Enhancing the Production of Pinene in Escherichia coli by Using a Combination of Shotgun, Product-Tolerance and I-SceI Cleavage Systems
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Primers
2.2. Construction of E. coli Genomic Library
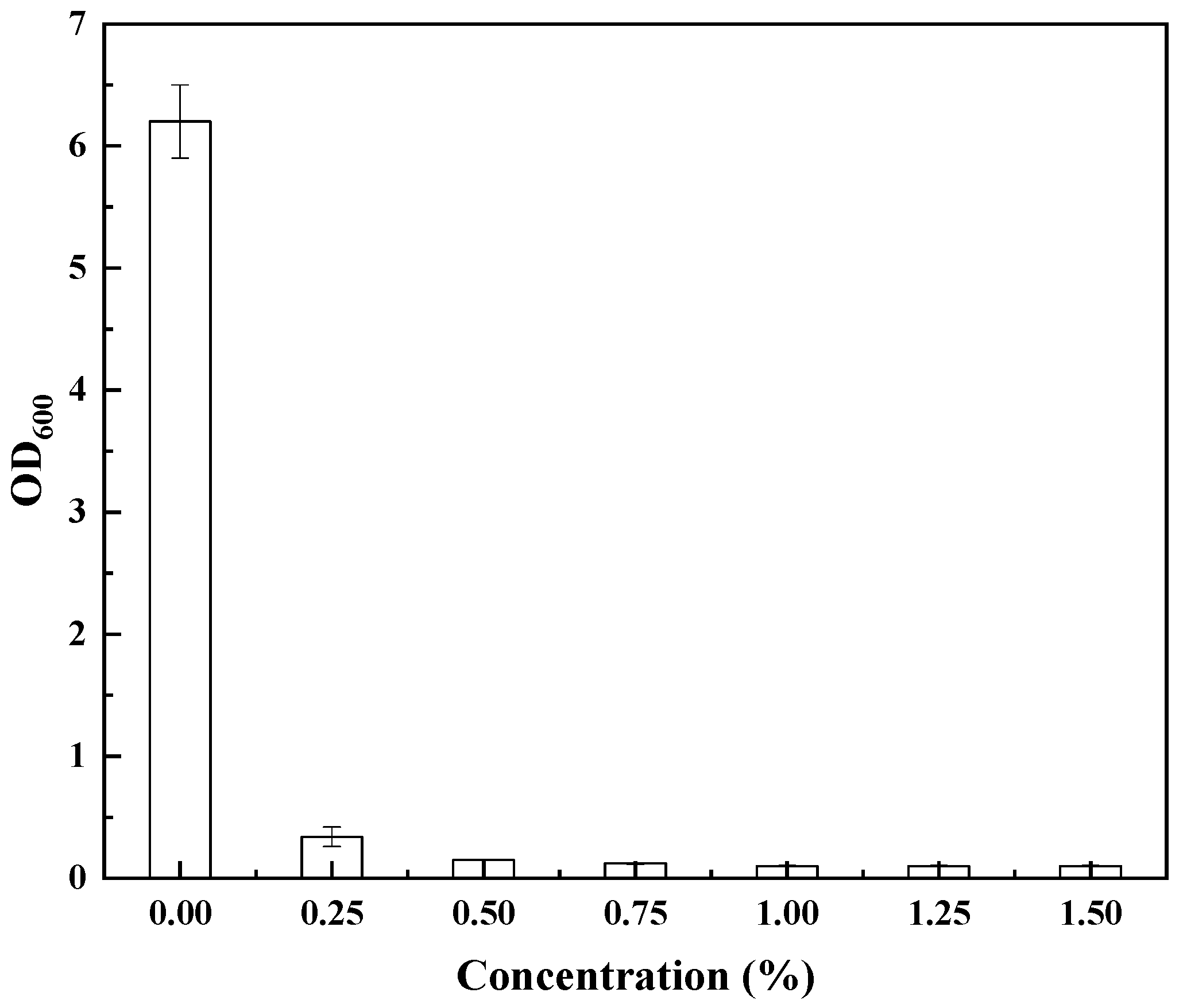
2.3. Limit Screening Method (LMS Method) Combined with Tolerance for Gene Isolation
2.4. DNA Sequencing and Analysis
2.5. Construction of Plasmids
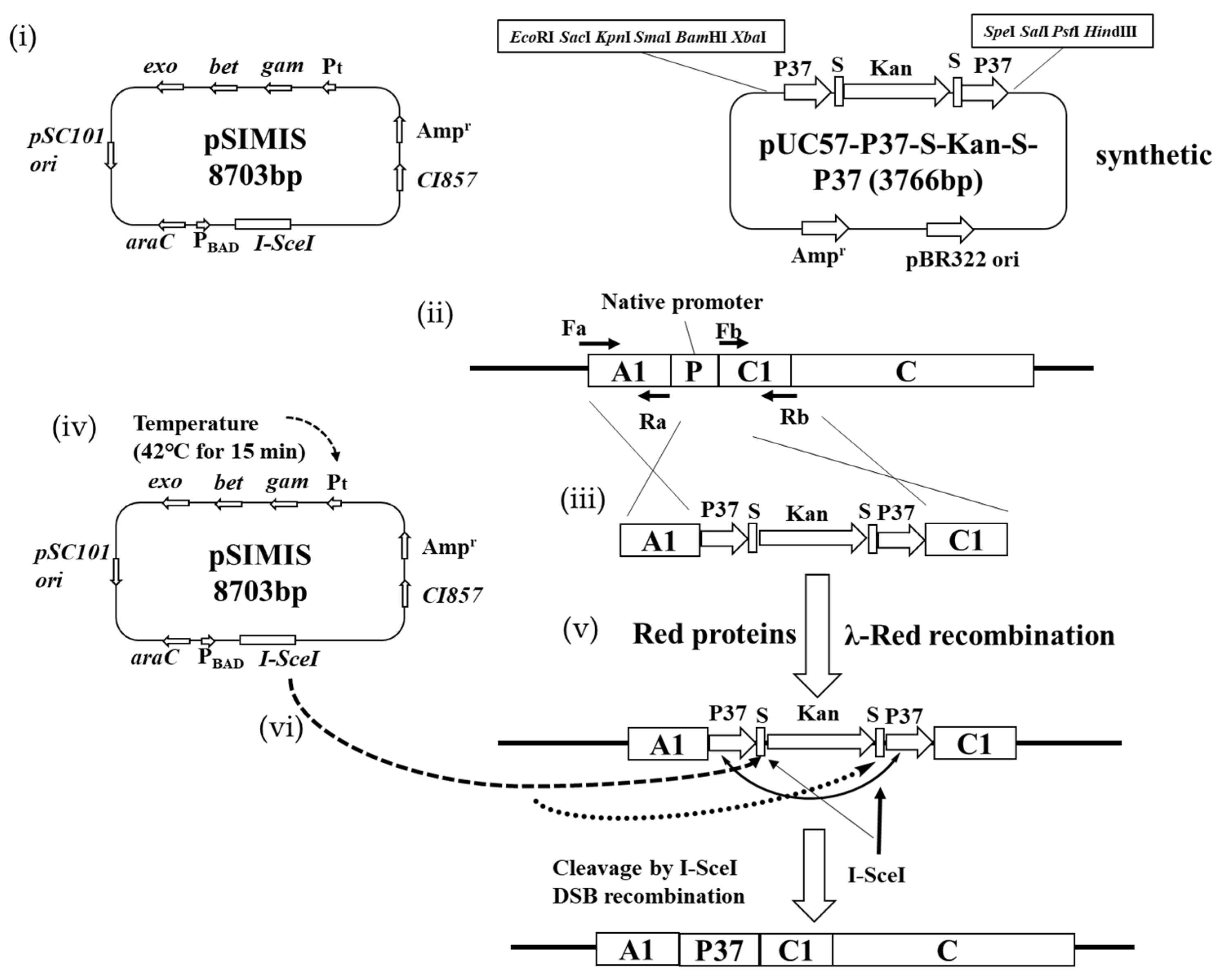
2.6. Promoter Replacement Based on I-SceI System
2.7. Pinene biosynthesis in shake flasks
2.8. GC analysis
2.9. Statistical analysis
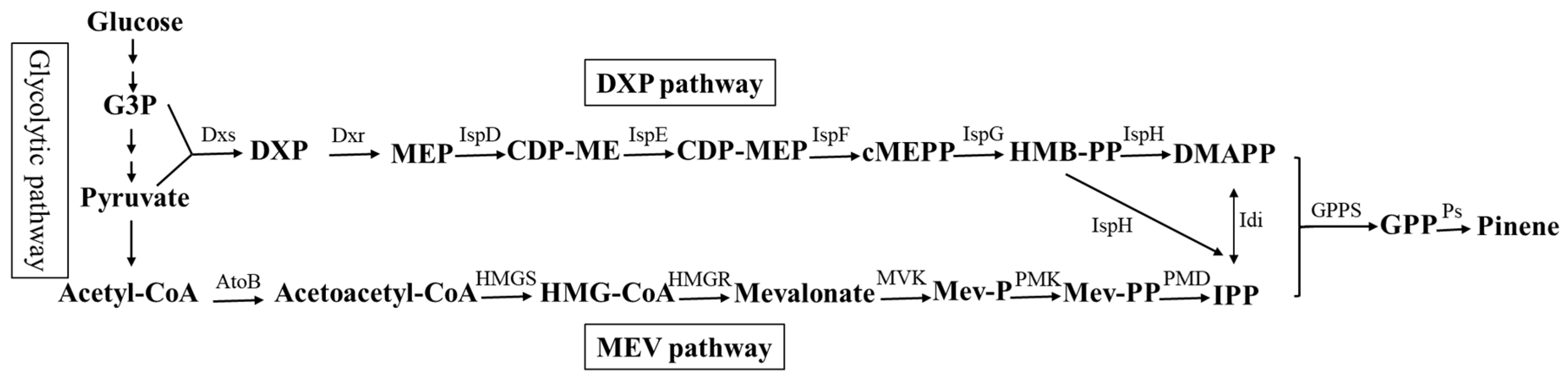
3. Results and Discussion
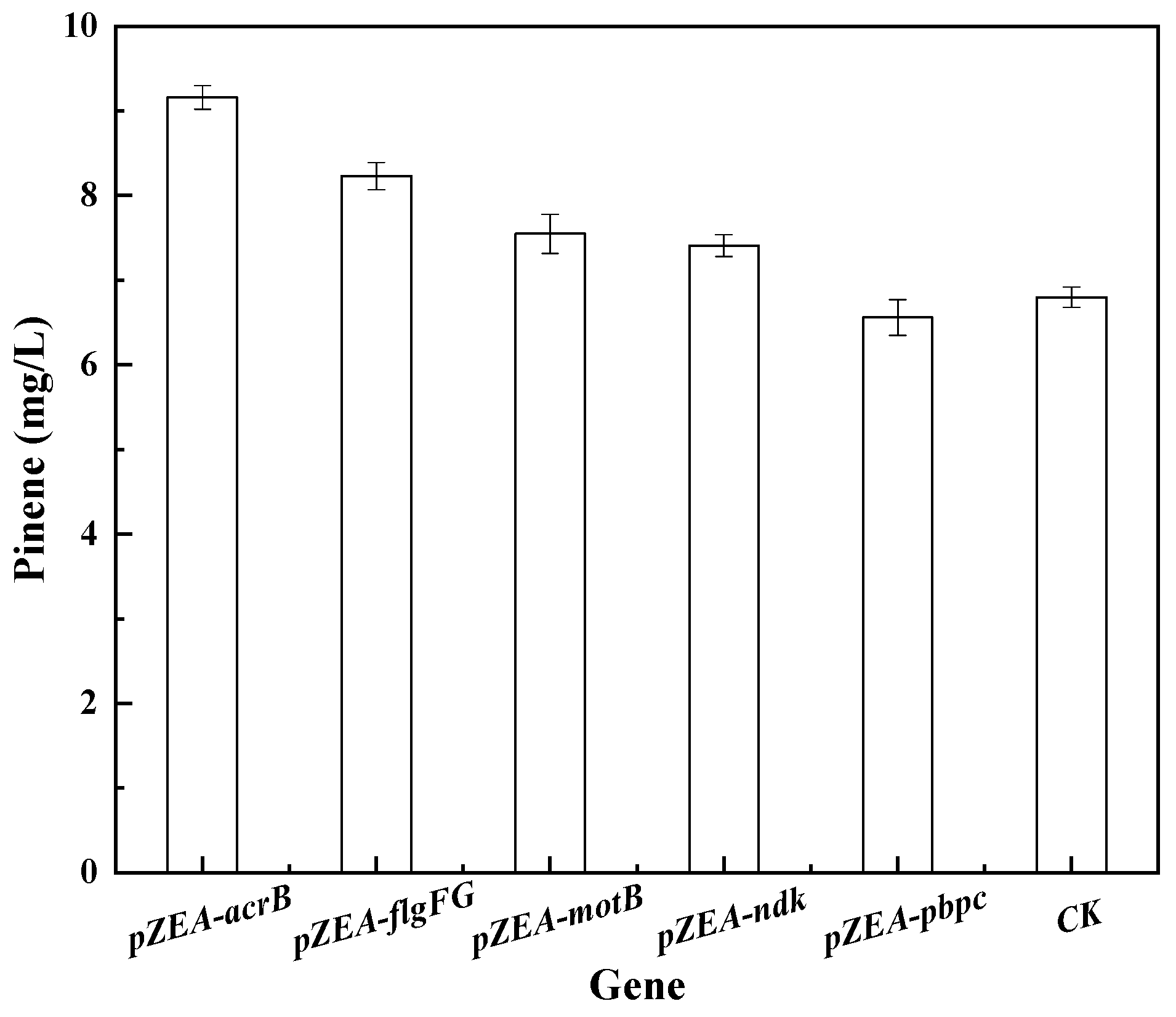
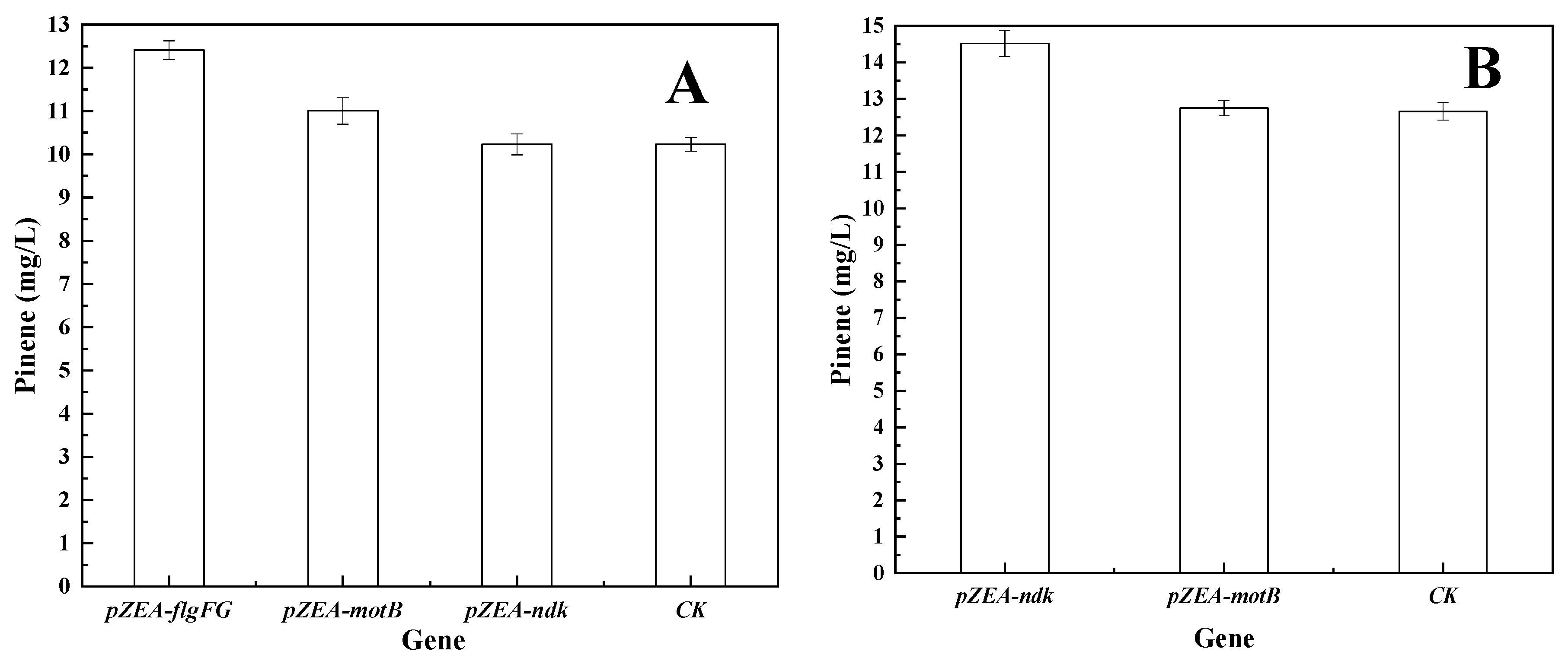
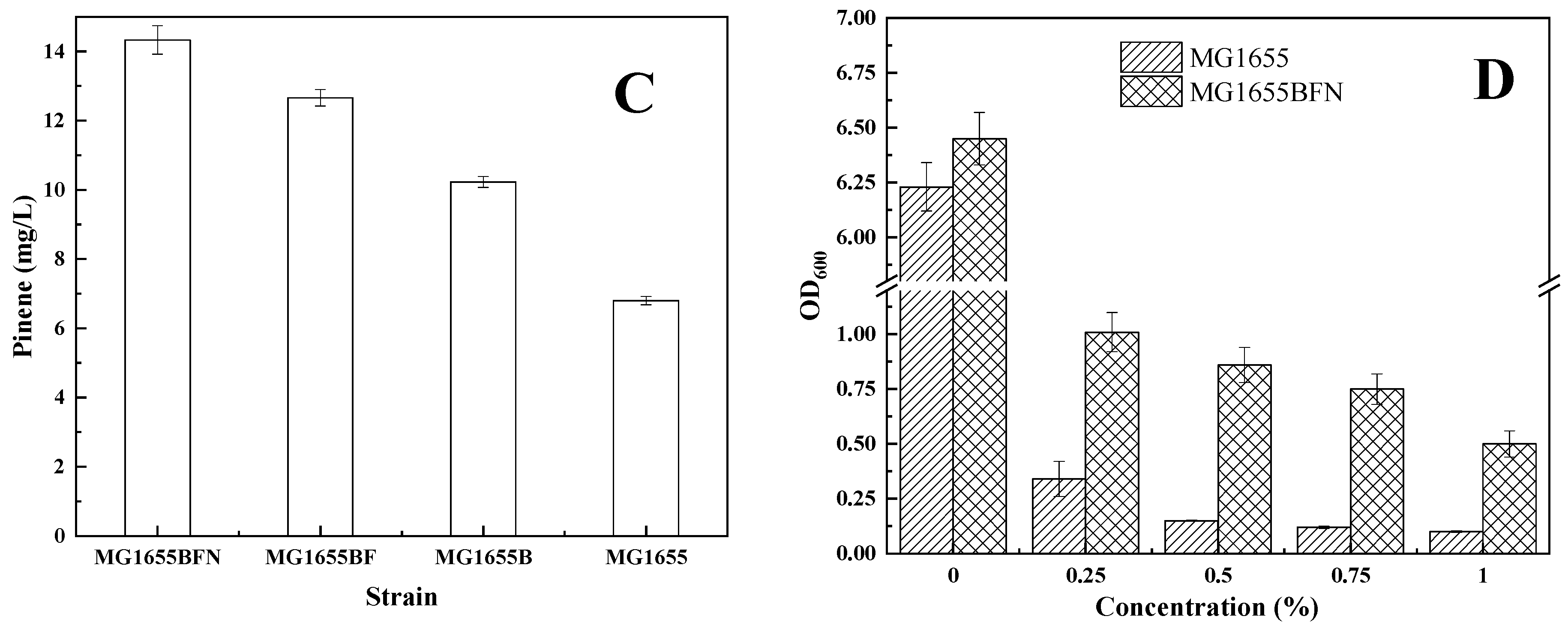
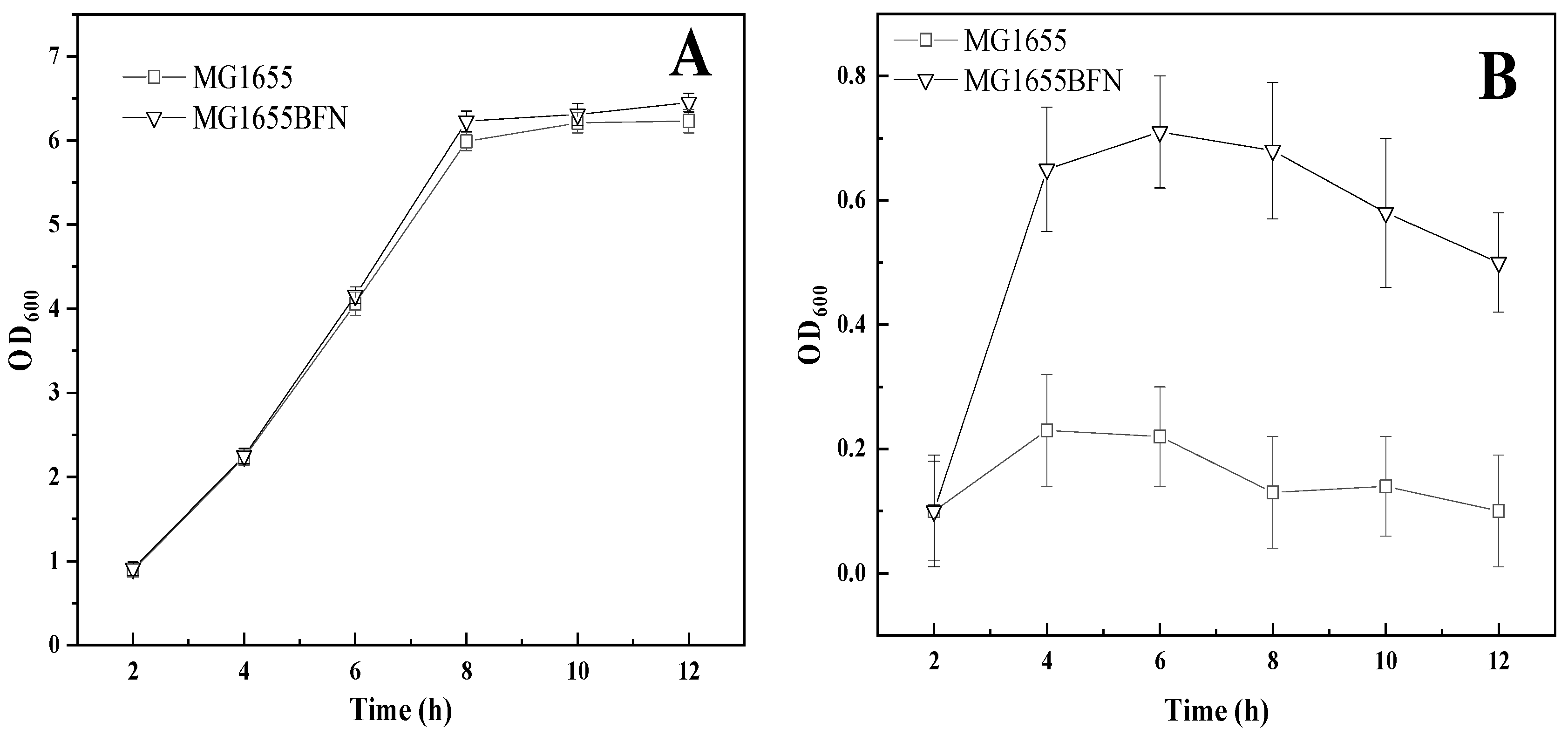
3.1. Isolation of Genes Conferring Increased Pinene Accumulation in E. coli Using the LMS Method
3.2. Effect of I-SceI-Induced Recombination Promoter Replacement on Pinene Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fargione, J.; Hill, J.; Tilman, D.; Polasky, S.; Hawthorne, P.; Land Fargione, J.; Hill, J.; Tilman, D.; Polasky, S.; Hawthorne, P. Land clearing and the biofuel carbon debt. Science 2008, 319, 1235–1238. [Google Scholar] [CrossRef] [PubMed]
- Phillips, E. Chapter 31—Nanotechnological Interventions in Biofuel Production. In Handbook of Biofuels; Sahay, S., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 593–604. ISBN 978-0-12-822810-4. [Google Scholar]
- Lamsen, E.N.; Atsumi, S. Recent progress in synthetic biology for microbial production of C3–C10 alcohols. Front. Microbiol. 2012, 3, 196. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Ranaivoarisoa, T.O.; Singh, R.; Rengasamy, K.; Bose, A. Sustainable production of the biofuel n -butanol by rhodopseudomonas palustris tie-1. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tseng, H.C.; Prather, K. Controlled biosynthesis of odd-chain fuels and chemicals via engineered modular metabolic pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17925–17930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cortez, J.D.; Hammer, S.K.; Carrasco-López, C.; Avalos, J.L. Biosensor for branched-chain amino acid metabolism in yeast and applications in isobutanol and isopentanol production. Nat. Commun. 2022, 13, 270. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, R.; Nielsen, D.R. Styrene biosynthesis from glucose by engineered E. coli. Metab. Eng. 2011, 13, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotech. 2014, 2, 21. [Google Scholar] [CrossRef]
- Tashiro, M.; Kiyota, H.; Kawai-Noma, S.; Saito, K.; Ikeuchi, M.; Iijima, Y.; Umeno, D. Bacterial Production of Pinene by a Laboratory-Evolved Pinene-Synthase. ACS Synth. Biol. 2016, 5, 1011–1020. [Google Scholar] [CrossRef]
- Niu, F.-X.; He, X.; Wu, Y.-Q.; Liu, J.-Z. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front. Microbiol. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Wang, C.; Yoon, S.-H.; Jang, H.-J.; Chung, Y.-R.; Kim, J.-Y.; Choi, E.-S.; Kim, S.-W. Metabolic engineering of Escherichia coli for α-farnesene production. Metab. Eng. 2011, 13, 648–655. [Google Scholar] [CrossRef]
- Yang, X.; Nambou, K.; Wei, L.; Hua, Q. Heterologous production of alpha-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour. Technol. 2016, 216, 1040–1048. [Google Scholar] [CrossRef]
- Mukhopadhyay, A. Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 2015, 23, 498–508. [Google Scholar] [CrossRef]
- Sawada, T.; Eguchi, M.; Asaki, S.; Kashiwagi, R.; Shimomura, K.; Taguchi, F.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; et al. MexEF-OprN multidrug efflux pump transporter negatively controls N-acyl-homoserine lactone accumulation in pseudomonas syringae pv. Tabaci 6605. Mol. Genet. Genom. 2018, 293, 907–917. [Google Scholar] [CrossRef]
- Almeida, J.R.M.; Modig, T.; Petersson, A.; Hahn-Hagerdal, B.; Liden, G.; Gorwa-Grauslund, M.F. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biot. 2007, 82, 340–349. [Google Scholar] [CrossRef]
- Alper, H.; Moxley, J.; Nevoigt, E.; Fink, G.R.; Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 2006, 314, 1565–1568. [Google Scholar] [CrossRef]
- Alsaker, K.V.; Paredes, C.; Papoutsakis, E.T. Metabolite stress and tolerance in the production of biofuels and chemicals: Gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 2010, 105, 1131–1147. [Google Scholar] [CrossRef]
- Atsumi, S.; Wu, T.Y.; Machado, I.; Huang, W.C.; Chen, P.Y.; Pellegrini, M.; Liao, J.C. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010, 6, 449. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.-F.; Li, H.-P.; Wang, L.-Y.; Zhang, C.; Xing, X.-H.; Bao, C.-Y. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl. Microbiol. Biot. 2014, 98, 5387–5396. [Google Scholar] [CrossRef]
- Elhussieny, N.I.; Bakri, M.M.; Ganash, M.; Ghany, T. Chemical mutagenesis of Saccharomyces cerevisiae for enhancing bioethanol production with fermentation at very high sugar concentration. Bioresources 2020, 15, 1354–1369. [Google Scholar] [CrossRef]
- Espeso, D.R.; Dvořák, P.; Aparicio, T.; de Lorenzo, V. An automated DIY framework for experimental evolution of Pseudomonas putida. Microb. Biotechnol. 2021, 14, 2679–2685. [Google Scholar] [CrossRef]
- Wen, Z.; Ledesma-Amaro, R.; Lin, J.; Jiang, Y.; Yang, S.; Atomi, H. Improved n-Butanol Production from Clostridium cellulovorans by Integrated Metabolic and Evolutionary Engineering. Appl. Environ. Microbiol. 2019, 85, e02560-18. [Google Scholar] [CrossRef]
- Shen, Y.-P.; Fong, L.S.; Yan, Z.-B.; Liu, J.-Z. Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli. Biotechnol. Biofuels 2019, 12, 94. [Google Scholar] [CrossRef]
- Niu, F.-X.; Huang, Y.-B.; Ji, L.-N.; Liu, J.-Z. Genomic and transcriptional changes in response to pinene tolerance and overproduction in evolved Escherichia coli. Synth. Syst. Biotechnol. 2019, 4, 113–119. [Google Scholar] [CrossRef]
- George, K.W.; Alonso-Gutierrez, J.; Keasling, J.D.; Lee, T.S. Isoprenoid Drugs, Biofuels, and Chemicals—Artemisinin, Farnesene, and Beyond. In Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Springer International Publishing: Cham, Switzerlad, 2015; pp. 355–389. ISBN 978-3-319-20107-8. [Google Scholar]
- Niu, F.-X.; Huang, Y.-B.; Shen, Y.-P.; Ji, L.-N.; Liu, J.-Z. Enhanced production of pinene by using a cell-free system with modular cocatalysis. J. Agric. Food. Chem. 2020, 68, 2139–2145. [Google Scholar] [CrossRef]
- Whited, G.M.; Feher, F.J.; Benko, D.A. Development of a gas-phase bioprocess for isoprene-monomer production using metabolic pathway engineering. Ind. Biot. 2010, 6, 152–163. [Google Scholar] [CrossRef]
- Zhu, F.; Zhong, X.; Hu, M.; Lu, L.; Deng, Z.; Liu, T. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol. Bioeng. 2014, 111, 1396–1405. [Google Scholar] [CrossRef]
- Li, X.R.; Tian, G.Q.; Shen, H.J.; Liu, J.Z. Metabolic engineering of Escherichia coli to produce zeaxanthin. J. Ind. Microbiol. Biot. 2015, 42, 627. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, B.Y.; Liu, J.Z. Genome engineering Escherichia coli for L-DOPA overproduction from glucose. Sci. Rep. 2016, 6, 30080. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Lee, Y.M.; Yoon, S.H.; Kim, J.H.; Ock, S.W.; Jung, K.H.; Shin, Y.C.; Keasling, J.D.; Kim, S.W. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol. Bioeng. 2005, 91, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.J.; Kang, K.H.; Hyoung, L.J.; Hyun, S.B.; Sun, K.M.; Chang, K.S. Rapid and efficient construction of markerless deletions in the Escherichia coli genome. Nucleic Acids Res. 2008, 36, e84. [Google Scholar] [CrossRef] [PubMed]
- Nordman, J.; Wright, A. Escherichia coli nucleoside diphosphate kinase mutants depend on translesion DNA synthesis to prevent mutagenesis. J. Bacteriol. 2011, 193, 4531–4533. [Google Scholar] [CrossRef]
- Nordman, J.; Wright, A. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc. Natl. Acad. Sci. USA 2008, 105, 10197–10202. [Google Scholar] [CrossRef]
- Suzuki, T.; Komeda, Y. Incomplete flagellar structures in Escherichia coli mutants. J. Bacteriol. 1981, 145, 1036–1041. [Google Scholar] [CrossRef]
- Saijo-Hamano, Y.; Uchida, N.; Namba, K.; Oosawa, K. In vitro characterization of FlgB, FlgC, FlgF, FlgG, and FliE, flagellar basal body proteins of Salmonella. J. Mol. Biol. 2004, 339, 423–435. [Google Scholar] [CrossRef]
- Braun, T.F.; Blair, D.F. Targeted disulfide cross-linking of the motb protein of Escherichia coli: Evidence for two H(+) channels in the stator complex. Biochemistry 2001, 40, 13051–13059. [Google Scholar] [CrossRef]
- Fisher, M.A.; Boyarskiy, S.; Yamada, M.R.; Kong, N.; Bauer, S.; Tullman-Ercek, D. Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli acrb efflux pump to secrete the non-native substrate n-butanol. ACS Synth. Biol. 2014, 3, 30–40. [Google Scholar] [CrossRef]
- Dunlop, M.J.; Dossani, Z.Y.; Szmidt, H.L.; Chu, H.C.; Lee, T.S.; Keasling, J.D.; Hadi, M.Z.; Mukhopadhyay, A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 2011, 7, 487. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, X.; Stephanopoulos, G.; Too, H.P. Efflux transporter engineering markedly improves amorphadiene production in Escherichia coli. Biotechnol. Bioeng. 2016, 113, 1755–1763. [Google Scholar] [CrossRef]
- Foo, J.L.; Leong, S. Directed evolution of an E. coli inner membrane transporter for improved efflux of biofuel molecules. Biotechnol. Biofuels 2013, 6, 81. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Strains/Plasmids | Description | Source/Purpose |
|---|---|---|
| Strain | ||
| E. coli DH5α | supE44 Δ(lacZYA-argF) U169 (Φ80lacZ ΔM15) hsdR17 recA endA1 gyrA96 thi-1 relA1 | Invitrogen |
| E. coli MG1655 | F- λ- ilvG- rfb-50 rph-1 | ATCC#700926 |
| E. coli MG1655B | E. coli MG1655, P37-acrB | This study |
| E. coli MG1655BF | E. coli MG1655B, P37-flgFG | This study |
| E. coli MG1655BFN | E. coli MG1655BF, P37-ndk | This study |
| Plasmid | ||
| pMEVIGPS | pBbA5c-MTSAe-T1f-MBI(f)-T1002i-trGPPSA.grandis-PSA.grandi coding for MEV pathway enzymes to produce pinene from glucose in E. coli, p15A ori, PlacUV5 promoter, CmR | [10] |
| pZEABP | Expression vector, P37 promoter, pBR322 ori, AmpR, BglBrick, ePathBrick contains four isocaudamer (AvrII, NheI, SpeI, and XbaI) | [29] |
| pZEA-acrB | pZEA*BP contains acrB of E. coli, P37 promoter, pBR322 ori, AmpR | This study |
| pZEA-flgFG | pZEA*BP contains flgFG of E. coli, P37 promoter, pBR322 ori, AmpR | This study |
| pZEA-motB | pZEA*BP contains motB of E. coli, P37 promoter, pBR322 ori, AmpR | This study |
| pZEA-ndk | pZEA*BP contains ndk of E. coli, P37 promoter, pBR322 ori, AmpR | This study |
| pZEA-pbpc | pZEA*BP contains pbpc of E. coli, P37 promoter, pBR322 ori, AmpR | This study |
| pUC57 | T vector, pBR322 ori, AmpR | Addgene |
| pUC57-P37-S-Kan-S-P37 | pUC57 contains P37-S-Kan-S-P37 cassette | This study |
| pUC-IsecI-acrB | pUC57-P37-S-Kan-S-P37 containing upstream and downstream homologous arms for promoter replacement of acrB | This study |
| pUC-IsecI-flgFG | pUC57-P37-S-Kan-S-P37 containing upstream and downstream homologous arms for promoter replacement of flgF | This study |
| pUC-IsecI-ndk | pUC57-P37-S-Kan-S-P37 containing upstream and downstream homologous arms for promoter replacement of ndk | This study |
| pSIMIS | pSC101 repliconts PL-gam-bet-exo cI857, arabinose-inducible I-SceI endonuclease gene, AmpR | [30] |
| Positive Clone | Gene | Protein | Reference |
|---|---|---|---|
| P1 | pbpc-ndk | peptidoglycan glycosyltransferase PbpC, nucleoside diphosphate kinase | [33,34] |
| P2 | flgF-flgG | flagellar basal-body rod protein FlgF, flagellar basal-body rod protein FlgG | [35,36] |
| P3 | motB | protein that enables flagellar motor rotation | [37] |
| P4 | acrB | multidrug efflux pump RND permease AcrB | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.-Y.; Wang, W.-Y.; Liang, Z.-Z.; Huang, Y.-C.; Yi, Y.; Niu, F.-X. Enhancing the Production of Pinene in Escherichia coli by Using a Combination of Shotgun, Product-Tolerance and I-SceI Cleavage Systems. Biology 2022, 11, 1484. https://doi.org/10.3390/biology11101484
Huang M-Y, Wang W-Y, Liang Z-Z, Huang Y-C, Yi Y, Niu F-X. Enhancing the Production of Pinene in Escherichia coli by Using a Combination of Shotgun, Product-Tolerance and I-SceI Cleavage Systems. Biology. 2022; 11(10):1484. https://doi.org/10.3390/biology11101484
Chicago/Turabian StyleHuang, Ming-Yue, Wei-Yang Wang, Zhen-Zhen Liang, Yu-Chen Huang, Yi Yi, and Fu-Xing Niu. 2022. "Enhancing the Production of Pinene in Escherichia coli by Using a Combination of Shotgun, Product-Tolerance and I-SceI Cleavage Systems" Biology 11, no. 10: 1484. https://doi.org/10.3390/biology11101484
APA StyleHuang, M.-Y., Wang, W.-Y., Liang, Z.-Z., Huang, Y.-C., Yi, Y., & Niu, F.-X. (2022). Enhancing the Production of Pinene in Escherichia coli by Using a Combination of Shotgun, Product-Tolerance and I-SceI Cleavage Systems. Biology, 11(10), 1484. https://doi.org/10.3390/biology11101484






