Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Generation of Stable RGD-Expressing Cell Line RGD-HEK 293FT
2.3. Extracellular Vesicle Isolation and Characterisation
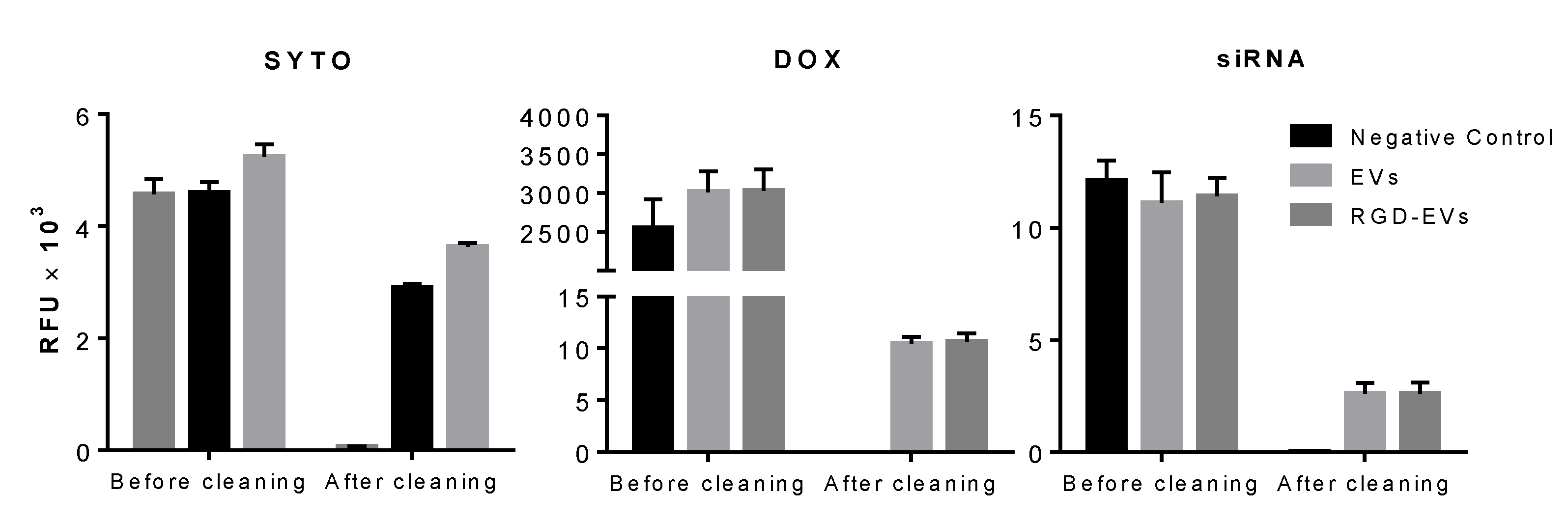
2.4. EV Uptake Assay
2.5. Doxorubicin Loading
2.6. Cell Viability Assay
2.7. siRNA Loading and Gene Expression Analysis
2.8. Statistical Analysis
3. Results
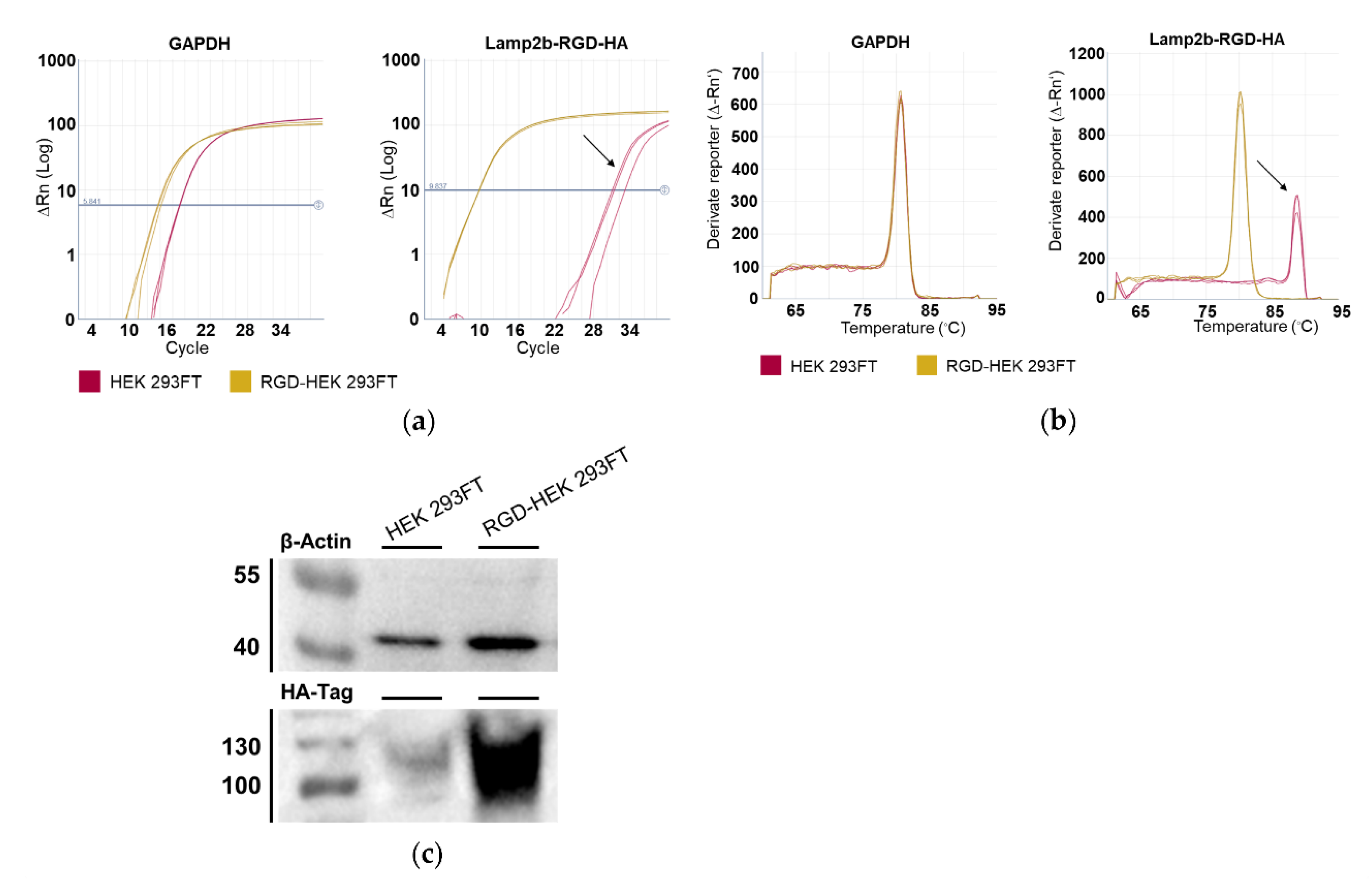
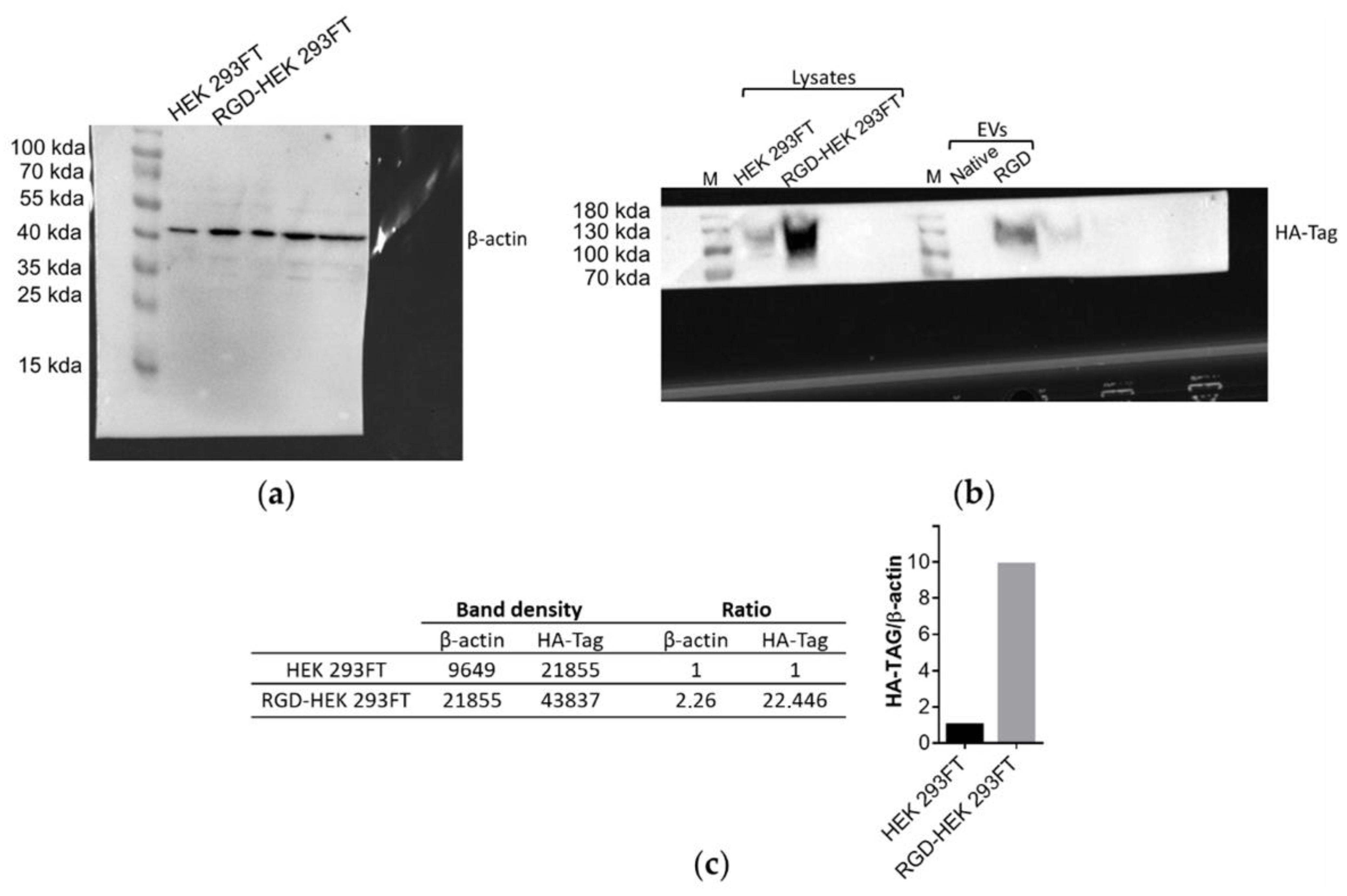
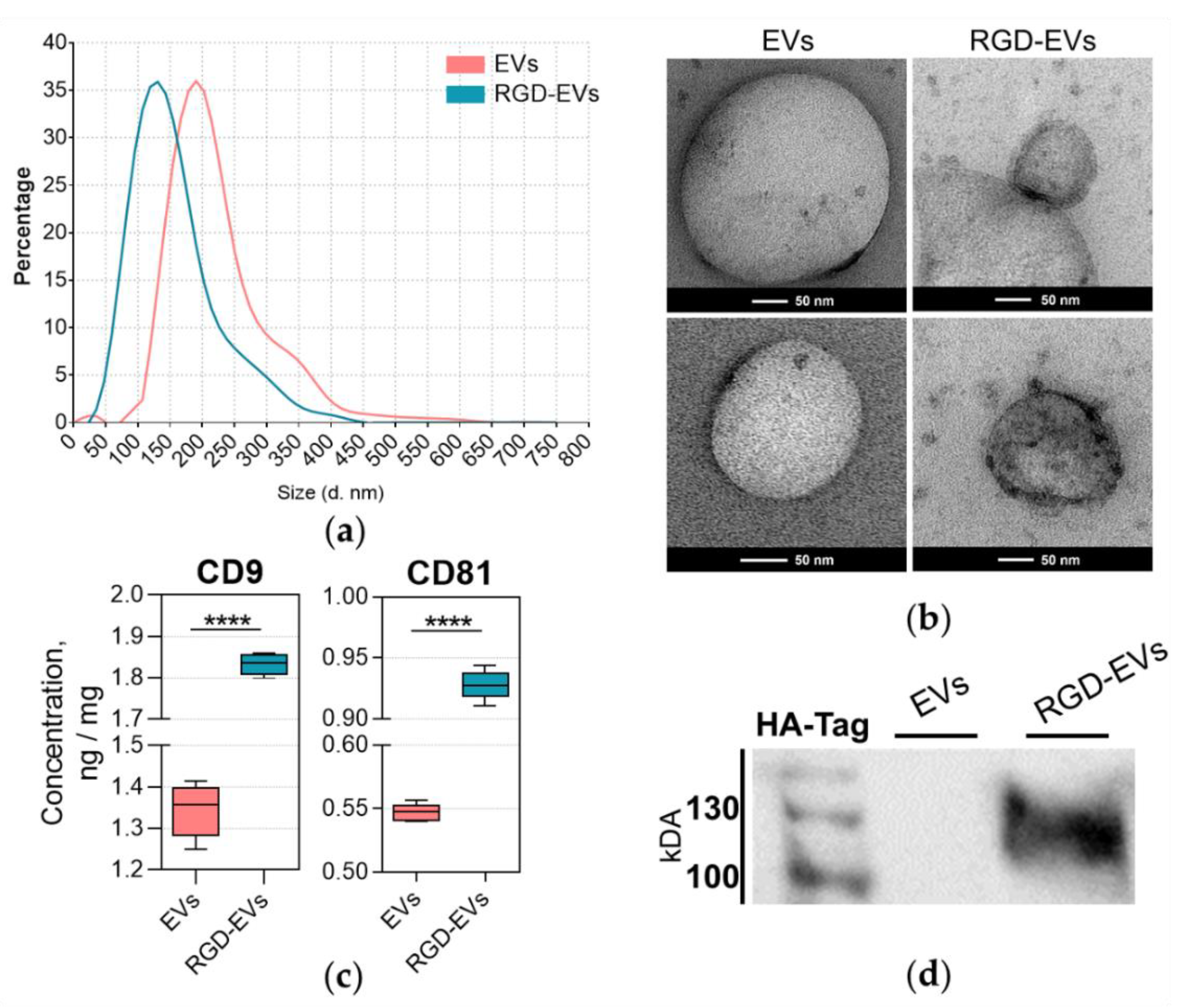
3.1. Lamp2b-RGD-HA Expression in RGD-HEK 293FT Cells and EVs
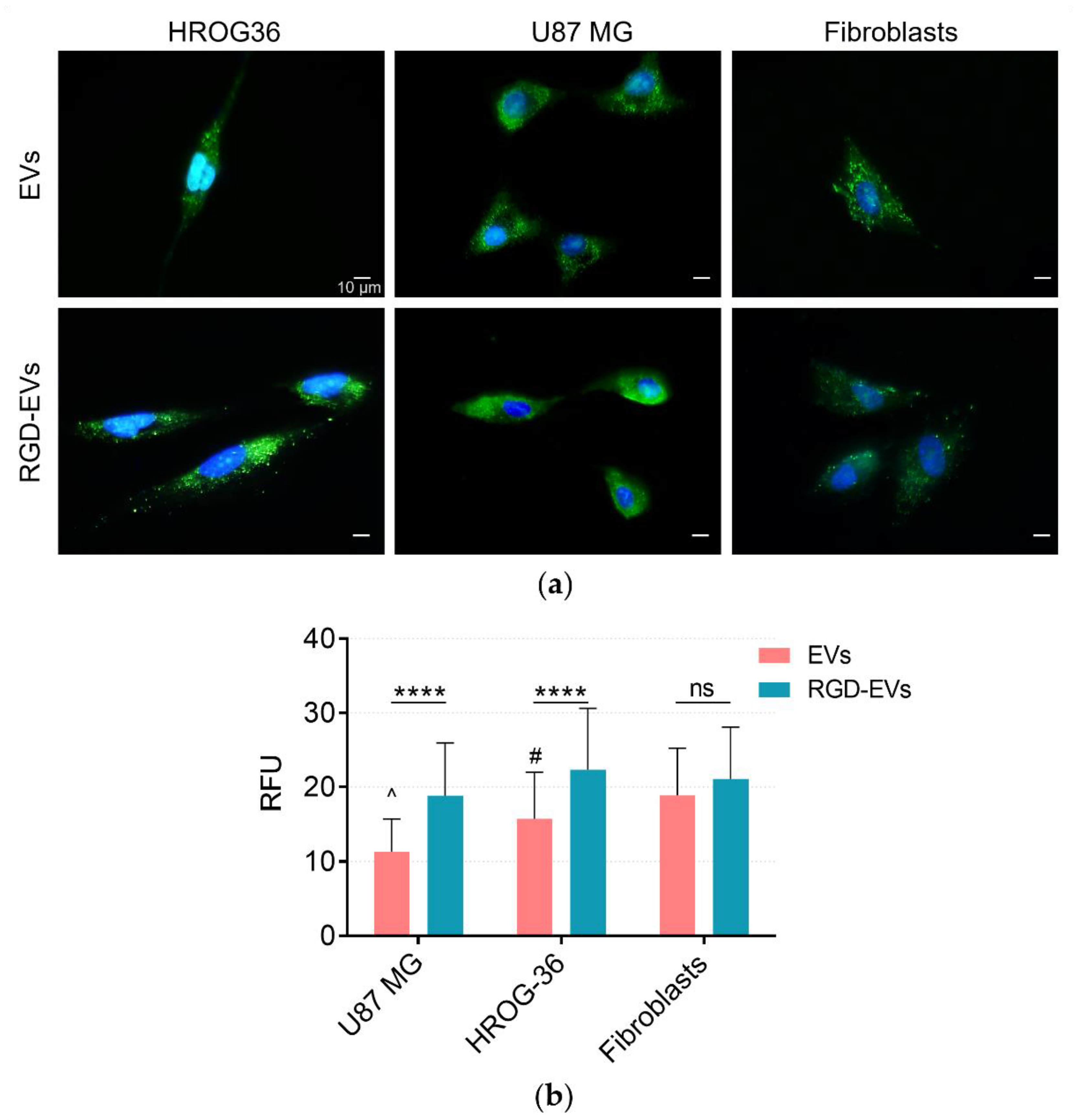
3.2. Tracing EV Uptake by HROG36, U87 MG and Human Fibroblast Cells
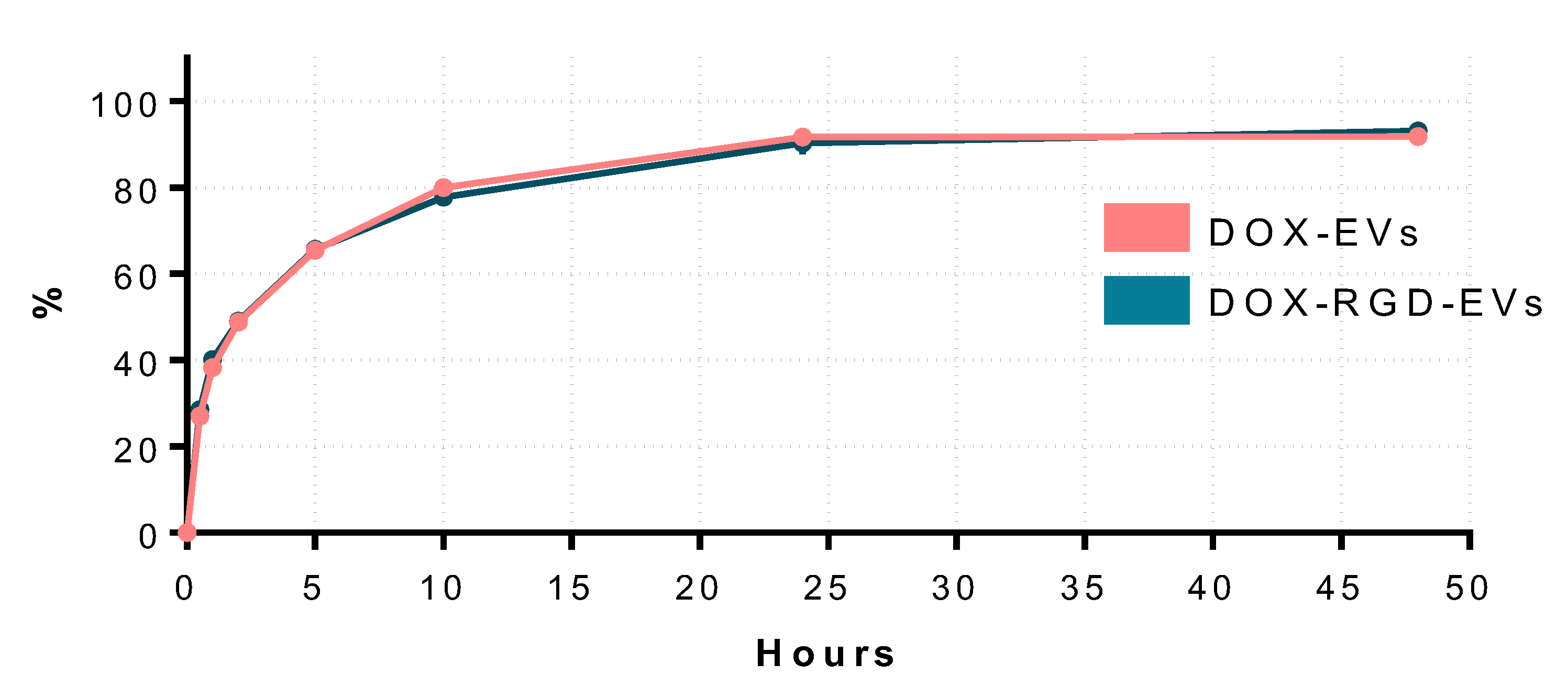
3.3. DOX Delivery by Exosome-Like Particles
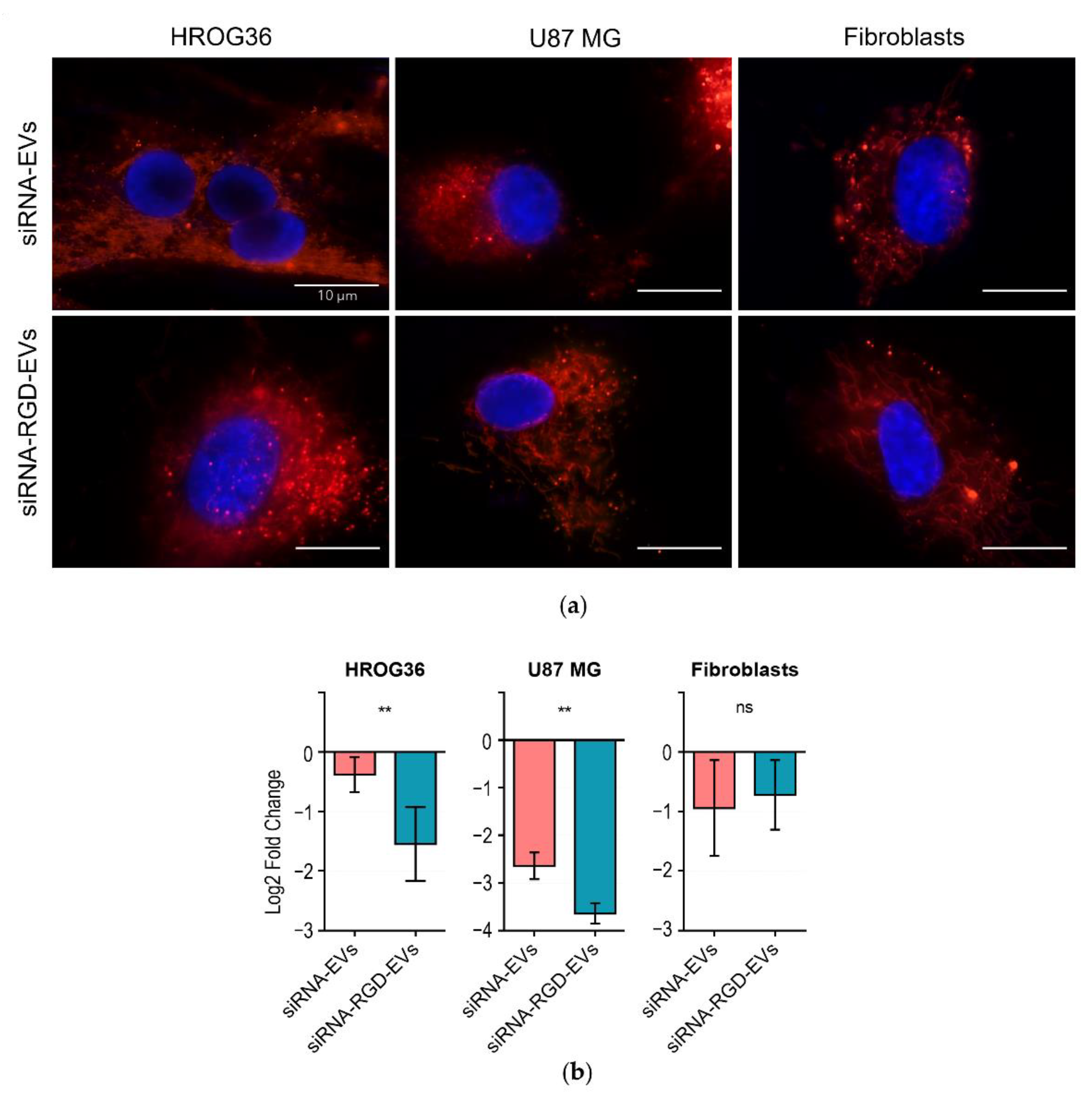
3.4. siRNA Delivery to GBM and Fibroblast Cells by EVs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Target | Sequence |
|---|---|
| β-Actin | F: AGAGCTACGAGCTGCCTGAC R: AGCACTGTGTTGGCGTACAG |
| GAPDH | F: TCAAGATCATCAGCAATGCCT R: CATGAGTCCCACGATACC |
| Lamp2b-RGD-HA | F: GGCGATAAAGGCCCGGATT R: CAGGAAAAGCCAGGTCCGAA |
References
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6977. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef]
- Isola, A.L.; Chen, S. Exosomes: The Messengers of Health and Disease. Curr. Neuropharmacol. 2017, 15, 157. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of Exosomes in Central Nervous System Diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef]
- Bellin, G.; Gardin, C.; Ferroni, L.; Chachques, J.C.; Rogante, M.; Mitrečić, D.; Ferrari, R.; Zavan, B. Exosome in Cardiovascular Diseases: A Complex World Full of Hope. Cells 2019, 8, 166. [Google Scholar] [CrossRef]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 91, 1–11. [Google Scholar] [CrossRef]
- Zhu, X.; Badawi, M.; Pomeroy, S.; Sutaria, D.S.; Xie, Z.; Baek, A.; Jiang, J.; Elgamal, O.A.; Mo, X.; Perle, K.; et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HET cells. J. Extracell. Vesicles 2017, 6, 1324730. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Zhang, X.; Huang, H.; Tang, S.; Chai, Y.; Xu, Z.; Li, M.; Chen, X.; Liu, J.; et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnol. 2022, 20, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Maisano, D.; Mimmi, S.; Russo, R.; Fioravanti, A.; Fiume, G.; Vecchio, E.; Nisticò, N.; Quinto, I.; Iaccino, E.; Nakada, M.; et al. Potential Strategies Uncovering the Exosomes Diversity: A Window of Opportunity for Tumor Progression Monitoring. Overcoming the Temozolomide Resistance for Glioblastoma. Pharmaceuticals 2020, 13, 180. [Google Scholar] [CrossRef]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Paolillo, M.; Serra, M.; Schinelli, S. Integrins in glioblastoma: Still an attractive target? Pharmacol. Res. 2016, 113, 55–61. [Google Scholar] [CrossRef]
- Belvisi, L.; Riccioni, T.; Marcellini, M.; Vesci, L.; Chiarucci, I.; Efrati, D.; Potenza, D.; Scolastico, C.; Manzoni, L.; Lombardo, K.; et al. Biological and molecular properties of a new A v B 3/A v B 5 integrin antagonist. Mol. Cancer Ther. 2005, 4, 1670–1680. [Google Scholar] [CrossRef]
- Schnell, O.; Krebs, B.; Wagner, E.; Romagna, A.; Beer, A.J.; Grau, S.J.; Thon, N.; Goetz, C.; Kretzschmar, H.A.; Tonn, J.C.; et al. Expression of Integrin αvβ3 in Gliomas Correlates with Tumor Grade and Is not Restricted to Tumor Vasculature. Brain Pathol. 2008, 18, 378. [Google Scholar] [CrossRef]
- Echavidre, W.; Picco, V.; Faraggi, M.; Montemagno, C. Integrin-αvβ3 as a Therapeutic Target in Glioblastoma: Back to the Future? Pharmaceutics 2022, 14, 1053. [Google Scholar] [CrossRef] [PubMed]
- El-Andaloussi, S.; Lee, Y.; Lakhal-Littleton, S.; Li, J.; Seow, Y.; Gardiner, C.; Alvarez-Erviti, L.; Sargent, I.L.; Wood, M.J.A. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012, 7, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Tian, J.; Wang, Z.; Gao, Y.; Wu, X.; Ding, X.; Qiang, L.; Li, G.; Han, Z.; Yuan, Y.; et al. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnol. 2019, 17, 1–18. [Google Scholar] [CrossRef]
- Zhou, Y.; Yuan, Y.; Liu, M.; Hu, X.; Quan, Y.; Chen, X. Tumor-specific delivery of KRAS siRNA with iRGD-exosomes efficiently inhibits tumor growth. ExRNA 2019, 1, 1–7. [Google Scholar] [CrossRef]
- Song, S.; Mao, G.; Du, J.; Zhu, X. Novel RGD containing, temozolomide-loading nanostructured lipid carriers for glioblastoma multiforme chemotherapy. Drug Deliv. 2016, 23, 1404–1408. [Google Scholar] [CrossRef]
- Quader, S.; Liu, X.; Chen, Y.; Mi, P.; Chida, T.; Ishii, T.; Miura, Y.; Nishiyama, N.; Cabral, H.; Kataoka, K. cRGD peptide-installed epirubicin-loaded polymeric micelles for effective targeted therapy against brain tumors. J. Control. Release 2017, 258, 56–66. [Google Scholar] [CrossRef]
- Jiang, X.; Sha, X.; Xin, H.; Xu, X.; Gu, J.; Xia, W.; Chen, S.; Xie, Y.; Chen, L.; Chen, Y.; et al. Integrin-facilitated transcytosis for enhanced penetration of advanced gliomas by poly(trimethylene carbonate)-based nanoparticles encapsulating paclitaxel. Biomaterials 2013, 34, 2969–2979. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Ling, X.; Yang, Y.; Zhang, J.; Li, Q.; Niu, X.; Hu, G.; Chen, B.; Li, H.; Wang, Y.; et al. Embryonic Stem Cells-Derived Exosomes Endowed with Targeting Properties as Chemotherapeutics Delivery Vehicles for Glioblastoma Therapy. Adv. Sci. 2019, 6, 1801899. [Google Scholar] [CrossRef]
- Kučuk, N.; Primožič, M.; Knez, Ž.; Leitgeb, M. Exosomes Engineering and Their Roles as Therapy Delivery Tools, Therapeutic Targets, and Biomarkers. Int. J. Mol. Sci. 2021, 22, 9543. [Google Scholar] [CrossRef]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585. [Google Scholar] [CrossRef]
- Che, Y.; Shi, X.; Shi, Y.; Jiang, X.; Ai, Q.; Shi, Y.; Gong, F.; Jiang, W. Exosomes Derived from miR-143-Overexpressing MSCs Inhibit Cell Migration and Invasion in Human Prostate Cancer by Downregulating TFF3. Mol. Ther. Nucleic Acids 2019, 18, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.E.; Leonard, J.N. Stabilization of Exosome-targeting Peptides via Engineered Glycosylation. J. Biol. Chem. 2015, 290, 8166–8172. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Ayed, Z.; Cuvillier, L.; Dobhal, G.; Goreham, R.V. Electroporation of outer membrane vesicles derived from Pseudomonas aeruginosa with gold nanoparticles. SN Appl. Sci. 2019, 1, 1600. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.; Gittleman, H.; Liao, P.; Rouse, C.; Chen, Y.; Dowling, J.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro-Oncology 2014, 16, iv1–iv63. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterise heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Ferreira, J.V.; Soares, A.D.R.; Ramalho, J.; Carvalho, C.M.; Cardoso, M.H.; Pintado, P.; Carvalho, A.S.; Beck, H.C.; Matthiesen, R.; Zuzarte, M.; et al. LAMP2A regulates the loading of proteins into exosomes. Sci. Adv. 2022, 8, 1140. [Google Scholar] [CrossRef]
- Stanke, K.M.; Wilson, C.; Kidambi, S. High Expression of Glycolytic Genes in Clinical Glioblastoma Patients Correlates With Lower Survival. Front. Mol. Biosci. 2021, 8, 1281. [Google Scholar] [CrossRef]
- French, K.C.; Antonyak, M.A.; Cerione, R.A. Extracellular vesicle docking at the cellular port: Extracellular vesicle binding and uptake. Semin. Cell Dev. Biol. 2017, 67, 48. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Dai, G.; Wu, Y.; Zhang, M.; Yang, M.; Wang, X.; Song, M.; Li, X.; Xia, R.; Wu, Z. iRGD-modified exosomes-delivered BCL6 siRNA inhibit the progression of diffuse large B-cell lymphoma. Front. Oncol. 2022, 12, 3763. [Google Scholar] [CrossRef]
- Cen, B.; Wei, Y.; Huang, W.; Teng, M.; He, S.; Li, J.; Wang, W.; He, G.; Bai, X.; Liu, X.; et al. An Efficient Bivalent Cyclic RGD-PIK3CB siRNA Conjugate for Specific Targeted Therapy against Glioblastoma In Vitro and In Vivo. Mol. Ther. Nucleic Acids 2018, 13, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-W.; Lo, J.-M.; Juan, H.-F.; Chang, H.-Y.; Chuang, C.-Y. Combination of RGD Compound and Low-Dose Paclitaxel Induces Apoptosis in Human Glioblastoma Cells. PLoS ONE 2012, 7, e37935. [Google Scholar] [CrossRef]
- Hou, J.; Diao, Y.; Li, W.; Yang, Z.; Zhang, L.; Chen, Z.; Wu, Y. RGD peptide conjugation results in enhanced antitumor activity of PD0325901 against glioblastoma by both tumor-targeting delivery and combination therapy. Int. J. Pharm. 2016, 505, 329–340. [Google Scholar] [CrossRef]
- Xin, L.; Yuan, Y.-W.; Liu, C.; Zhou, L.-Q.; Liu, L.; Zhou, Q.; Li, S.-H. Preparation of Internalizing RGD-Modified Recombinant Methioninase Exosome Active Targeting Vector and Antitumor Effect Evaluation. Am. J. Dig. Dis. 2020, 66, 1045–1053. [Google Scholar] [CrossRef]
- Zhao, Z.; Shuang, T.; Gao, Y.; Lu, F.; Zhang, J.; He, W.; Qu, L.; Chen, B.; Hao, Q. Targeted delivery of exosomal miR-484 reprograms tumor vasculature for chemotherapy sensitisation. Cancer Lett. 2022, 530, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Roth, P.; Silginer, M.; Goodman, S.L.; Hasenbach, K.; Thies, S.; Maurer, G.; Schraml, P.; Tabatabai, G.; Moch, H.; Tritschler, I.; et al. Integrin control of the transforming growth factor-β pathway in glioblastoma. Brain 2013, 136, 564–576. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gečys, D.; Kazlauskas, A.; Gečytė, E.; Paužienė, N.; Kulakauskienė, D.; Lukminaitė, I.; Jekabsone, A. Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells. Biology 2022, 11, 1483. https://doi.org/10.3390/biology11101483
Gečys D, Kazlauskas A, Gečytė E, Paužienė N, Kulakauskienė D, Lukminaitė I, Jekabsone A. Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells. Biology. 2022; 11(10):1483. https://doi.org/10.3390/biology11101483
Chicago/Turabian StyleGečys, Dovydas, Arūnas Kazlauskas, Emilija Gečytė, Neringa Paužienė, Deimantė Kulakauskienė, Indrė Lukminaitė, and Aistė Jekabsone. 2022. "Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells" Biology 11, no. 10: 1483. https://doi.org/10.3390/biology11101483
APA StyleGečys, D., Kazlauskas, A., Gečytė, E., Paužienė, N., Kulakauskienė, D., Lukminaitė, I., & Jekabsone, A. (2022). Internalisation of RGD-Engineered Extracellular Vesicles by Glioblastoma Cells. Biology, 11(10), 1483. https://doi.org/10.3390/biology11101483






