About the Analysis of 18S rDNA Sequence Data from Trypanosomes in Barcoding and Phylogenetics: Tracing a Continuation Error Occurring in the Literature
Abstract
:Simple Summary
Abstract
1. Introduction
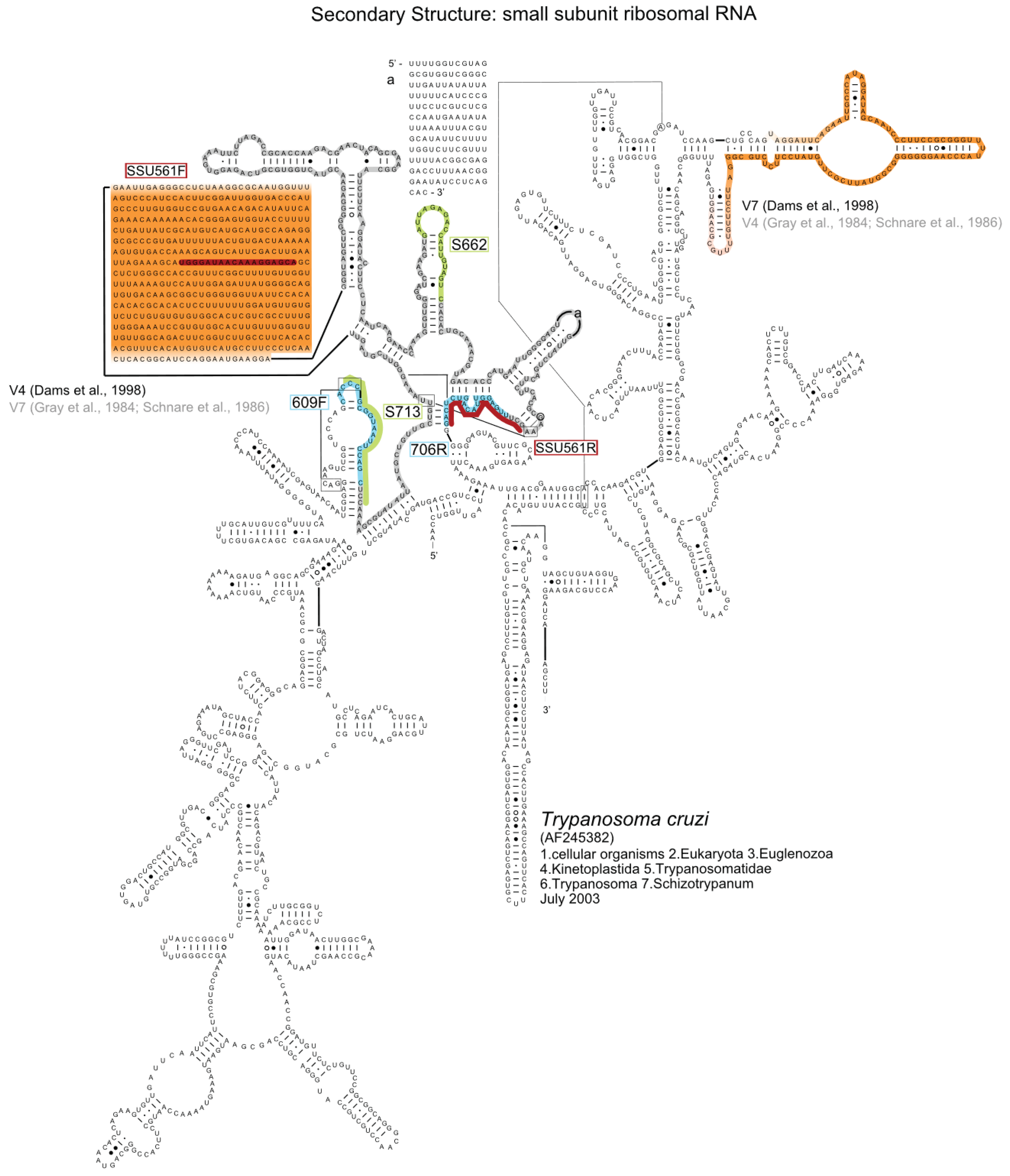
2. Results and Discussion

Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothschild, L.J.; Ragan, M.A.; Coleman, A.W.; Heywood, P.; Gerbi, S.A. Are rRNA sequence comparisons the Rosetta stone of phylogenetics? Cell 1986, 47, 640. [Google Scholar] [CrossRef]
- Olsen, G.J.; Woese, C.R. Ribosomal RNA: A key to phylogeny. FASEB J. 1993, 7, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, J.S. Comparative analyses of the V4 and V9 regions of 18S rDNA for the extant eukaryotic community using the Illumina platform. Sci. Rep. 2020, 10, 6519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiegler, P.; Carbon, P.; Ebel, J.-P.; Ehresmann, C. A General Secondary-Structure Model for Procaryotic and Eucaryotic RNAs of the Small Ribosomal Subunits. JBIC J. Biol. Inorg. Chem. 1981, 120, 487–495. [Google Scholar] [CrossRef]
- Spencer, D.F.; Schnare, M.N.; Gray, M.W. Pronounced structural similarities between the small subunit ribosomal RNA genes of wheat mitochondria and Escherichia coli. Proc. Natl. Acad. Sci. USA 1984, 81, 493–497. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.; Sankoff, D.; Cedergren, R.J. On the evolutionary descent of organisms and organelles: A global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984, 12, 5837–5852. [Google Scholar] [CrossRef]
- Schnare, M.N.; Collings, J.C.; Gray, M.W. Structure and evolution of the small subunit ribosomal RNA gene of Crithidia fasciculata. Curr. Genet. 1986, 10, 405–410. [Google Scholar] [CrossRef]
- Hernández, R.; Rios, P.; Valdes, A.; Piñero, D. Primary structure of Trypanosoma cruzi small-subunit ribosomal RNA coding region: Comparison with other trypanosomatids. Mol. Biochem. Parasitol. 1990, 41, 207–212. [Google Scholar] [CrossRef]
- Van de Peer, Y.; De Rijk, P.; Wuyts, J.; Winkelmans, T.; De Wachter, R. The European Small Subunit Ribosomal RNA database. Nucleic Acids Res. 2002, 28, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Huysmans, E.; De Wachter, R. Compilation of small ribosomal sobunit RNA sequences. Nucleic Acids Res. 1986, 14, r73–r118. [Google Scholar] [CrossRef]
- Dams, E.; Hendriks, L.; Van De Peer, Y.; Neefs, J.-M.; Smits, G.; Vandenbempt, I.; De Wachter, R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988, 16, r87–r173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neefs, J.-M.; De Wachter, R. A proposal for the secondary structure of a variable area of eukaryotic small ribosomal subunit RNA involving the existence of a pseudoknot. Nucleic Acids Res. 1990, 18, 5695–5704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neefs, J.-M.; Van de Peer, Y.; Hendriks, L.; De Wachter, R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990, 18, 2237–2317. [Google Scholar] [CrossRef] [PubMed]
- Neefs, J.-M.; Van de Peer, Y.; De Rijk, P.; Chapelle, S.; De Wachter, R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993, 21, 3025–3049. [Google Scholar] [CrossRef] [PubMed]
- Neefs, J.-M.; Van de Peer, Y.; De Rijk, P.; Goris, A.; De Wachter, R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991, 19, 1987–2015. [Google Scholar] [CrossRef] [Green Version]
- De Rijk, P.; Neefs, J.-M.; Van De Peer, Y.; De Wachter, R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1992, 20, 2075–2089. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Lin, J.; Qin, Y.; Zhou, J.; Bu, W. Structural diversity of eukaryotic 18S rRNA and its impact on alignment and phylogenetic reconstruction. Protein Cell 2011, 2, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Ki, J.-S. Hypervariable regions (V1–V9) of the dinoflagellate 18S rRNA using a large dataset for marker considerations. J. Appl. Phycol. 2011, 24, 1035–1043. [Google Scholar] [CrossRef]
- Bininda-Emonds, O.R.P. 18S rRNA variability maps reveal three highly divergent, conserved motifs within Rotifera. BMC Ecol. Evol. 2021, 21, 118. [Google Scholar] [CrossRef]
- Bradley, I.M.; Pinto, A.J.; Guest, J.S. Design and Evaluation of Illumina MiSeq-Compatible, 18S rRNA Gene-Specific Primers for Improved Characterization of Mixed Phototrophic Communities. Appl. Environ. Microbiol. 2016, 82, 5878–5891. [Google Scholar] [CrossRef]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.-W.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Harder, C.B.; Rønn, R.; Brejnrod, A.; Bass, D.; Abu Al-Soud, W.; Ekelund, F. Local diversity of heathland Cercozoa explored by in-depth sequencing. ISME J. 2016, 10, 2488–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taib, N.; Mangot, J.-F.; Domaizon, I.; Bronner, G.; Debroas, D. Phylogenetic Affiliation of SSU rRNA Genes Generated by Massively Parallel Sequencing: New Insights into the Freshwater Protist Diversity. PLoS ONE 2013, 8, e58950. [Google Scholar] [CrossRef]
- Hadziavdic, K.; Lekang, K.; Lanzén, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA Gene for Designing Universal Eukaryote Specific Primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.K.; Liu, Z.; Lie, A.A.Y.; Countway, P.D.; Kim, D.Y.; Jones, A.C.; Gast, R.J.; Cary, S.C.; Sherr, E.B.; Sherr, B.F.; et al. Estimating Protistan Diversity Using High-Throughput Sequencing. J. Eukaryot. Microbiol. 2015, 62, 688–693. [Google Scholar] [CrossRef]
- Latz, M.A.C.; Grujcic, V.; Brugel, S.; Lycken, J.; John, U.; Karlson, B.; Andersson, A.; Andersson, A.F. Short- and long-read metabarcoding of the eukaryotic rRNA operon: Evaluation of primers and comparison to shotgun metagenomics sequencing. Mol. Ecol. Resour. 2022, 22, 2304–2318. [Google Scholar] [CrossRef] [PubMed]
- DA Silva, F.M.; Noyes, H.; Campaner, M.; Junqueira, A.C.V.; Coura, J.R.; Añez, N.; Shaw, J.J.; Stevens, J.R.; Teixeira, M.M.G. Phylogeny, taxonomy and grouping of Trypanosoma rangeli isolates from man, triatomines and sylvatic mammals from widespread geographical origin based on SSU and ITS ribosomal sequences. Parasitology 2004, 129, 549–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Votýpka, J.; Svobodová, M. Trypanosoma avium: Experimental transmission from black flies to canaries. Parasitol Res. 2004, 92, 147–151. [Google Scholar] [CrossRef]
- Votýpka, J.; Oborník, M.; Volf, P.; Svobodová, M.; Lukeš, J. Trypanosoma avium of raptors (Falconiformes): Phylogeny and identification of vectors. Parasitology 2002, 125, 253–263. [Google Scholar] [CrossRef]
- Nickrent, D.L.; Sargent, M.L. An overview of the secondary structure of the V4 region of eukaryotic small-subunit ribosomal RNA. Nucleic Acids Res. 1991, 19, 227–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchinson, R.; Stevens, J. Barcoding in trypanosomes. Parasitology 2017, 145, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noyes, H.A.; Camps, A.P.; Chance, M.L. Leishmania herreri (Kinetoplastida; Trypanosomatidae) is more closely related to Endotrypanum (Kinetoplastida; Trypanosomatidae) than to Leishmania. Mol. Biochem. Parasitol. 1996, 80, 119–123. [Google Scholar] [CrossRef]
| Authors | Year of Publication | Digital Object Identifier |
|---|---|---|
| Maia da Silva et al. | 2004 | 10.1017/S0031182004005931 |
| Rodrigues et al. | 2006 | 10.1017/S0031182005008929 |
| Cortez et al. | 2006 | 10.1017/S0031182006000254 |
| Ferreira et al. | 2007 | 10.1017/S0031182007003058 |
| Maia da Silva et al. | 2007 | 10.1111/j.1365-294X.2007.03371.x |
| Martins et al. | 2008 | 10.4269/ajtmh.2008.79.427 |
| Viola et al. | 2008 | 10.1017/S0031182008004253 |
| Rodrigues et al. | 2008 | 10.1017/S0031182008004848 |
| Marcili et al. | 2009 | 10.1017/S0031182009005861 |
| Viola et al. | 2009 | 10.1017/S003118200800512X |
| Marcili et al. | 2009 | 10.1016/j.meegid.2009.07.003 |
| Marcili et al. | 2009 | 10.1016/j.ijpara.2008.09.015 |
| Averis et al. | 2009 | 10.1017/S0031182009990801 |
| Maia da Silva et al. | 2009 | 10.1016/j.actatropica.2008.11.005 |
| Maia da Silva et al. | 2010 | 10.1016/j.meegid.2010.02.005 |
| Cavazzana et al. | 2010 | 10.1016/j.ijpara.2009.08.015 |
| Teixeira et al. | 2011 | 10.1016/j.protis.2011.01.001 |
| Garcia et al. | 2011 | 10.1016/j.ijpara.2011.09.001 |
| Lima et al. | 2012 | 10.1016/j.protis.2011.12.003 |
| Martinković et al. | 2012 | 10.1111/j.1550-7408.2011.00599.x |
| Hamilton et al. | 2012 | 10.1016/j.ympev.2012.01.007 |
| Ramirez et al. | 2012 | 10.1016/j.exppara.2012.09.017 |
| Borghesan et al. | 2013 | 10.1016/j.protis.2012.06.001 |
| Marcili et al. | 2013 | 10.5402/2013/328794 |
| Lima et al. | 2013 | 10.1186/1756-3305-6-221 |
| Fermino et al. | 2013 | 10.1186/1756-3305-6-313 |
| Silva-Iturriza et al. | 2013 | 10.1016/j.parint.2012.10.003 |
| Marcili et al. | 2013 | 10.1645/12-156.1 |
| Guhl et al. | 2013 | 10.1016/j.meegid.2013.08.028 |
| Acosta et al. | 2014 | 10.1603/ME13177 |
| Marcili et al. | 2014 | 10.1016/j.meegid.2014.04.001 |
| Da Costa et al. | 2014 | 10.4172/ijbbd.1000120 |
| Lemos et al. | 2015 | 10.1186/s13071-015-1193-7 |
| Fermino et al. | 2015 | 10.1016/j.ijppaw.2015.10.005 |
| Juliana et al. | 2015 | 10.1007/s11230-015-9558-z |
| Lima et al. | 2015 | 10.1186/s13071-015-1255-x |
| Da Costa et al. | 2015 | 10.1089/vbz.2015.1771 |
| Da Costa et al. | 2015 | 10.1089/vbz.2015.1866 |
| Lima et al. | 2015 | 10.1016/j.actatropica.2015.07.015 |
| Martins et al. | 2015 | 10.1515/ap-2015-0009 |
| Dario et al. | 2016 | 10.1186/s13071-016-1754-4 |
| Attias et al. | 2016 | 10.1111/jeu.12310 |
| Zanetti et al. | 2016 | 10.1016/j.ejop.2016.09.004 |
| Szpeiter et al. | 2017 | 10.1590/s1984-29612017022 |
| Galvis-Ovallos | 2017 | 10.1186/s13071-017-2211-8 |
| Da Costa et al. | 2018 | 10.1590/0037-8682-0098-2018 |
| Ribeiro et al. | 2018 | 10.4269/ajtmh.16-0200 |
| Pacheco et al. | 2018 | 10.1590/s1984-296120180049 |
| Dos Santos et al. | 2018 | 10.1017/S0031182017001834 |
| Espinosa et al. | 2018 | 10.1017/S0031182016002092 |
| Borghesan et al. | 2018 | 10.3389/fmicb.2018.00131 |
| Espinosa-Álvarez et al. | 2018 | 10.1016/j.ijpara.2017.12.008 |
| Suganuma et al. | 2019 | 10.1007/s00436-019-06313-x |
| Borges et al. | 2019 | 10.1111/jeu.12678 |
| Barros et al. | 2019 | 10.1016/j.ijppaw.2018.12.009 |
| Fermino et al. | 2019 | 10.1186/s13071-019-3463-2 |
| Pérez et al. | 2019 | 10.1186/s13071-019-3726-y |
| Garcia et al. | 2019 | 10.1007/s10393-019-01440-4 |
| Latif et al. | 2019 | 10.4102/ojvr.v86i1.1634 |
| Kuhls et al. | 2019 | 10.1007/978-1-4939-9210-2_2 |
| Barros et al. | 2020 | 10.3390/pathogens9090736 |
| Garcia et al. | 2020 | 10.1186/s13071-020-04169-0 |
| Rodrigues et al. | 2020 | 10.1016/j.meegid.2019.104143 |
| Boucinha et al. | 2020 | 10.1590/0074-02760200504 |
| e Azevedo et al. | 2020 | 10.1590/0103-8478cr20200262 |
| Marcili et al. | 2020 | 10.1089/vbz.2020.2638 |
| Jaimes-Dueñez et al. | 2020 | 10.1016/j.prevetmed.2020.105159 |
| Dario et al. | 2021 | 10.3390/pathogens10060736 |
| Rosyadi et al. | 2021 | 10.1017/S0031182021001360 |
| Mule et al. | 2021 | 10.1038/s42003-021-01762-6 |
| Dario et al. | 2021 | 10.1016/j.ijppaw.2021.04.003 |
| Ardila et al. | 2022 | 10.1007/s12639-021-01459-x |
| Yasein et al. | 2022 | 10.29261/pakvetj/2022.034 |
| Chiariello et al. | 2022 | 10.1016/j.ijppaw.2021.11.006 |
| Kostygov et al. | 2022 | 10.1186/s13071-022-05212-y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rackevei, A.S.; Borges, A.; Engstler, M.; Dandekar, T.; Wolf, M. About the Analysis of 18S rDNA Sequence Data from Trypanosomes in Barcoding and Phylogenetics: Tracing a Continuation Error Occurring in the Literature. Biology 2022, 11, 1612. https://doi.org/10.3390/biology11111612
Rackevei AS, Borges A, Engstler M, Dandekar T, Wolf M. About the Analysis of 18S rDNA Sequence Data from Trypanosomes in Barcoding and Phylogenetics: Tracing a Continuation Error Occurring in the Literature. Biology. 2022; 11(11):1612. https://doi.org/10.3390/biology11111612
Chicago/Turabian StyleRackevei, Antonia S., Alyssa Borges, Markus Engstler, Thomas Dandekar, and Matthias Wolf. 2022. "About the Analysis of 18S rDNA Sequence Data from Trypanosomes in Barcoding and Phylogenetics: Tracing a Continuation Error Occurring in the Literature" Biology 11, no. 11: 1612. https://doi.org/10.3390/biology11111612
APA StyleRackevei, A. S., Borges, A., Engstler, M., Dandekar, T., & Wolf, M. (2022). About the Analysis of 18S rDNA Sequence Data from Trypanosomes in Barcoding and Phylogenetics: Tracing a Continuation Error Occurring in the Literature. Biology, 11(11), 1612. https://doi.org/10.3390/biology11111612








