SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Yeast Strains, and Plasmids
2.2. DNA Amplification with the Polymerase Chain Reaction and Sequencing
2.3. Plasmid Construction
2.4. Bacterial Culture
2.5. Yeast Two-Hybrid Methods
2.6. Cell Culture, Lysis, and Transfection
2.7. GST and 6His Fusion Proteins, Electrophoresis, and Immunoblot
2.8. Mutagenesis
2.9. In Vitro Ubiquitination Assays
2.10. Analysis of SNRPD2 Ubiquitination Sites by MALDI-MS(/MS)
2.11. Quantification of Protein Bands and Statistics
3. Results
3.1. Identification of Mammalian Binding Partners for Salmonella SlrP through a Yeast Two-Hybrid Screen
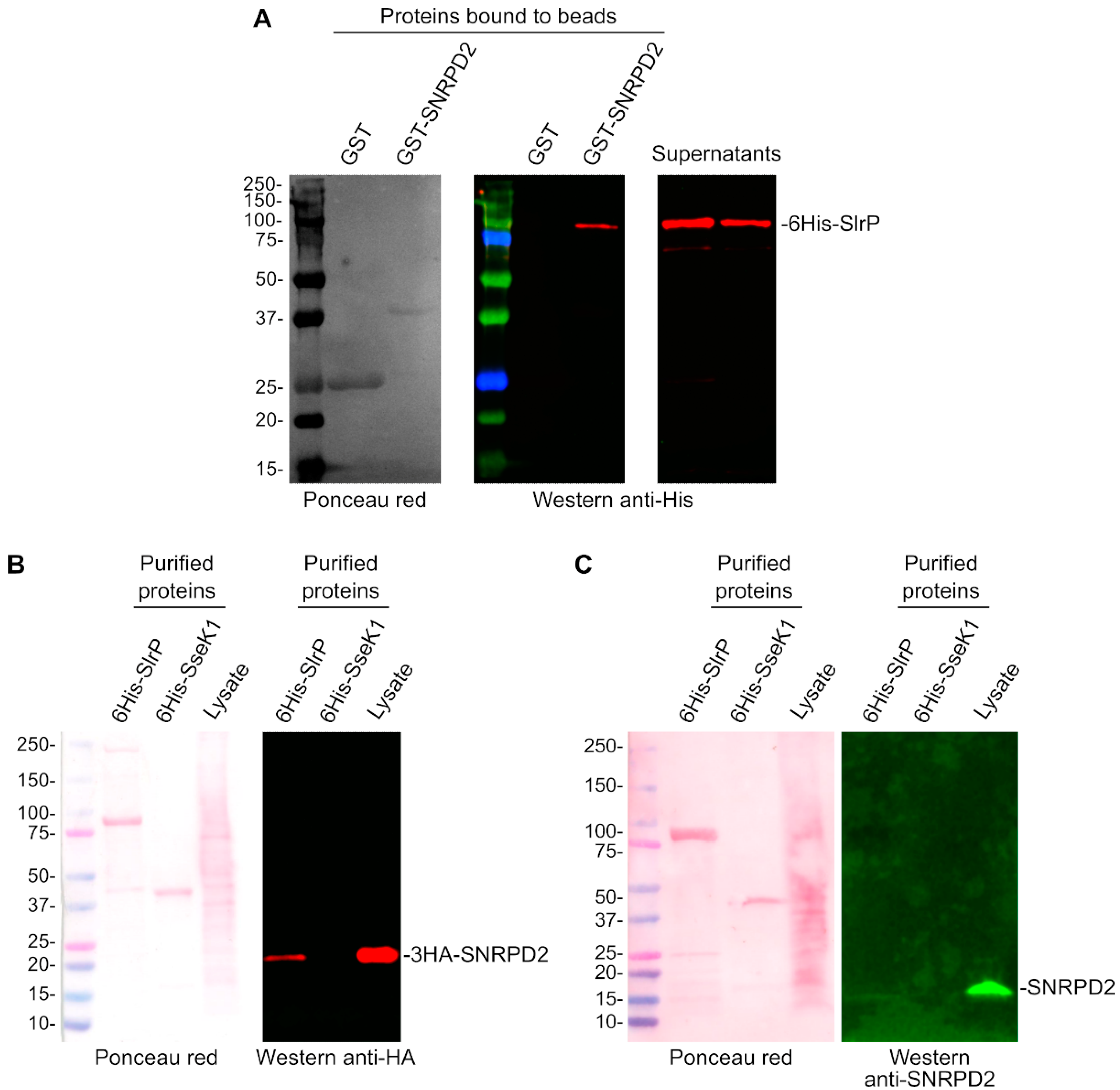
3.2. Confirmation of the Interaction of SlrP with SNRPD2
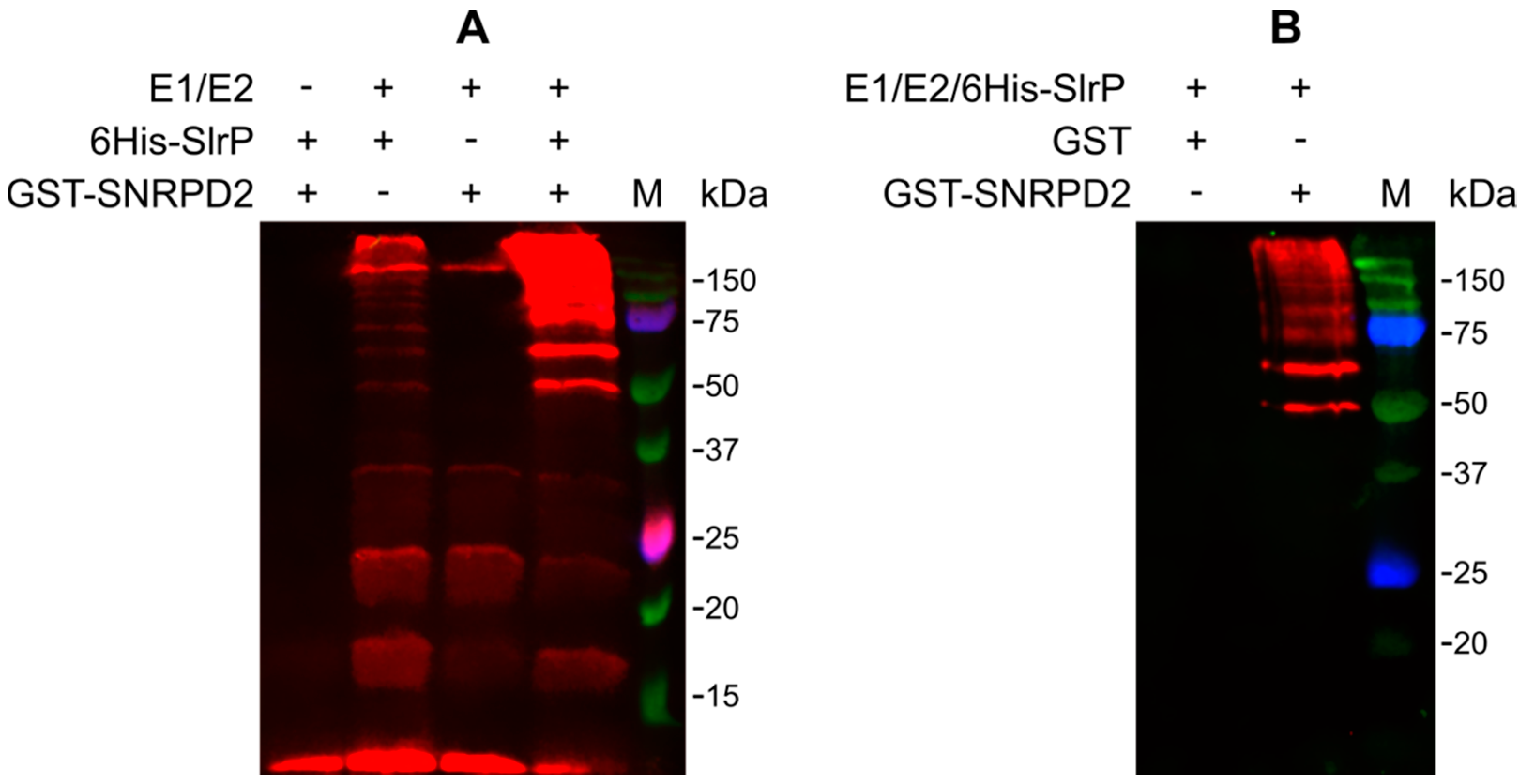
3.3. SNRPD2 Is a Target of the E3 Ubiquitin Ligase Activity of SlrP
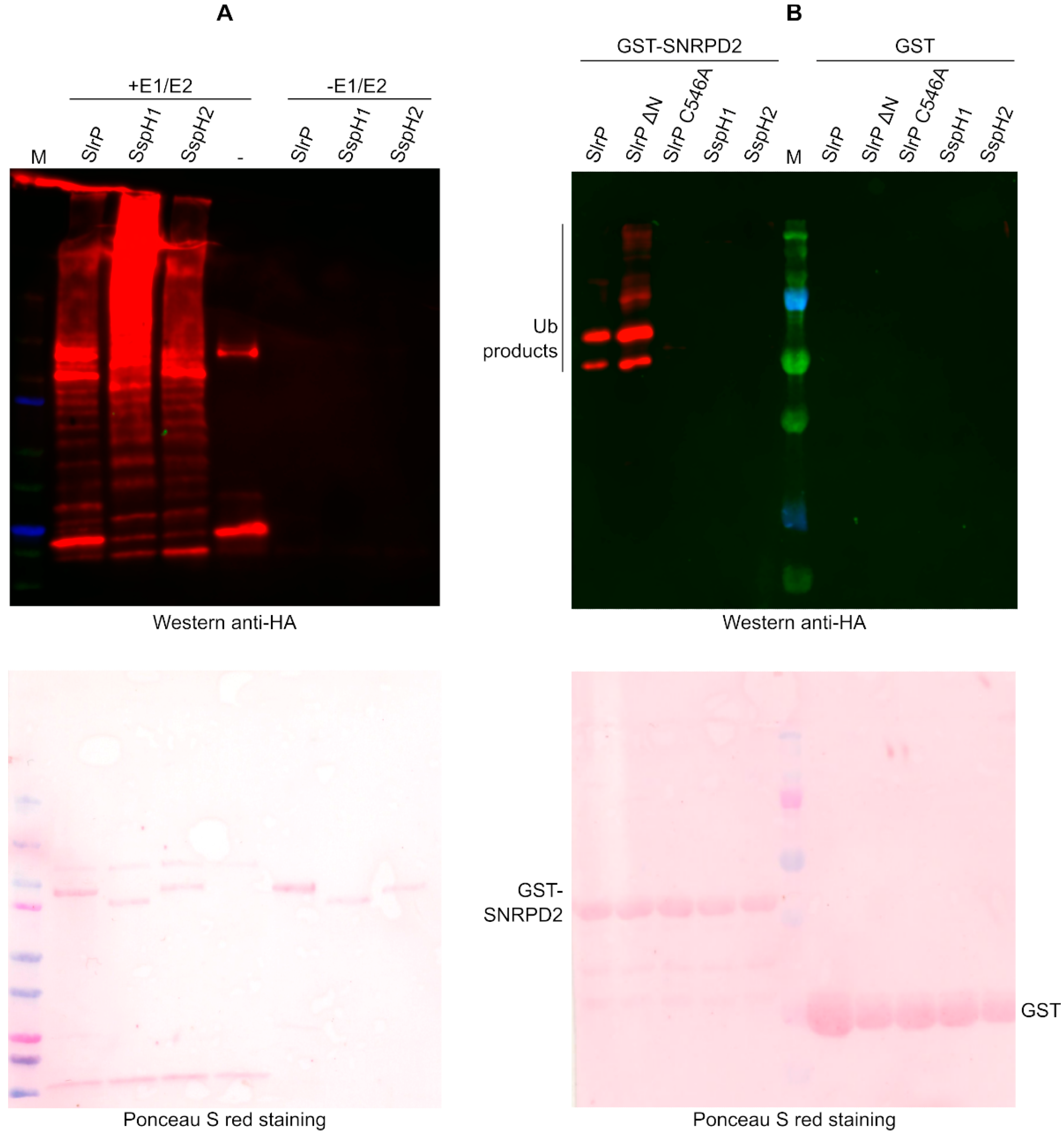
3.4. Specificity of the Interaction and Ubiquitination of SNRPD2
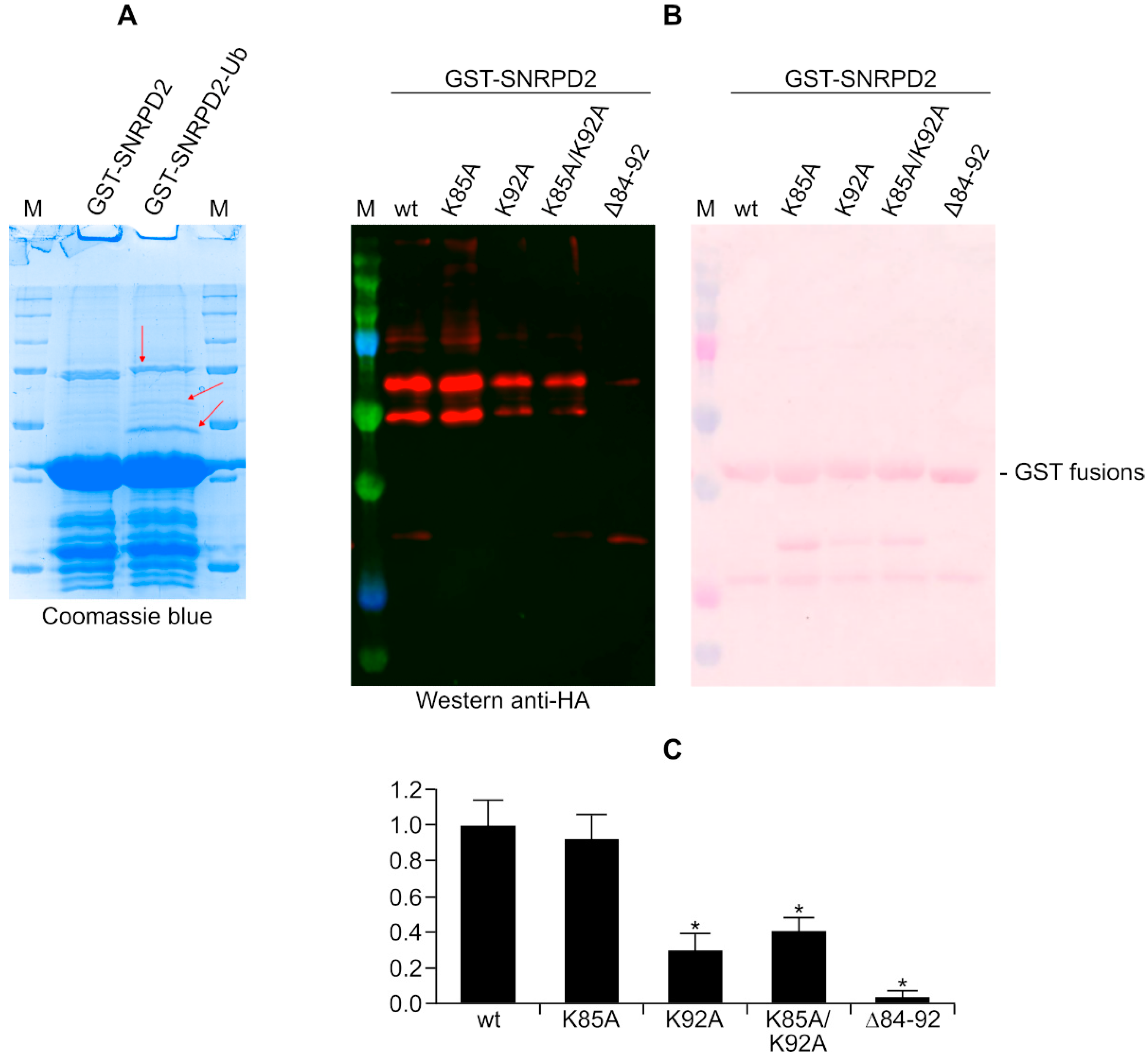
3.5. Analysis of SNRPD2 Ubiquitination by Mass Spectrometry and Mutagenesis
3.6. Lack of Effect of SlrP on SNRPD2 Levels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jajere, S.M. A Review of Salmonella Enterica with Particular Focus on the Pathogenicity and Virulence Factors, Host Specificity and Antimicrobial Resistance Including Multidrug Resistance. Vet. World 2019, 12, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Agbor, T.A.; Mccormick, B.A. Salmonella Effectors: Important Players Modulating Host Cell Function during Infection. Cell. Microbiol. 2011, 13, 1858–1869. [Google Scholar] [CrossRef] [PubMed]
- Egan, F.; Barret, M.; O’Gara, F. The SPI-1-like Type III Secretion System: More Roles than You Think. Front. Plant Sci. 2014, 5, 34. [Google Scholar] [CrossRef]
- Jennings, E.; Thurston, T.L.M.; Holden, D.W. Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences. Cell Host Microbe 2017, 22, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Lara-Tejero, M.; Galán, J.E. The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Hume, P.J.; Singh, V.; Davidson, A.C.; Koronakis, V. Swiss Army Pathogen: The Salmonella Entry Toolkit. Front. Cell. Infect. Microbiol. 2017, 7, 348. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Morales, F. Impact of Salmonella Enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. ISRN Cell. Biol. 2012, 2012, 1–36. [Google Scholar] [CrossRef]
- Pinaud, L.; Sansonetti, P.J.; Phalipon, A. Host Cell Targeting by Enteropathogenic Bacteria T3SS Effectors. Trends Microbiol. 2018, 26, 266–283. [Google Scholar] [CrossRef] [PubMed]
- Cerny, O.; Holden, D.W. Salmonella SPI-2 Type III Secretion System-Dependent Inhibition of Antigen Presentation and T Cell Function. Immunol. Lett. 2019, 215, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Tsolis, R.M.; Townsend, S.M.; Miao, E.A.; Miller, S.I.; Ficht, T.A.; Adams, L.G.; Bäumler, A.J. Identification of a Putative Salmonella Enterica Serotype Typhimurium Host Range Factor with Homology to IpaH and YopM by Signature-Tagged Mutagenesis. Infect. Immun. 1999, 67, 6385–6393. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Alba, M.; Ramos-Morales, F. Patterns of Expression and Translocation of the Ubiquitin Ligase SlrP in Salmonella Enterica Serovar Typhimurium. J. Bacteriol. 2014, 196, 3912–3922. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, C.D.; Slauch, J.M. RtsA and RtsB Coordinately Regulate Expression of the Invasion and Flagellar Genes in Salmonella Enterica Serovar Typhimurium. J. Bacteriol. 2003, 185, 5096–5108. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, C.D.; Slauch, J.M. RtsA Coordinately Regulates DsbA and the Salmonella Pathogenicity Island 1 Type III Secretion System. J. Bacteriol. 2004, 186, 68–79. [Google Scholar] [CrossRef]
- Miao, E.A.; Miller, S.I. A Conserved Amino Acid Sequence Directing Intracellular Type III Secretion by Salmonella Typhimurium. Proc. Natl. Acad. Sci. USA 2000, 97, 7539–7544. [Google Scholar] [CrossRef]
- Norkowski, S.; Schmidt, M.A.; Rüter, C. The Species-Spanning Family of LPX-Motif Harbouring Effector Proteins. Cell. Microbiol. 2018, 20, e12945. [Google Scholar] [CrossRef]
- Rohde, J.R.; Breitkreutz, A.; Chenal, A.; Sansonetti, P.J.; Parsot, C. Type III Secretion Effectors of the IpaH Family Are E3 Ubiquitin Ligases. Cell Host Microbe 2007, 1, 77–83. [Google Scholar] [CrossRef]
- Bernal-Bayard, J.; Ramos-Morales, F. Salmonella Type III Secretion Effector SlrP Is an E3 Ubiquitin Ligase for Mammalian Thioredoxin. J. Biol. Chem. 2009, 284, 27587–27595. [Google Scholar] [CrossRef]
- Bernal-Bayard, J.; Cardenal-Muñoz, E.; Ramos-Morales, F. The Salmonella Type III Secretion Effector, Salmonella Leucine-Rich Repeat Protein (SlrP), Targets the Human Chaperone ERdj3. J. Biol. Chem. 2010, 285, 16360–16368. [Google Scholar] [CrossRef]
- Schmieger, H. Phage P22-Mutants with Increased or Decreased Transduction Abilities. Mol. Gen. Genet. 1972, 119, 75–88. [Google Scholar] [CrossRef]
- Maloy, S.R. Experimental Techniques in Bacterial Genetics; Jones and Bartlett Learning: Burlington, MA, USA, 1990; ISBN 0867201185. [Google Scholar]
- Hanahan, D. Studies on Transformation of Escherichia Coli with Plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Boyer, H.W.; Roulland-Dussoix, D. A Complementation Analysis of the Restriction and Modification of DNA in Escherichia Coli. J. Mol. Biol. 1969, 41, 459–472. [Google Scholar] [CrossRef]
- Bullock, W.O.; Fernandez, J.M.; Short, J.M. XL1-Blue: A High Efficiency Plasmid Transforming RecA Escherichia Coli Strain with Betagalactosidase Selection. Bio. Tech. 1987, 5, 376–379. [Google Scholar]
- Vojtek, A.B.; Hollenberg, S.M.; Cooper, J.A. Mammalian Ras Interacts Directly with the Serine/Threonine Kinase Raf. Cell 1993, 74, 205–214. [Google Scholar] [CrossRef]
- Van Aelst, L. Two-Hybrid Analysis of Ras-Raf Interactions. Methods Mol. Biol. 1998, 84, 201–222. [Google Scholar] [CrossRef]
- Selig, L.; Benichou, S.; Rogel, M.E.; Wu, L.I.; Vodicka, M.A.; Sire, J.; Benarous, R.; Emerman, M. Uracil DNA Glycosylase Specifically Interacts with Vpr of Both Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus of Sooty Mangabeys, but Binding Does Not Correlate with Cell Cycle Arrest. J. Virol. 1997, 71, 4842–4846. [Google Scholar] [CrossRef]
- Zouhir, S.; Bernal-Bayard, J.; Cordero-Alba, M.; Cardenal-Muñoz, E.; Guimaraes, B.; Lazar, N.; Ramos-Morales, F.; Nessler, S. The Structure of the Slrp-Trx1 Complex Sheds Light on the Autoinhibition Mechanism of the Type III Secretion System Effectors of the NEL Family. Biochem. J. 2014, 464, 135–144. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Sherman, F.; Fink, G.R.; Hicks, J.B. Laboratory Course Manual for Methods in Yeast Genetics; Cold Spring Harbor Laboratory: New York, NY, USA, 1987; ISBN 0879691972. [Google Scholar]
- Fuxman Bass, J.I.; Reece-Hoyes, J.S.; Walhout, A.J.M. Colony Lift Colorimetric Assay for β-Galactosidase Activity. Cold Spring Harb. Protoc. 2016, 2016, pdb. prot088963. [Google Scholar] [CrossRef][Green Version]
- Van Criekinge, W.; Beyaert, R. Yeast Two-Hybrid: State of the Art. Biol. Proced. Online 1999, 2, 1–38. [Google Scholar] [CrossRef]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Agafonov, D.E.; Deckert, J.; Wolf, E.; Odenwälder, P.; Bessonov, S.; Will, C.L.; Urlaub, H.; Lührmann, R. Semiquantitative Proteomic Analysis of the Human Spliceosome via a Novel Two-Dimensional Gel Electrophoresis Method. Mol. Cell. Biol. 2011, 31, 2667–2682. [Google Scholar] [CrossRef] [PubMed]
- Hegele, A.; Kamburov, A.; Grossmann, A.; Sourlis, C.; Wowro, S.; Weimann, M.; Will, C.L.; Pena, V.; Lührmann, R.; Stelzl, U. Dynamic Protein-Protein Interaction Wiring of the Human Spliceosome. Mol. Cell. 2012, 45, 567–580. [Google Scholar] [CrossRef]
- Jurica, M.S.; Licklider, L.J.; Gygi, S.P.; Grigorieff, N.; Moore, M.J. Purification and Characterization of Native Spliceosomes Suitable for Three-Dimensional Structural Analysis. RNA 2002, 8, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yan, C.; Hang, J.; Finci, L.I.; Lei, J.; Shi, Y. An Atomic Structure of the Human Spliceosome. Cell 2017, 169, 918–929.e14. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Schneider, C.; Hossbach, M.; Urlaub, H.; Rauhut, R.; Elbashir, S.; Tuschl, T.; Lührmann, R. The Human 18S U11/U12 SnRNP Contains a Set of Novel Proteins Not Found in the U2-Dependent Spliceosome. RNA 2004, 10, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Koedoot, E.; van Steijn, E.; Vermeer, M.; González-Prieto, R.; Vertegaal, A.C.O.; Martens, J.W.M.; le Dévédec, S.E.; van de Water, B. Splicing Factors Control Triple-Negative Breast Cancer Cell Mitosis through SUN2 Interaction and Sororin Intron Retention. J. Exp. Clin. Cancer Res. 2021, 40, 82. [Google Scholar] [CrossRef] [PubMed]
- Azam, S.; Hou, S.; Zhu, B.; Wang, W.; Hao, T.; Bu, X.; Khan, M.; Lei, H. Nuclear Retention Element Recruits U1 SnRNP Components to Restrain Spliced LncRNAs in the Nucleus. RNA Biol. 2019, 16, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhao, C.; Zhang, N.; Cui, X.; Ren, Y.; Su, C.; Wu, S.; Yao, Z.; Yang, J. Genetic Expression and Mutational Profile Analysis in Different Pathologic Stages of Hepatocellular Carcinoma Patients. BMC Cancer 2021, 21, 786. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, J.; Guo, D.; Chen, B.; Liu, P.; Xiao, Y.; Yang, K.; Liu, Z.; Liu, Q. Identification of 13 Key Genes Correlated With Progression and Prognosis in Hepatocellular Carcinoma by Weighted Gene Co-Expression Network Analysis. Front. Genet. 2020, 11, 153. [Google Scholar] [CrossRef]
- Tao, Y.; Han, Y.; Yu, L.; Wang, Q.; Leng, S.X.; Zhang, H. The Predicted Key Molecules, Functions, and Pathways That Bridge Mild Cognitive Impairment (MCI) and Alzheimer’s Disease (AD). Front. Neurol. 2020, 11, 233. [Google Scholar] [CrossRef]
- Bertram, K.; Agafonov, D.E.; Dybkov, O.; Haselbach, D.; Leelaram, M.N.; Will, C.L.; Urlaub, H.; Kastner, B.; Lührmann, R.; Stark, H. Cryo-EM Structure of a Pre-Catalytic Human Spliceosome Primed for Activation. Cell 2017, 170, 701–713.e11. [Google Scholar] [CrossRef]
- Pai, A.A.; Baharian, G.; Pagé Sabourin, A.; Brinkworth, J.F.; Nédélec, Y.; Foley, J.W.; Grenier, J.C.; Siddle, K.J.; Dumaine, A.; Yotova, V.; et al. Widespread Shortening of 3′ Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS Genet. 2016, 12, e1006338. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.R.; Scott, H.M.; West, K.O.; Vail, K.J.; Fitzsimons, T.C.; Coleman, A.K.; Carter, K.E.; Watson, R.O.; Patrick, K.L. Global Transcriptomics Uncovers Distinct Contributions From Splicing Regulatory Proteins to the Macrophage Innate Immune Response. Front. Immunol. 2021, 12, 2365. [Google Scholar] [CrossRef]
- Huang, F.; Yamaguchi, A.; Tsuchiya, N.; Ikawa, T.; Tamura, N.; Virtala, M.M.K.; Granfors, K.; Yasaei, P.; Yu, D.T.Y. Induction of Alternative Splicing of HLA-B27 by Bacterial Invasion. Arthritis Rheum. 1997, 40, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Toyotome, T.; Kataoka, N.; Ohno, M.; Abe, H.; Shimura, Y.; Seyedarabi, A.; Pickersgill, R.; Sasakawa, C. Shigella Effector IpaH9.8 Binds to a Splicing Factor U2AF(35) to Modulate Host Immune Responses. Biochem. Biophys. Res. Commun. 2005, 333, 531–539. [Google Scholar] [CrossRef]
- Walch, P.; Selkrig, J.; Knodler, L.A.; Rettel, M.; Stein, F.; Fernandez, K.; Viéitez, C.; Potel, C.M.; Scholzen, K.; Geyer, M.; et al. Global Mapping of Salmonella Enterica-Host Protein-Protein Interactions during Infection. Cell Host Microbe 2021, 29, 1316–1332.e12. [Google Scholar] [CrossRef]
- Mattiroli, F.; Sixma, T.K. Lysine-Targeting Specificity in Ubiquitin and Ubiquitin-like Modification Pathways. Nat. Struct. Mol. Biol. 2014, 21, 308–316. [Google Scholar] [CrossRef]
- Carroll, E.C.; Marqusee, S. Site-Specific Ubiquitination: Deconstructing the Degradation Tag. Curr. Opin. Struct. Biol. 2022, 73, 102345. [Google Scholar] [CrossRef]
- Tekaia, F.; Yeramian, E.; Dujon, B. Amino Acid Composition of Genomes, Lifestyles of Organisms, and Evolutionary Trends: A Global Picture with Correspondence Analysis. Gene 2002, 297, 51–60. [Google Scholar] [CrossRef]
- Salas-Lloret, D.; González-Prieto, R. Insights in Post-Translational Modifications: Ubiquitin and SUMO. Int. J. Mol. Sci. 2022, 23, 3281. [Google Scholar] [CrossRef]
- Kambach, C.; Walke, S.; Young, R.; Avis, J.M.; de La Fortelle, E.; Raker, V.A.; Lührmann, R.; Li, J.; Nagai, K. Crystal Structures of Two Sm Protein Complexes and Their Implications for the Assembly of the Spliceosomal SnRNPs. Cell 1999, 96, 375–387. [Google Scholar] [CrossRef]
- Weber, G.; Trowitzsch, S.; Kastner, B.; Lührmann, R.; Wahl, M.C. Functional Organization of the Sm Core in the Crystal Structure of Human U1 SnRNP. EMBO J. 2010, 29, 4172–4184. [Google Scholar] [CrossRef] [PubMed]
- Bullones-Bolaños, A.; Bernal-Bayard, J.; Ramos-Morales, F. The NEL Family of Bacterial E3 Ubiquitin Ligases. Int. J. Mol. Sci. 2022, 23, 7725. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.W.; Galán, J.E. Hijacking the Host Ubiquitin Pathway: Structural Strategies of Bacterial E3 Ubiquitin Ligases. Curr. Opin. Microbiol. 2010, 13, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Keszei, A.F.; Tang, X.; McCormick, C.; Zeqiraj, E.; Rohde, J.R.; Tyers, M.; Sicheri, F. Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase. Mol. Cell. Biol. 2014, 34, 362–373. [Google Scholar] [CrossRef]
- Bhavsar, A.P.; Brown, N.F.; Stoepel, J.; Wiermer, M.; Martin, D.D.O.; Hsu, K.J.; Imami, K.; Ross, C.J.; Hayden, M.R.; Foster, L.J.; et al. The Salmonella Type III Effector SspH2 Specifically Exploits the NLR Co-Chaperone Activity of SGT1 to Subvert Immunity. PLoS Pathog. 2013, 9, e1003518. [Google Scholar] [CrossRef]



| Strain/Plasmid | Relevant Characteristics | Source/Reference |
|---|---|---|
| Escherichia coli | ||
| BL21(DE3) | F- ompT gal dcm lon hsdSB (r- m-; E. coli B strain), with DE3, a λ prophage carrying the T7 RNA pol gene | Stratagene |
| DH5α | supE44 ∆lacU169 (Ø80 lacZ∆M15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | [21] |
| HB101 | F- mcrB mrr hsdS20 (rB- mB-) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(SmR) glnV44λ- | [22] |
| M15 | lac ara gal mtl | |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 ∆lac-pro/F’ proAB lacIq lacZ∆M15 Tn10 (Tetr) | [23] |
| Salmonella enterica serovar Typhimuriuma | ||
| 14028 | Wild type | ATCC |
| SV5193 | slrP::3xFLAG, Kmr | [17] |
| Saccharomyces cerevisiae | ||
| L40 | MATα trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ | [24] |
| Plasmids | ||
| pCS2-3xHA | Mammalian expression vector | Laboratory stock |
| pGEX-4T-1 | GST fusion vector, Apr | GE Healthcare |
| pGEX-4T-2 | GST fusion vector, Apr | GE Healthcare |
| pGAD1318 | Yeast two-hybrid vector, Apr | [25] |
| pLEX10 | Yeast two-hybrid vector, Apr | [26] |
| pIZ1628 | pLEX10-SlrP | This work |
| pIZ1720 | pCS2-SlrP-3xFLAG | [17] |
| pIZ1725 | pcDNA3-SlrP-3xFLAG | [17] |
| pIZ1749 | pQE30-SlrP | This work |
| pIZ1784 | pQE30-SlrP(140-765) | [27] |
| pIZ2177 | pQE80L-SseK1 | Laboratory stock |
| pIZ2370 | pGAD1318-SNRPD2 | This work |
| pIZ3403 | pGEX-4T-2-SNRPD2 | This work |
| pIZ3407 | pLEX10-SspH1 | This work |
| pIZ3408 | pLEX10-SspH2 | This work |
| pIZ3542 | pQE80L-SlrP(C546A) | This work |
| pIZ3551 | pCS2-3HA-SNRPD2 | This work |
| pIZ3557 | pGEX-4T-2-SNRPD2(K85A) | This work |
| pIZ3558 | pGEX-4T-2-SNRPD2(K92A) | This work |
| pIZ3562 | pGEX-4T-2-SNRPD2(K85A/K92A) | This work |
| pIZ3591 | pGEX-4T-2-SNRPD2(∆84-92) | This work |
| pIZ3597 | pQE80L-SspH1 | This work |
| pIZ3598 | pQE80L-SspH2 | This work |
| pQE80L | 6His fusion vector, Apr | Qiagen |
| pREP4 | lacI Kmr | Qiagen |
| Oligonucleotide/Use | Sequence 5′-3′ |
|---|---|
| Construction of pIZ3407 | |
| sspH1bamfw | ATGCGGATCCATGTTTAATATCCGCAATAC |
| sspH1xhorv | TGACCTCGAGTCAGTTAAGACGCCACCGGG |
| Construction of pIZ3408 | |
| sspH2ecofw | ATGCGAATTCATGCCCTTTCATATTGGAAG |
| sspH2salrv | GATCGTCGACTCAGTTACGACGCCACTGAAC |
| Construction of pIZ3509 | |
| TBCBecoRIfw | ATCGGAATTCGAGGTGACGGGGGTGTCGGC |
| TBCBSTOPxhoIrev | ATCGCTCGAGGTCATATCTCGTCCAACCCG |
| Construction of pIZ3551 | |
| SNRPD2ecofw | ATGCGAATTCAGCCTCCTCAACAAGCCCAAG |
| SNRPD2bamrv | ATCGTCTAGACTACTTGCCGGCGATGAGC |
| Construction of pIZ3557 | |
| SNRPD2K85Afw | GTGGCAAGGGCAAGGCGAAGTCCAAGCCAG |
| SNRPD2K85Arv | CTGGCTTGGACTTCGCCTTGCCCTTGCCAC |
| Construction of pIZ3558 | |
| SNRPD2K92Afw | CCAAGCCAGTCAACGCAGACCGCTACATCTC |
| SNRPD2K92Arv | GAGATGTAGCGGTCTGCGTTGACTGGCTTGG |
| Construction of pIZ3591 | |
| SNRPD2-84-92delfw | CAAGAGTGGCAAGGGCGACCGCTACATCTCC |
| SNRPD2-84-92delrv | GGAGATGTAGCGGTCGCCCTTGCCACTCTTG |
| Amplification of pQE80L | |
| pQE80fw | CTGAGCTTGGACTCCTGTTG |
| pQE80rev | GTGATGGTGATGGTGATGCG |
| Construction of pIZ3597 | |
| P1-pQE80-sspH1-fw | CACCATCACCATCACATGTTTAATATCCGCAATACACAACC |
| P2-pQE80-sspH1-rv | GGAGTCCAAGCTCAGTCAGTTAAGACGCCACCGGG |
| Construction of pIZ3598 | |
| P1-pQE80-sspH2-fw | CACCATCACCATCACATGCCCTTTCATATTGGAAGC |
| P2-pQE80-sspH2-rv | GGAGTCCAAGCTCAGTCAGTTACGACGCCACTGAAC |
| Checking of SNRPD2 mutations | |
| SNRPD2bamHIfw | GATCGGATCCATGAGCCTCCTCAACAAGCC |
| SNRPD2comp-K85A-rv | GTTGACTGGCTTGGACTTCGC |
| SNRPD2comp-K92A-rv | CTTGGAGATGTAGCGGTCTGC |
| SNRPD2comp-Del84-92-rv | CTTTGTTGACTGGCTTGGAC |
| Identification of candidates carrying LSM2 | |
| LSM2fw | TCAAGTCCCTTGTGGGCAAG |
| LSM2rev | TCACTGTTTCTGCTGCAGGG |
| Identification of candidates carrying PPP1R7 | |
| PPP1R7fw | CTAAACTTCAGAACCTGGATG |
| PPP1R7rev | TCAGAACCTGACGAACGTGG |
| Identification of candidates carrying RABIF | |
| RABIFfw | CGTTGCGGCTCCCGGGTGCTG |
| RABIFrev | TTACTCATGGGAAACTCGTTC |
| Identification of candidates carrying SNRPD2 | |
| SNRPD2fw | AGGAGCTGCAGAAGCGAGAG |
| SNRPD2rev | CTACTTGCCGGCGATGAGCG |
| Identification of candidates carrying TRX | |
| tio5′ | GTCAGAATTCGCCGCCACGATGGTGAAGCAGATC |
| tio3′ | GTCAGAATTCGCCGCCACGATGGTGAAGCAGATC |
| Sequencing of two-hybrid screen candidates | |
| Gal4AD | TACCACTACAATGGATG |
| Gene | Number of Clones | Description of the Product | Amino Acids Encoded in Different Clones 1 |
|---|---|---|---|
| ABHD14B | 5 | Serine hydrolase with lysine deacetylase activity | 1–210 |
| ANP32A | 2 | Acidic leucine-rich nuclear phosphoprotein | 25–249 |
| CEP97 | 5 | Centrosomal protein | 120–865 |
| EXOSC7 | 4 | Exosome complex component | 40–291 |
| LSM2 | 29 | Sm-like protein with a role in pre-mRNA splicing | 1–95/6–95 |
| MITD1 | 9 | Required for efficient abscission at the end of cytokinesis | 1–249/8–249/72–249 |
| NOP58 | 4 | Nucleolar protein required for 60S ribosomal subunit biogenesis | 360–529 |
| PLK4 | 11 | Serine/threonine protein kinase | 698–970/727–970/777–970/837–970 |
| PPP1R7 | 9 | Regulatory subunit of protein phosphatase 1 | 196–360/203–360 |
| RABIF | 8 | Guanine nucleotide exchange factor | 1–123/4–123 |
| SNRPD2 | 236 | Small nuclear ribonucleoprotein with a role in pre-mRNA splicing | 1–118/10–118/13–118/15–118/19–118 |
| TBCE | 4 | Tubulin-folding protein | 129–527 |
| TXN | 220 | Thioredoxin | 1–105 |
| XRCC6 | 6 | Single-stranded DNA-dependent ATP-dependent helicase | 315–609/402–609/405–609/464–609 |
| ZFPM1 | 3 | Zinc finger protein | 895–1006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bullones-Bolaños, A.; Araujo-Garrido, J.L.; Fernández-García, J.; Romero, F.; Bernal-Bayard, J.; Ramos-Morales, F. SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP. Biology 2022, 11, 1517. https://doi.org/10.3390/biology11101517
Bullones-Bolaños A, Araujo-Garrido JL, Fernández-García J, Romero F, Bernal-Bayard J, Ramos-Morales F. SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP. Biology. 2022; 11(10):1517. https://doi.org/10.3390/biology11101517
Chicago/Turabian StyleBullones-Bolaños, Andrea, Juan Luis Araujo-Garrido, Jesús Fernández-García, Francisco Romero, Joaquín Bernal-Bayard, and Francisco Ramos-Morales. 2022. "SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP" Biology 11, no. 10: 1517. https://doi.org/10.3390/biology11101517
APA StyleBullones-Bolaños, A., Araujo-Garrido, J. L., Fernández-García, J., Romero, F., Bernal-Bayard, J., & Ramos-Morales, F. (2022). SNRPD2 Is a Novel Substrate for the Ubiquitin Ligase Activity of the Salmonella Type III Secretion Effector SlrP. Biology, 11(10), 1517. https://doi.org/10.3390/biology11101517






