfaye, D.; Looft, C.; Juengst, H.; et al. Investigation on interferon alpha-inducible protein 6 (IFI6) gene as a candidate for meat and carcass quality in pig. Meat Sci. 2011, 88, 755–760. [Google Scholar] [CrossRef]
- Hirose, K.; Ito, T.; Fukawa, K.; Arakawa, A.; Mikawa, S.; Hayashi, Y.; Tanaka, K. Evaluation of effects of multiple candidate genes (LEP, LEPR, MC4R, PIK3C3, and VRTN) on production traits in Duroc pigs. Anim. Sci. J. 2014, 85, 198–206. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, C.; Wang, X.; Gong, Z. Leptin induces muscle wasting in a zebrafish kras-driven hepatocellular carcinoma (HCC) model. Dis. Model. Mech. 2019, 12, dmm038240. [Google Scholar] [CrossRef]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT Pathway in Skeletal Muscle Pathophysiology. Front. Physiol. 2019, 10, 500. [Google Scholar] [CrossRef]
- Elabd, C.; Cousin, W.; Upadhyayula, P.; Chen, R.Y.; Chooljian, M.S.; Li, J.; Kung, S.; Jiang, K.P.; Conboy, I.M. Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 2014, 5, 4082. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Black, B.L.; Martin, J.F.; Olson, E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 1995, 83, 1125–1136. [Google Scholar] [CrossRef]
- Haberland, M.; Arnold, M.A.; McAnally, J.; Phan, D.; Kim, Y.; Olson, E.N. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol. Cell. Biol. 2007, 27, 518–525. [Google Scholar] [CrossRef] [PubMed]
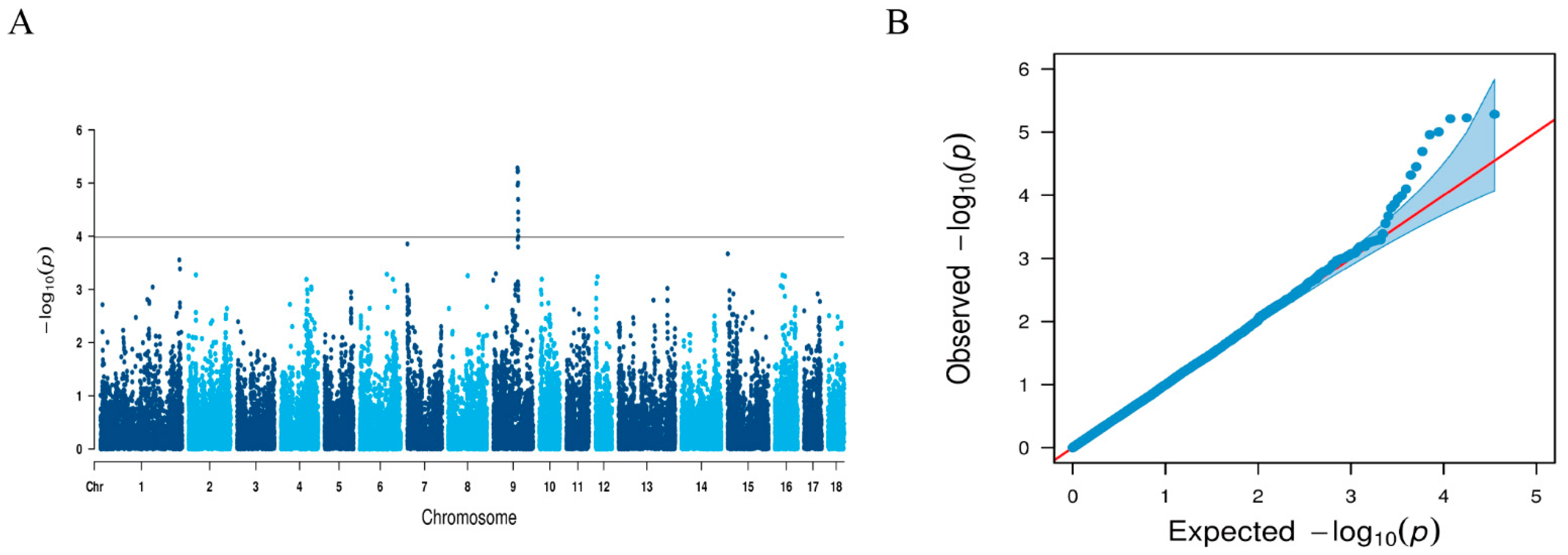
| Marker | Position a | Rs b | p-Value | Var (%) c |
|---|---|---|---|---|
| WU_10.2_9_93503815 | 84,937,219 | rs336943465 | 5.19 × 10−6 | 2.08 |
| ALGA0054101 | 85,233,732 | rs81413966 | 1.10 × 10−5 | 1.94 |
| ASGA0043938 | 85,259,596 | rs81413967 | 6.12 × 10−6 | 2.05 |
| ASGA0043959 | 86,791,976 | rs81414045 | 7.94 × 10−5 | 1.56 |
| MARC0073290 | 86,843,385 | rs81259452 | 5.91 × 10−6 | 2.06 |
| MARC0046912 | 86,880,793 | rs81238869 | 2.02 × 10−5 | 1.82 |
| MARC0065750 | 86,920,421 | rs81253220 | 9.91 × 10−6 | 1.96 |
| MARC0055652 | 87,156,018 | rs81245907 | 3.52 × 10−5 | 1.71 |
| ASGA0043969 | 87,327,648 | rs81414101 | 4.76 × 10−5 | 1.66 |
| MARC0069139 | 87,837,573 | rs81256749 | 1.01 × 10−4 | 1.51 |
| Sample_Name | Raw_Reads | Clean_Reads | Q30 (%) | GC_Content (%) | Mapped (%) |
|---|---|---|---|---|---|
| H1 | 42,921,002 | 41,411,838 | 98.03 | 52.00 | 96.48 |
| H2 | 51,521,550 | 50,088,198 | 98.55 | 51.50 | 97.22 |
| H3 | 49,671,858 | 47,485,598 | 97.96 | 52.00 | 95.60 |
| H4 | 45,497,868 | 44,432,658 | 97.49 | 52.00 | 97.66 |
| H5 | 40,988,148 | 39,848,532 | 97.98 | 52.00 | 97.22 |
| H6 | 39,841,846 | 38,699,270 | 97.97 | 52.00 | 97.13 |
| H7 | 47,118,756 | 45,666,934 | 98.21 | 51.00 | 96.92 |
| H8 | 50,646,242 | 49,107,270 | 97.97 | 52.00 | 96.96 |
| L1 | 54,959,674 | 52,970,650 | 98.22 | 52.00 | 96.38 |
| L2 | 52,534,364 | 51,224,866 | 98.06 | 52.00 | 97.51 |
| L3 | 45,241,998 | 43,696,868 | 98.32 | 51.00 | 96.58 |
| L4 | 42,172,534 | 41,077,200 | 98.25 | 51.50 | 97.40 |
| L5 | 41,125,930 | 39,957,600 | 98.17 | 52.00 | 97.16 |
| L6 | 38,604,910 | 37,465,804 | 98.13 | 52.00 | 97.05 |
| L7 | 52,999,242 | 51,634,384 | 98.00 | 53.00 | 97.42 |
| L8 | 38,195,034 | 37,346,508 | 98.25 | 52.00 | 97.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, R.; Chen, L.; Liu, X.; Liu, H.; Shi, G.; Hou, X.; Zhang, R.; Yang, M.; Niu, N.; Wang, L.; et al. Integrating Genome-Wide Association Study with RNA-Sequencing Reveals HDAC9 as a Candidate Gene Influencing Loin Muscle Area in Beijing Black Pigs. Biology 2022, 11, 1635. https://doi.org/10.3390/biology11111635
Hou R, Chen L, Liu X, Liu H, Shi G, Hou X, Zhang R, Yang M, Niu N, Wang L, et al. Integrating Genome-Wide Association Study with RNA-Sequencing Reveals HDAC9 as a Candidate Gene Influencing Loin Muscle Area in Beijing Black Pigs. Biology. 2022; 11(11):1635. https://doi.org/10.3390/biology11111635
Chicago/Turabian StyleHou, Renda, Li Chen, Xiance Liu, Hai Liu, Guohua Shi, Xinhua Hou, Run Zhang, Man Yang, Naiqi Niu, Lixian Wang, and et al. 2022. "Integrating Genome-Wide Association Study with RNA-Sequencing Reveals HDAC9 as a Candidate Gene Influencing Loin Muscle Area in Beijing Black Pigs" Biology 11, no. 11: 1635. https://doi.org/10.3390/biology11111635
APA StyleHou, R., Chen, L., Liu, X., Liu, H., Shi, G., Hou, X., Zhang, R., Yang, M., Niu, N., Wang, L., & Zhang, L. (2022). Integrating Genome-Wide Association Study with RNA-Sequencing Reveals HDAC9 as a Candidate Gene Influencing Loin Muscle Area in Beijing Black Pigs. Biology, 11(11), 1635. https://doi.org/10.3390/biology11111635





