Molecular Diversity and Evolutionary Relatedness of Paulownia Witches’-Broom Phytoplasma in Different Geographical Distributions in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
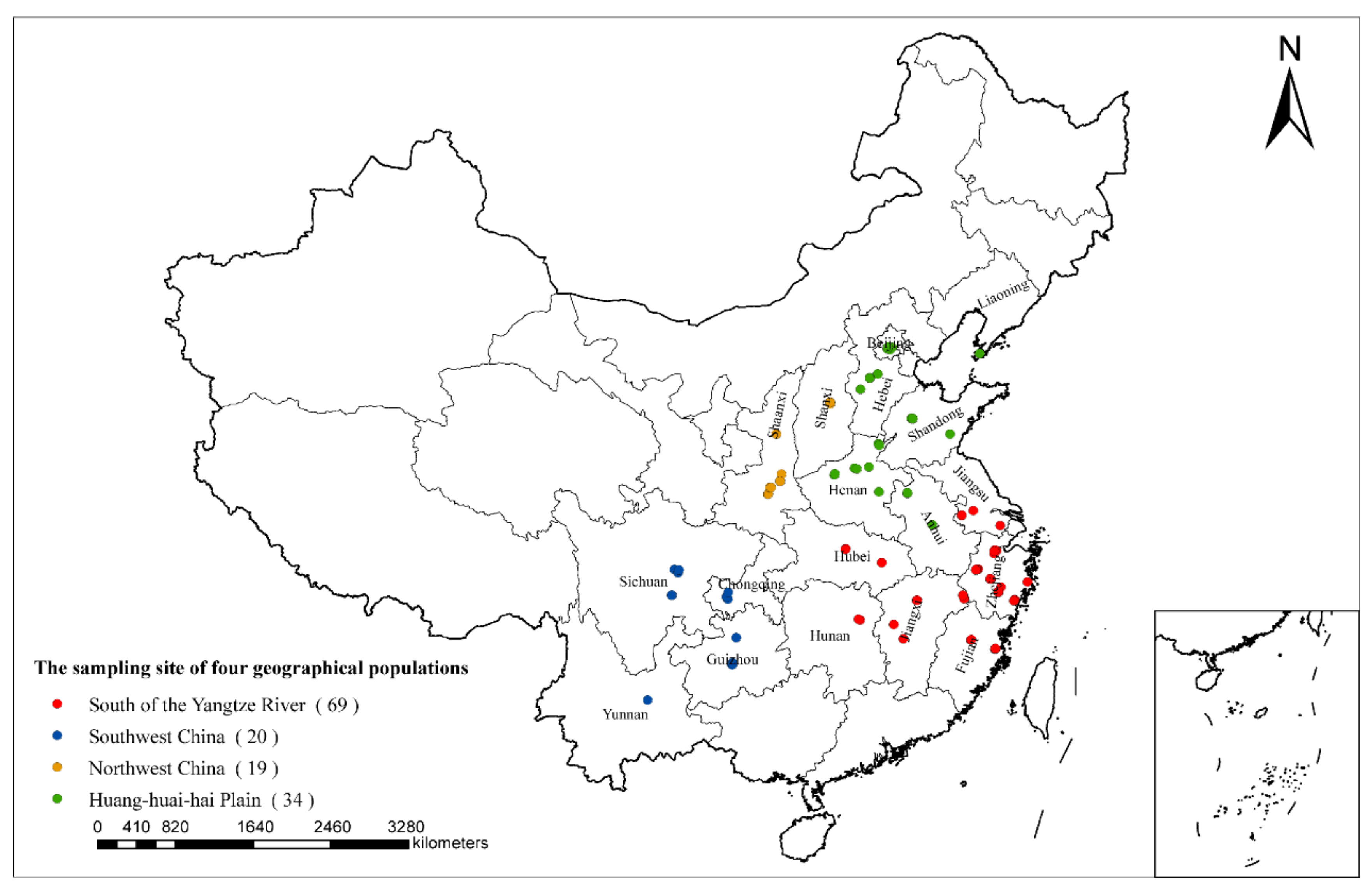
2.1. Plant Material Collection and DNA Preparation
2.2. Selection and Amplification of Housekeeping Genes
2.3. PaWB Phytoplasma Diversity Analysis
2.4. Split Network and Recombination Analysis
2.5. Molecular Typing and Phylogeny of PaWB Phytoplasmas
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Genetic Diversity of 16S rRNA Gene and Different Housekeeping Genes in PaWB Phytoplasma
3.2. Geographical Distribution and Population Genetics Analysis of PaWB Phytoplasma
3.2.1. Genetic Diversity of PaWB Phytoplasma in Different Geographical Populations
3.2.2. Population Genetics and Mismatch Distribution Analysis of PaWB Phytoplasma
3.3. Allelic Sequences Recombination Analysis
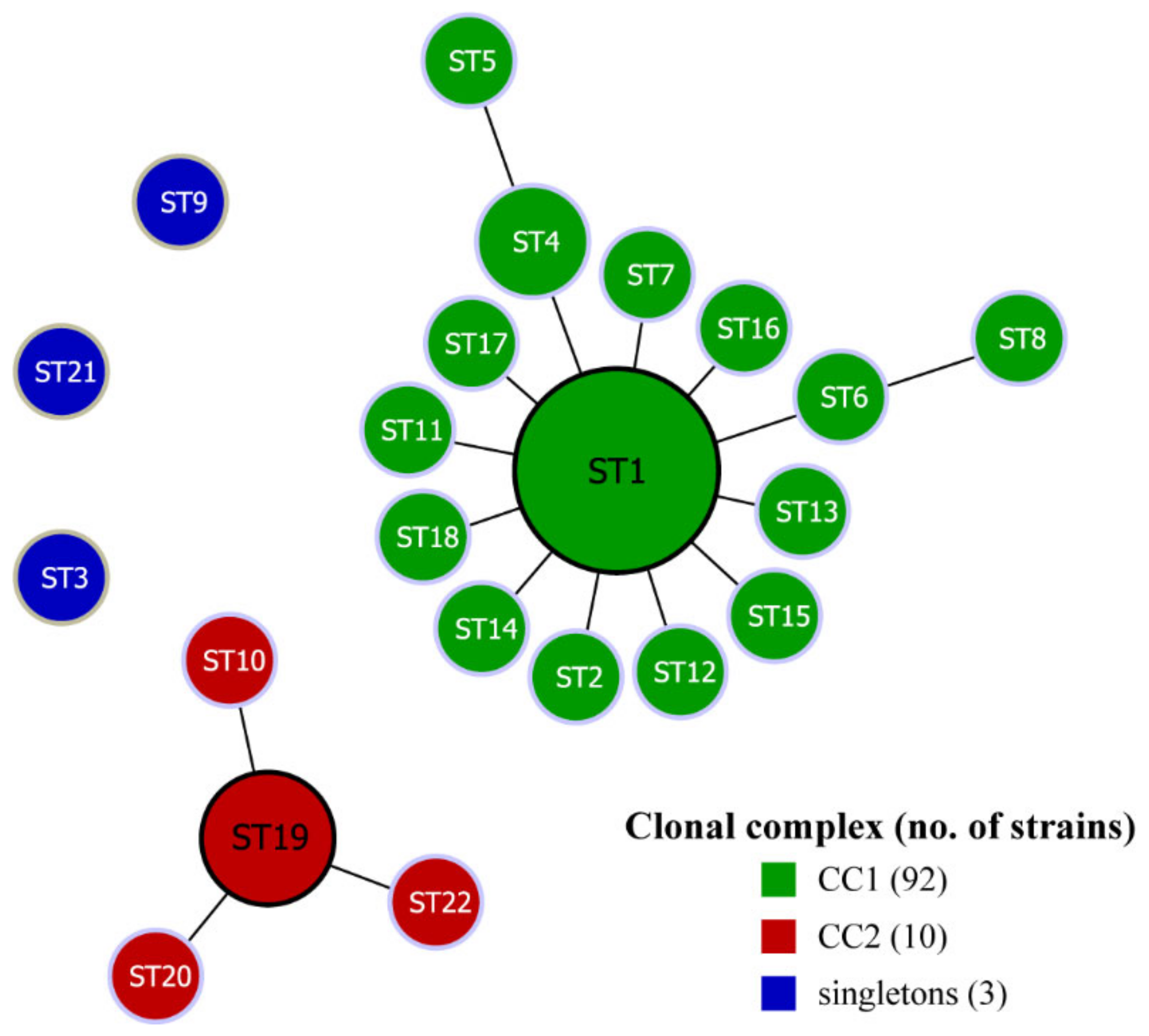
3.4. Evolutionary Analysis with goeBURST Algorithm
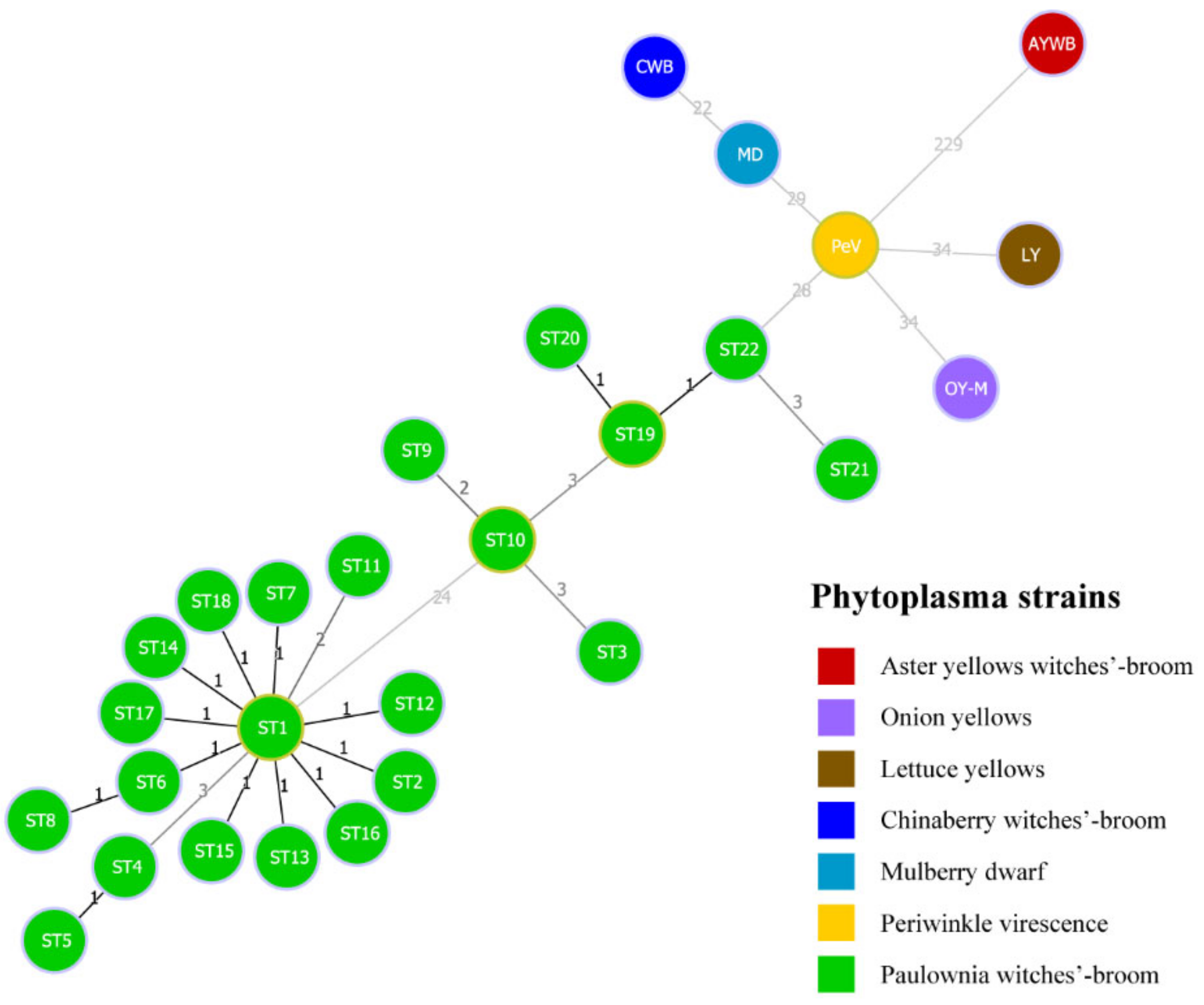
3.5. Minimum Spanning Tree Analysis
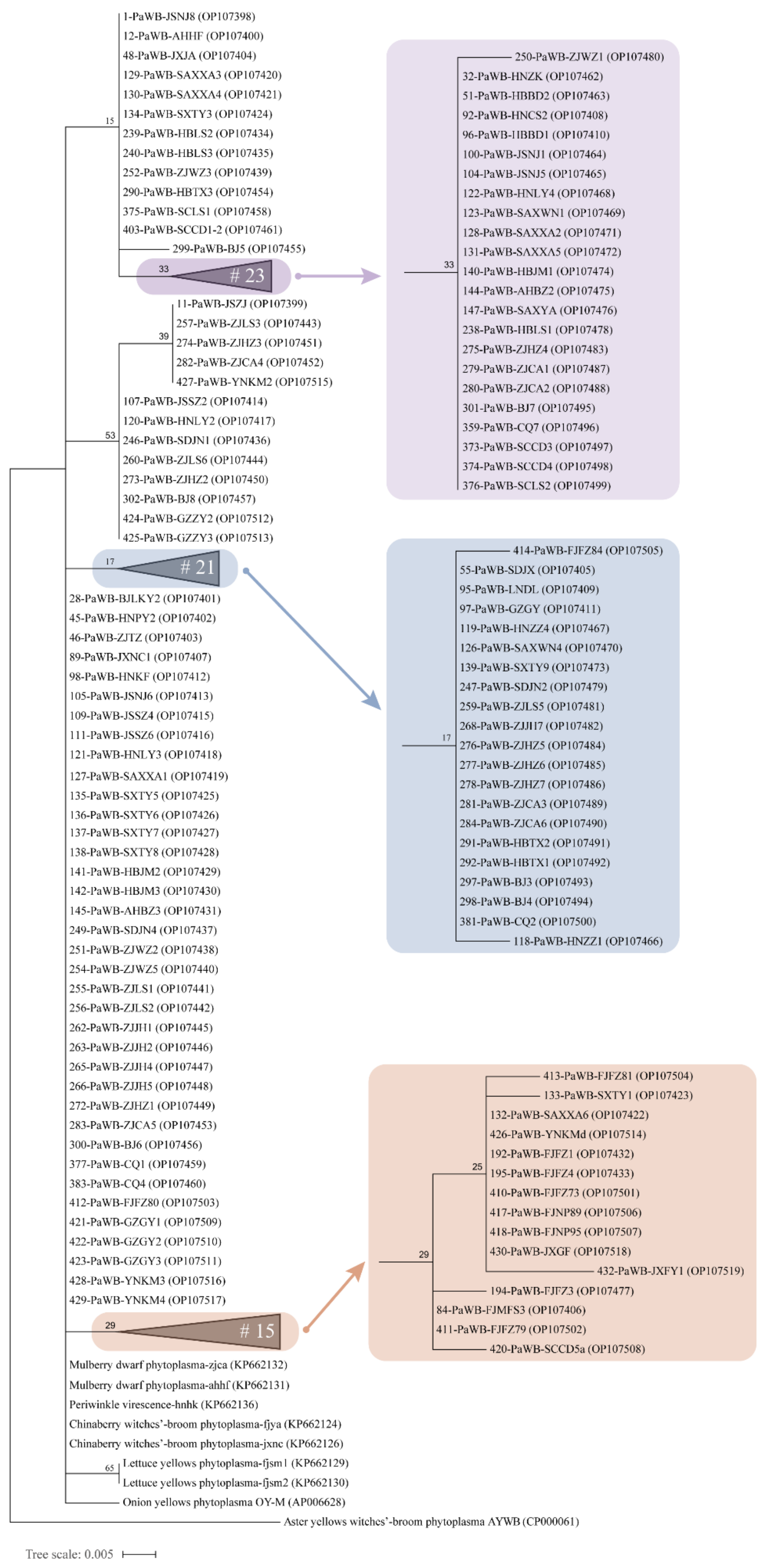
3.6. Phylogenetic Analysis of 16S rRNA Gene Sequences and Concatenated Sequences
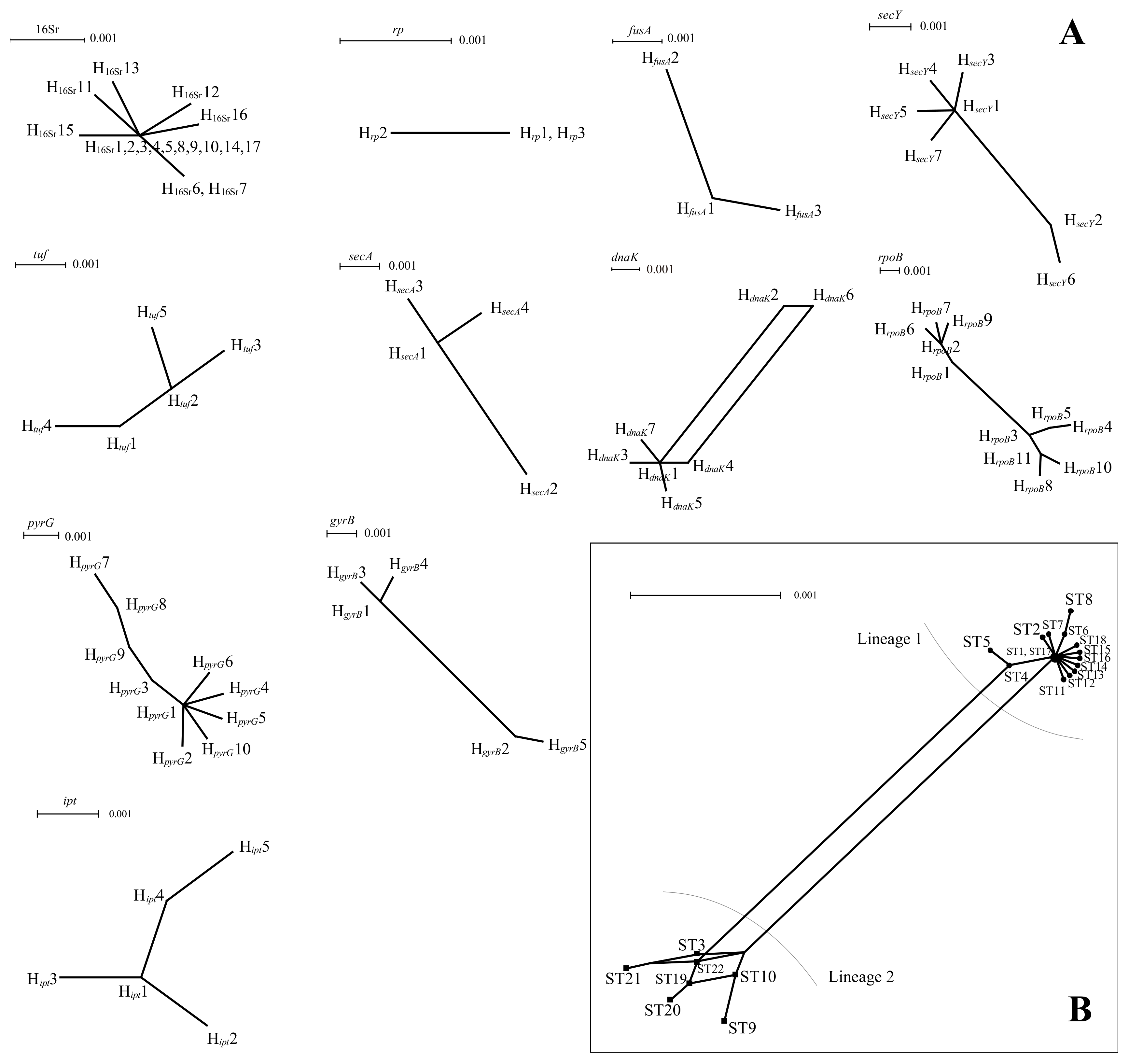
3.7. Phylogenetic Analysis of Each Housekeeping Gene in PaWB Phytoplasma
4. Discussion
4.1. PaWB Phytoplasma as a Unique Phylogenetic Branch of Group 16SrI That Is Closely Related to 16SrI-B
4.2. Valuable Methods for PaWB Phytoplasma Traceability and Disease Epidemics Based on MLST Scheme
4.3. Variation in PaWB Phytoplasma Housekeeping Genes Related to Geographical and Environmental Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.Q.; Fu, J.M.; Qiao, J.; Yan, Z.S.; Du, K.Y.; Dong, S.Q. Capacity of Halyomorpha picus transmitting phytoplasma associated with paulownia witches’ broom. For. Res. China 1999, 12, 606–611. [Google Scholar] [CrossRef]
- Kong, D.Z.; Tian, G.Z.; Zhang, W.X.; Wang, S.J.; Song, C.S.; Deng, B.H.; Guo, M.W.; Piao, C.G.; Lin, C.L. Affection factors of the regional difference of paulownia witches’-broom disease in urban and rural districts of China. Zhiwu Baohu 2022, 48, 280–290. [Google Scholar] [CrossRef]
- Doi, Y.; Teranaka, M.; Yora, K.; Asuyama, H. Mycoplasma-or PLT group-like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches’ broom, aster yellows, or paulownia witches’-broom. Ann. Phytopathol. Soc. Jpn. 1967, 33, 259–266. [Google Scholar] [CrossRef]
- Tian, G.Z.; Raychaudhuri, S.P. Paulownia witches’-broom disease in China: Present status. In Forest Trees and Palms: Diseases and Control; Raychaudhuri, S.P., Maramorosch, K., Eds.; Oxford and IBH Publishing: New Delhi, India, 1996; pp. 227–251. [Google Scholar]
- Mou, H.Q.; Lu, J.; Zhu, S.F.; Lin, C.L.; Tian, G.Z.; Xu, X.; Zhao, W.J. Transcriptomic Analysis of Paulownia Infected by Paulownia Witches’-Broom Phytoplasma. PLoS ONE 2013, 8, e77217. [Google Scholar] [CrossRef]
- Cao, Y.B.; Sun, G.L.; Zhai, X.Q.; Xu, P.L.; Ma, L.M.; Deng, M.J.; Zhao, Z.L.; Yang, H.B.; Dong, Y.P.; Shang, Z.H.; et al. Genomic insights into the fast growth of paulownias and the formation of Paulownia witches’ broom. Mol. Plant. 2021, 14, 1668–1682. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.Z.; Zhang, X.J. Recent advances in paulownia witches’-broom disease research. World For. Res. China 1996, 2, 33–38. [Google Scholar]
- Lee, I.-M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seemüller, E. ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tian, G.Z.; Piao, C.G.; Zhu, S.F. Rapid molecular differentiation and identification of different phytoplasmas from several plants in China. Acta Phytopathol. Sin. 2005, 35, 293–299. [Google Scholar] [CrossRef]
- Wei, W.; Zhao, Y. Phytoplasma Taxonomy: Nomenclature, Classification, and Identification. Biology 2022, 11, 1119. [Google Scholar] [CrossRef]
- Bertaccini, A.; Arocha-Rosete, Y.; Contaldo, N.; Duduk, B.; Fiore, N.; Montano, H.G.; Kube, M.; Kuo, C.-H.; Martini, M.; Oshima, K.; et al. Revision of the ‘Candidatus Phytoplasma’ species description guidelines. Int. J. Syst. Evol. Microbiol. 2022, 72, 005353. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.L.; Zhou, T.; Li, H.F.; Fan, Z.F.; Li, Y.; Piao, C.G.; Tian, G.Z. Molecular characterisation of two plasmids from paulownia witches’-broom phytoplasma and detection of a plasmid-encoded protein in infected plants. Eur. J. Plant Pathol. 2009, 123, 321–330. [Google Scholar] [CrossRef]
- Wang, J.; Tian, G.Z.; Xu, Q.C.; Liu, Y.G.; Gao, R.; Li, X.D.; Zhu, X.P. Molecular detection of phytoplasma strains from several plants around diseased paulownia infected with paulownia witches’-broom phytoplasma. Sci. Agric. Sin. 2010, 43, 304–312. [Google Scholar] [CrossRef]
- Song, C.S.; Hu, J.X.; Lin, C.L.; Ren, Z.G.; Geng, X.S.; Tian, G.Z. Prokaryotic Expression, Purification and Enzyme Activity Assay of Thymidylate Kinase of the Paulownia Witches’-broom Phytoplasma. For. Res. China 2014, 27, 786–793. [Google Scholar] [CrossRef]
- Yu, S.S. Molecular Characterization of Phytoplasmal tuf Gene Promoter and Phytoplasma-Resistant Substances from Jujube. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2016. [Google Scholar]
- Maiden, M.C.J. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006, 60, 561–588. [Google Scholar] [CrossRef]
- Danet, J.L.; Balakishiyeva, G.; Cimerman, A.; Sauvion, N.; Marie-Jeanne, V.; Labonne, G.; Lavina, A.; Batlle, A.; Krizanac, I.; Skoric, D.; et al. Multilocus sequence analysis reveals the genetic diversity of European fruit tree phytoplasmas and supports the existence of inter-species recombination. Microbiology 2011, 157, 438–450. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Piao, C.G.; Tian, G.Z.; Liu, Z.X.; Guo, M.W.; Lin, C.L.; Wang, X.Z. Multilocus sequences confirm the close genetic relationship of four phytoplasmas of peanut witches’-broom group 16SrII-A. J. Basic Microbiol. 2014, 54, 818–827. [Google Scholar] [CrossRef]
- Martini, M.; Quaglino, F.; Bertaccini, A. Multilocus Genetic Characterization of Phytoplasmas. In Phytoplasmas: Plant Pathogenic Bacteria-III; Bertaccini, A., Oshima, K., Eds.; Springer: Singapore, 2019; pp. 161–200. [Google Scholar] [CrossRef]
- Ćurčić, Ž.; Kosovac, A.; Stepanović, J.; Rekanović, E.; Kube, M.; Duduk, B. Multilocus Genotyping of ‘Candidatus Phytoplasma solani’ Associated with Rubbery Taproot Disease of Sugar Beet in the Pannonian Plain. Microorganisms 2021, 9, 1950. [Google Scholar] [CrossRef]
- Rodrigues Jardim, B.; Kinoti, W.M.; Tran-Nguyen, L.T.T.; Gambley, C.; Rodoni, B.; Constable, F.E. ‘Candidatus Phytoplasma stylosanthis’, a novel taxon with a diverse host range in Australia, characterised using multilocus sequence analysis of 16S rRNA, secA, tuf, and rp genes. Int. J. Syst. Evol. Microbiol. 2021, 71, 4589. [Google Scholar] [CrossRef]
- Jamshidi, E.; Murolo, S.; Ravari, S.B.; Salehi, M.; Romanazzi, G. Multilocus Genotyping of ‘Candidatus Phytoplasma Solani’ Associated with Grapevine Bois Noir in Iran. Biology 2022, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Pilet, F.; Mendes, C.D.; Yankey, E.N.; Lopes Parruque, M.; Attivor, I.N.; Nkansah-Poku, J.; Vaz, A. Genetic diversity of ‘Candidatus Phytoplasma palmicola’ in Ghana and Mozambique. Phytopathogenic Mollicutes 2022, 12, 69. [Google Scholar] [CrossRef]
- Arnaud, G.; Malembic-Maher, S.; Salar, P.; Bonnet, P.; Maixner, M.; Marcone, C.; Boudon-Padieu, E.; Foissac, X. Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescencedorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl. Environ. Microbiol. 2007, 73, 4001–4010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plavec, J.; Križanac, I.; Budinšćak, Ž.; Škorić, D.; Musić, M.Š. A case study of FD and BN phytoplasma variability in Croatia: Multigene sequence analysis approach. Eur. J. Plant Pathol. 2015, 142, 591–601. [Google Scholar] [CrossRef]
- Plavec, J.; Budinšćak, Ž.; Križanac, I.; Škorić, D.; Foissac, X.; Šeruga Musić, M. Multilocus sequence typing reveals the presence of three distinct flavescencedorée phytoplasma genetic clusters in Croatian vineyards. Plant Pathol. 2019, 68, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Jernej, P.; Nataša, M.; Petra, N.; Marina, D. Molecular diversity of ‘Candidatus Phytoplasma pyri’ isolates in Slovenia. Eur. J. Plant Pathol. 2014, 139, 801–809. [Google Scholar] [CrossRef]
- Abeysinghe, S.; Abeysinghe, P.D.; Kanatiwela-De Silva, C.; Udagama, P.; Warawichanee, K.; Aljafar, N.; Kawicha, P.; Dickinson, M. Refinement of the taxonomic structure of 16SrXI and 16SrXIV phytoplasmas of gramineous plants using multilocus sequence typing. Plant Dis. 2016, 100, 2001–2010. [Google Scholar] [CrossRef] [Green Version]
- Mou, D.F.; Helmick, E.; Bahder, B. Multi-locus sequence analysis reveals new hosts of palm lethal decline phytoplasmas in Florida, U.S.A. Plant Health Prog. 2022, in press. [Google Scholar] [CrossRef]
- Yu, S.S.; Li, Y.; Ren, Z.G.; Song, C.S.; Lin, C.L.; Piao, C.G.; Tian, G.Z. Multilocus Sequence Analysis for Revealing Finer Genetic Variation and Phylogenetic Interrelatedness of Phytoplasma Strains in 16SrI Group in China. Sci. Silvae Sin. 2017, 53, 105–118. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Dickinson, M.; Hodgetts, J. Phytoplasma: Methods and Protocols; Humana Press: New York, NY, USA, 2013; Volume 938. [Google Scholar]
- Lee, I.-M.; Martini, M.; Bottner, K.D.; Dane, R.A.; Black, M.C.; Troxclair, N. Ecological implications from a molecular analysis of phytoplasmas involved in an aster yellows epidemic in various crops in Texas. Phytopathology 2003, 93, 1368–1377. [Google Scholar] [CrossRef]
- Schneider, B.; Gibb, K.S.; Seemüller, E. Sequence and RFLP analysis of the elongation factor Tu gene used in differentiation and classification of phytoplasmas. Microbiology 1997, 143, 3381–3389. [Google Scholar] [CrossRef] [Green Version]
- Hodgetts, J.; Boonham, N.; Mumford, R.; Harrison, N.; Dickinson, M. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int. J. Syst. Evol. Microbiol. 2008, 58, 1826–1837. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.X.; Tian, G.Z.; Lin, C.L.; Song, C.S.; Mou, H.Q.; Ren, Z.G.; Guo, S.; Zhou, T.; Fan, Z.F.; Li, H.F. Cloning, expression and characterization of tRNA-isopentenyltransferase genes (tRNA-ipt) from paulownia witches’-broom phytoplasma. Acta Microbiol. Sin. 2013, 53, 832–841. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pair-wise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef]
- Smith, J.M.; Smith, N.H.; O’Rourke, M.; Spratt, B.G. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 1993, 90, 4384–4388. [Google Scholar] [CrossRef] [Green Version]
- Haubold, B.; Hudson, R.R. Lian 3.0: Detecting linkage disequilibrium in multilocus data. Bioinformatics 2000, 16, 847–849. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2020, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinf. 2009, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 1992, 132, 583–589. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Bila, J.; Mondjana, A.; Samils, B.; Högberg, N. High diversity, expanding populations and purifying selection in phytoplasmas causing coconut lethal yellowing in Mozambique. Plant Pathol. 2015, 64, 597–604. [Google Scholar] [CrossRef]
- Pilet, F.; Quaicoe, R.N.; Osagie, I.J.; Freire, M.; Foissac, X. Multilocus Sequence Analysis Reveals Three Distinct Populations of “Candidatus Phytoplasma palmicola” with a Specific Geographical Distribution on the African Continent. Appl. Environ. Microbiol. 2019, 85, e02716-18. [Google Scholar] [CrossRef] [Green Version]
- Panda, P.; Nigam, A.; Rao, G.P. Multilocus gene analysis reveals the presence of two phytoplasma groups in Impatiens balsamina showing flat stem and phyllody. 3 Biotech 2021, 11, 122. [Google Scholar] [CrossRef]
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Lee, I.-M.; Jomantiene, R.; Douglas, S.M. ‘Candidatus Phytoplasma pruni’, a novel taxon associated with X-disease of stone fruits, Prunus spp.: Multilocus characterization based on 16SrRNA, secY, and ribosomal protein genes. Int. J. Syst. Evol. Microbiol. 2013, 63, 766–776. [Google Scholar] [CrossRef]
- Siampour, M.; Izadpanah, K.; Martini, M.; Salehi, M. Multilocus sequence analysis of phytoplasma strains of 16SrII group in Iran and their comparison with related strains. Ann. Appl. Biol. 2019, 175, 83–97. [Google Scholar] [CrossRef]
- Noorizadeh, S.; Khakvar, R.; Golmohammadi, M.; Hashemian, S.M.B.; Faghihi, M.M. Multilocus genotyping of ‘Candidatus Phytoplasma aurantifolia’ associated with witches’ broom disease in Citrus spp. Trop. Plant Pathol. 2021, 46, 218–231. [Google Scholar] [CrossRef]
- Tian, G.Z.; Xiong, Y.G.; Wang, Y.; Zhao, D.N.; Li, F.; Xu, G. Resistances of different clones of paulownia to witches’ broom agent mycoplasma-like organism (MLO). For. Res. China 1994, 7, 155–161. [Google Scholar]
- Wang, J.W.; Liu, Q.Z.; Wei, W.; Davis, R.E.; Tan, Y.; Lee, I.-M.; Zhu, D.Z.; Wei, H.R.; Zhao, Y. Multilocus genotyping identifies a highly homogeneous phytoplasma lineage associated with sweet cherry virescence disease in China and its carriage by an erythroneurine leafhopper. Crop Prot. 2018, 106, 13–22. [Google Scholar] [CrossRef]
- Su, H.J.; Tsai, L.S. The Resistance of Paulownia against Witches’ Broom Diseaes. Q. J. Chin. For. 1983, 16, 187–202. [Google Scholar]
- Fan, J.F.; Zhou, Y.X.; Lian, W.H. Reviews and Prospects of Paulownia Breeding in Shaanxi Province. J. Northwest For. Univ. China 2005, 20, 80–84. [Google Scholar] [CrossRef]
- Suzuki, S.; Oshima, K.; Kakizawa, S.; Arashida, R.; Jung, H.Y.; Yamaji, Y.; Nishigawa, H.; Ugaki, M.; Namba, S. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 4252–4257. [Google Scholar] [CrossRef] [Green Version]
- Kakizawa, S.; Oshima, K.; Jung, H.Y.; Suzuki, S.; Nishigawa, H.; Arashida, R.; Miyata, S.I.; Ugaki, M.; Kishino, H.; Namba, S. Positive Selection Acting on a Surface Membrane Protein of the Plant-Pathogenic Phytoplasmas. J. Bacteriol. 2006, 188, 3424–3428. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.J.; Wang, S.K.; Lin, C.L.; Yu, S.S.; Wang, L.F.; Piao, C.G.; Guo, M.W.; Tian, G.Z. Loop-mediated isothermal amplification assay for detection of five phytoplasmas belonging to 16SrI group based on target tuf gene. Sci. Silvae Sin. China 2017, 53, 54–63. [Google Scholar] [CrossRef]
- Nishigawa, H.; Oshima, K.; Kakizawa, S.; Jung, H.Y.; Kuboyama, T.; Miyata, S.I.; Ugaki, M.; Namba, S. Evidence of intermolecular recombination between extrachromosomal DNAs in phytoplasma: A trigger for the biological diversity of phytoplasma? Microbiology 2002, 148, 1389–1396. [Google Scholar] [CrossRef] [PubMed]



| Primer Pairs | Primer Sequences (5′-3′) | Amplicon Length (bp) | References | |
|---|---|---|---|---|
| 16S rRNA | P1 | AAGAATTTGATCCTGGCTCAGGATT | 1437–1440 | Dickinson et al., 2013 [34] |
| P7 | CGTCCTTCATCGGCTCTT | |||
| rp | rp(I)FIA | TTTTCCCCTACACGTACTTA | 1080–1093 | Lee et al., 2003 [35] |
| rp(I)RIA | GTTCTTTTTGGCATTAACAT | |||
| fusA | YufusAf | GTTGTTGACTACCTTCCTGCTC | 914–926 | Yu et al., 2017 [32] |
| YufusAr | TCGCCAATAACATTTCCTAC | |||
| secY | AYsecYF1 | CAGCCATTTTAGCAGTTGGTGG | 1215–1258 | Dickinson et al., 2013 [34] |
| AYsecYR1 | CAGAAGCTTGAGTGCCTTTACC | |||
| tuf | fTuf1 | CACATTGACCACGGTAAAAC | 941–964 | Schneider et al., 1997 [36] |
| rTuf1 | CCACCTTCACGAATAGAGAAC | |||
| secA | secAfor1 | GARATGAAAACTGGRGAAGG | 713–734 | Hodgetts et al., 2008 [37] |
| secArev3 | GTTTTRGCAGTTCCTGTCATNCC | |||
| dnaK | YudnaKf | TGCTCTTTCTTATGGCGTTGA | 1135–1164 | Yu et al., 2017 [32] |
| YudnaKr | CATTGCGATTCCTTGAGATTC | |||
| rpoB | YurpoBf | TTCCCACTACGGCAGATTATG | 1093–1118 | Yu et al., 2017 [32] |
| YurpoBr | TGGACGATGCCTCCTTCAC | |||
| pyrG | YupyrGf | CCTGGAACAATGAGCCCTTA | 1038–1055 | Yu et al., 2017 [32] |
| YupyrGr | TGGCACGAATAAGAACCTAA | |||
| gyrB | YugyrBf | TATTCACCCCAAAACAGG | 1248–1279 | Yu et al., 2017 [32] |
| YugyrBr | AGTAAAGTTCTTATGTGGGC | |||
| ipt | Iptf-BamHI | CGGGATCCATGAAAAAAGTAATCGCTAT | 739–779 | Hu et al., 2013 [38] |
| iptr-SalI | ACGCGTCGACATCAGTTTTAAAAAATCGT | |||
| Genes | 16S rRNA | rp | fusA | secY | tuf | secA | dnaK | rpoB | pyrG | gyrB | ipt |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of strains | 122 | 138 | 133 | 134 | 133 | 132 | 130 | 127 | 105 | 86 | 83 |
| Fragment length (bp) | 1252 | 956 | 735 | 1138 | 782 | 757 | 979 | 955 | 867 | 1113 | 749 |
| cG + C (%) | 47.0 | 34.7 | 37.5 | 31.8 | 37.7 | 34.7 | 33.9 | 33.7 | 33.0 | 32.4 | 30.3 |
| Sequence similarity (%) | 99.44 | 99.58 | 99.73 | 99.47 | 99.62 | 99.47 | 99.18 | 99.06 | 99.42 | 99.19 | 99.73 |
| No. of variable sites | 10 | 1 | 3 | 9 | 4 | 5 | 11 | 14 | 9 | 10 | 4 |
| Percentage of variable sites (%) | 0.80 | 0.10 | 0.41 | 0.79 | 0.51 | 0.66 | 1.12 | 1.47 | 1.04 | 0.90 | 0.53 |
| No. of haplotypes | 17 | 2 | 3 | 7 | 5 | 4 | 7 | 11 | 10 | 5 | 5 |
| Haplotype diversity | 0.84 | 0.08 | 0.24 | 0.30 | 0.25 | 0.25 | 0.27 | 0.29 | 0.37 | 0.25 | 0.30 |
| Nucleotide diversity | 0.00128 | 0.00009 | 0.00063 | 0.00090 | 0.00036 | 0.00094 | 0.00163 | 0.00166 | 0.00120 | 0.00137 | 0.00046 |
| Ka/Ks ratio a | 0.35 | 0 | 0.29 | 0.13 | 2.63 | 0.54 | 0.11 | 1.72 | 0.39 | * | 0.14 |
| Tajima’s D | −0.342 ns | −0.581 ns | −0.264 ns | −0.900 ns | −1.166 ns | −0.462 ns | −0.526 ns | −1.022 ns | −0.975 ns | −0.610 ns | −1.178 ns |
| Fu and Li’s D | −2.269 ns | 0.473 ns | −0.649 ns | −1.838 ns | −2.812 b | −1.182 ns | −0.666 ns | −1.420 ns | −1.711 ns | −0.676 ns | −0.222 ns |
| Fu and Li’s F | −1.888 ns | 0.177 ns | −0.619 ns | −1.794 ns | −2.683 b | −1.114 ns | −0.734 ns | −1.520 ns | −1.727 ns | −0.775 ns | 0.618 ns |
| phi-test | 0.773 | / | 1.0 | 1.0 | / | 1.0 | 0.042 b | 1.0 | 1.0 | 1.0 | 1.0 |
| Populations | No. of Strains | No. of Variable Sites | MLST STs | Diversity of STs | Nucleotide Diversity | Tajima’s D | Fu and Li’s D | Fu and Li’s F |
|---|---|---|---|---|---|---|---|---|
| Northwest China | 15 | 29 | ST1, ST2, ST3, ST4, ST5, ST6, ST8 | 0.724 | 0.00073 | −2.030 a | −2.604 a | −2.814 b |
| Huang-huai-hai Plain | 25 | 11 | ST1, ST4, ST11, ST12, ST14, ST15, ST16, ST17, ST18 | 0.547 | 0.00014 | −2.341 c | −3.775 b | −3.900 b |
| Southwest China | 19 | 0 | ST1 | 0 | 0 | / | / | / |
| South of the Yangtze River | 46 | 32 | ST1, ST7, ST9, ST10, ST13, ST19, ST20, ST21, ST22 | 0.500 | 0.00153 | 1.089 ns | −0.137 ns | 0.349 ns |
| Total | 105 | 42 | 22 | 0.474 | 0.0009 | −0.914 ns | −1.792 ns | −1.724 ns |
| Population | Northwest China | Huang-Huai-Hai Plain | Southwest China | South of the Yangtze River |
|---|---|---|---|---|
| Northwest China | 0 | |||
| Huang-huai-hai Plain | 0.01315 | 0 | ||
| Southwest China | 0.03968 | 0 | 0 | |
| South of the Yangtze River | 0.07562 | 0.20514 | 0.23148 | 0 |
| Population | Northwest China | Huang-Huai-Hai Plain | Southwest China | South of the Yangtze River |
|---|---|---|---|---|
| Northwest China | 0 | |||
| Huang-huai-hai Plain | 37.53 | 0 | ||
| Southwest China | 12.10 | / | 0 | |
| South of the Yangtze River | 6.11 | 1.94 | 1.66 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, D.-Z.; Lin, C.-L.; Yu, S.-S.; Tian, G.-Z.; Ma, H.-B.; Wang, S.-J. Molecular Diversity and Evolutionary Relatedness of Paulownia Witches’-Broom Phytoplasma in Different Geographical Distributions in China. Biology 2022, 11, 1611. https://doi.org/10.3390/biology11111611
Kong D-Z, Lin C-L, Yu S-S, Tian G-Z, Ma H-B, Wang S-J. Molecular Diversity and Evolutionary Relatedness of Paulownia Witches’-Broom Phytoplasma in Different Geographical Distributions in China. Biology. 2022; 11(11):1611. https://doi.org/10.3390/biology11111611
Chicago/Turabian StyleKong, De-Zhi, Cai-Li Lin, Shao-Shuai Yu, Guo-Zhong Tian, Hai-Bin Ma, and Sheng-Jie Wang. 2022. "Molecular Diversity and Evolutionary Relatedness of Paulownia Witches’-Broom Phytoplasma in Different Geographical Distributions in China" Biology 11, no. 11: 1611. https://doi.org/10.3390/biology11111611
APA StyleKong, D.-Z., Lin, C.-L., Yu, S.-S., Tian, G.-Z., Ma, H.-B., & Wang, S.-J. (2022). Molecular Diversity and Evolutionary Relatedness of Paulownia Witches’-Broom Phytoplasma in Different Geographical Distributions in China. Biology, 11(11), 1611. https://doi.org/10.3390/biology11111611








