Paramutation-like Epigenetic Conversion by piRNA at the Telomere of Drosophila virilis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks and Maintenance
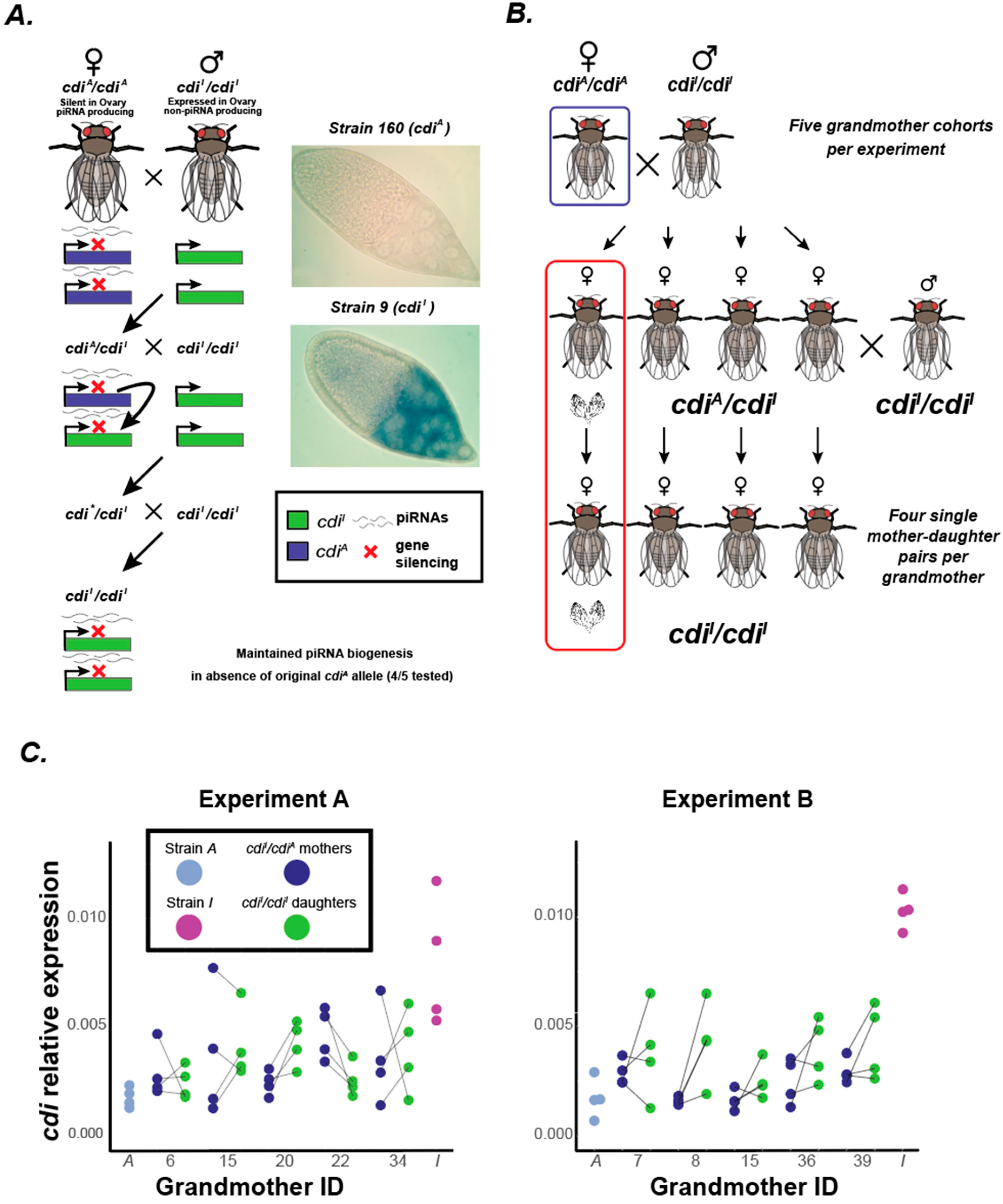
2.2. Cross Scheme
2.3. Replicate A
2.4. Replicate B
2.5. Daughter cdiI/cdiI Indel PCR Genotyping Assay and Sanger Sequencing Verification
2.6. Ovary RNA Extractions
2.7. DNase Treatment
2.8. Reverse Transcription
2.9. qPCR Analysis
2.10. Statistical Analysis
2.11. Nanopore Sequencing. TE Library and Telomere Annotation
2.12. piRNA Sequences and Mapping
2.13. Whole Mount In Situ RNA Hybridization
3. Results
3.1. Gene Silencing of cdi in the Ovary Is Maintained in the Absence of the Original Silencing cdiA Allele
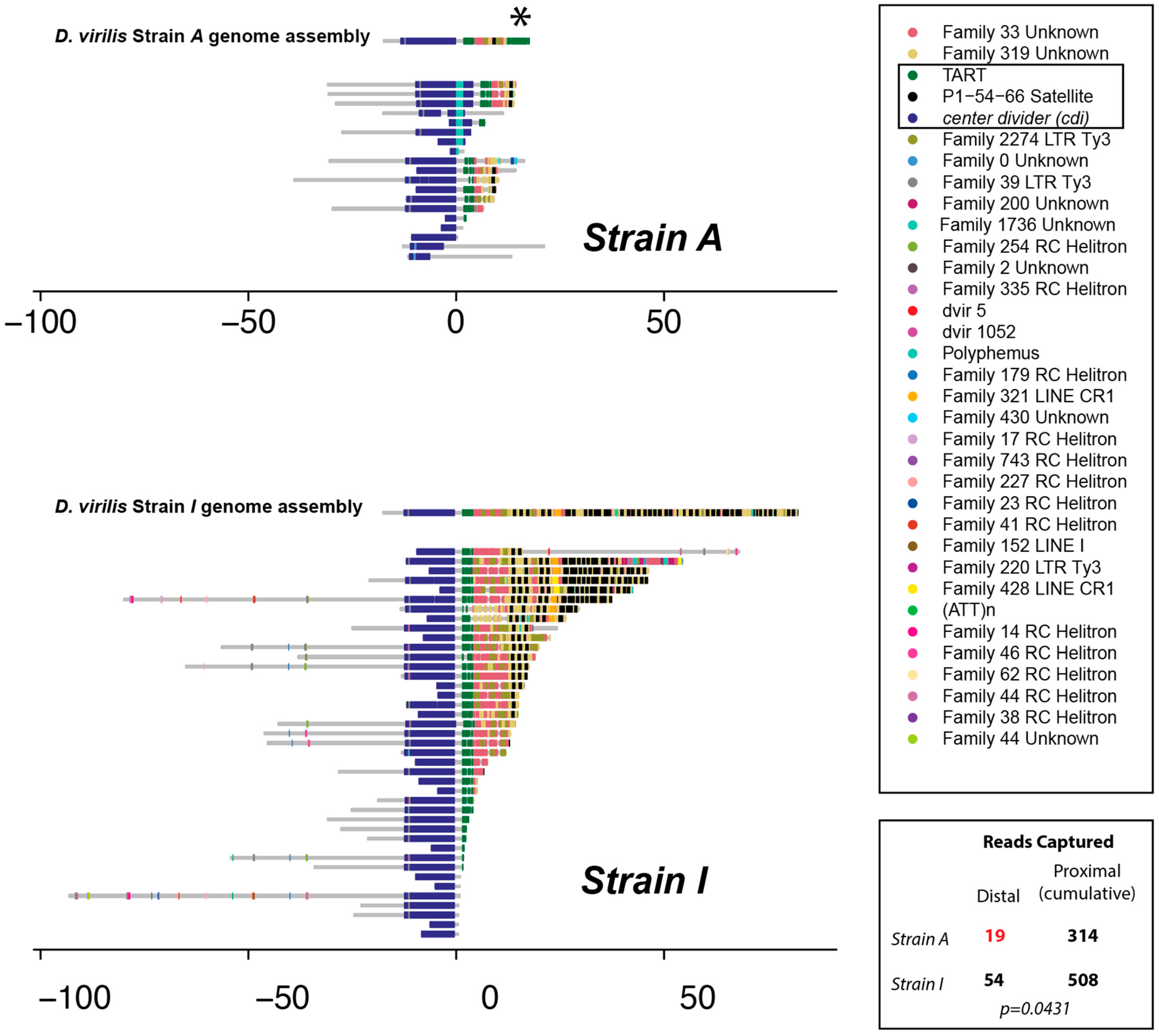
3.2. Single-Read Nanopore Sequencing Analysis of the Subtelomeric Regions of cdiA and cdiA Strains
3.3. piRNA Mapping to Individual Parental Genome Assemblies of the Telomere
4. Discussion
4.1. Paramutation or a Maternal Effect?
4.2. What Drives Epigenetic Conversion of a Sub-Telomeric Genic piRNA Cluster?
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chandler, V.L.; Stam, M.; Sidorenko, L.V. 7 Long-distance Cis and Trans interactions mediate paramutation. Adv Genet. 2002, 46, 215–234. [Google Scholar] [CrossRef]
- Coe, E.H. A regular and continuing conversion-type phenomenon at the b locus in maize. Proc. Natl. Acad. Sci. USA 1959, 45, 828–832. [Google Scholar] [CrossRef]
- Belele, C.L.; Sidorenko, L.; Stam, M.; Bader, R.; Arteaga-Vazquez, M.A.; Chandler, V.L. Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing. PLoS Genet. 2013, 9, e1003773. [Google Scholar] [CrossRef] [PubMed]
- Alleman, M.; Sidorenko, L.; McGinnis, K.; Seshadri, V.; Dorweiler, J.E.; White, J.; Sikkink, K.; Chandler, V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 2006, 442, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Arteaga-Vazquez, M.; Sidorenko, L.; Rabanal, F.A.; Shrivistava, R.; Nobuta, K.; Green, P.J.; Meyers, B.C.; Chandler, V.L. RNA-mediated trans -communication can establish paramutation at the b1 locus in maize. Proc. Natl. Acad. Sci. USA 2010, 107, 12986–12991. [Google Scholar] [CrossRef] [PubMed]
- Hollick, J.B. Paramutation and related phenomena in diverse species. Nat. Rev. Genet. 2017, 18, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Minkina, O.; Hunter, C.P. Stable Heritable Germline Silencing Directs Somatic Silencing at an Endogenous Locus. Mol. Cell 2017, 65, 659–670.e5. [Google Scholar] [CrossRef]
- Sapetschnig, A.; Sarkies, P.; Lehrbach, N.J.; Miska, E.A. Tertiary siRNAs Mediate Paramutation in C. elegans. PLoS Genet. 2015, 11, e1005078. [Google Scholar] [CrossRef] [PubMed]
- de Vanssay, A.; Bouge, A.L.; Boivin, A.; Hermant, C.; Teysset, L.; Delmarre, V.; Antoniewski, C.; Ronsseray, S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 2012, 490, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B 2020, 96, 32–42. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science 2008, 322, 1387–1392. [Google Scholar] [CrossRef]
- Akkouche, A.; Mugat, B.; Barckmann, B.; Varela-Chavez, C.; Li, B.; Raffel, R.; Pélisson, A.; Chambeyron, S. Piwi Is Required during Drosophila Embryogenesis to License Dual-Strand piRNA Clusters for Transposon Repression in Adult Ovaries. Mol. Cell 2017, 66, 411–419.e4. [Google Scholar] [CrossRef]
- Le Thomas, A.; Marinov, G.K.; Aravin, A.A. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep. 2014, 8, 1617–1623. [Google Scholar] [CrossRef]
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.-C.A.; Luo, Y.; Sachidanandam, R.; Toth, K.F.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680. [Google Scholar] [CrossRef]
- Ronsseray, S.; Boivin, A.; Anxolabehere, D. P-element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics 2001, 159, 1631–1642. [Google Scholar] [CrossRef]
- Rozhkov, N.V.; Aravin, A.A.; Zelentsova, E.S.; Schostak, N.G.; Sachidanandam, R.; McCombie, W.R.; Hannon, G.J.; Evgen’Ev, M.B. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 2010, 16, 1634–1645. [Google Scholar] [CrossRef]
- Evgen’ev, M.B.; Zelentsova, H.; Shostak, N.; Kozitsina, M.; Barskyi, V.; Lankenau, D.H.; Corces, V.G. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1997, 94, 196–201. [Google Scholar] [CrossRef]
- Funikov, S.Y.; Kulikova, D.A.; Krasnov, G.S.; Rezvykh, A.P.; Chuvakova, L.N.; Shostak, N.G.; Zelentsova, E.S.; Blumenstiel, J.P.; Evgen’Ev, M.B. Spontaneous gain of susceptibility suggests a novel mechanism of resistance to hybrid dysgenesis in Drosophila virilis. PLoS Genet. 2018, 14, e1007400. [Google Scholar] [CrossRef]
- Rezvykh, A.; Funikov, S.; Protsenko, L.; Kulikova, D.; Zelentsova, E.; Chuvakova, L.; Blumenstiel, J.; Evgen’Ev, M. Evolutionary Dynamics of the Pericentromeric Heterochromatin in Drosophila virilis and Related Species. Genes 2021, 12, 175. [Google Scholar] [CrossRef]
- Lyozin, G.T.; Makarova, K.S.; Velikodvorskaja, V.V.; Zelentsova, H.S.; Khechumian, R.R.; Kidwell, M.G.; Koonin, E.V.; Evgen’ev, M.B. The structure and evolution of Penelope in the virilis species group of Drosophila: An ancient lineage of retroelements. J. Mol. Evol. 2001, 52, 445–456. [Google Scholar] [CrossRef]
- Petrov, D.A.; Schutzman, J.L.; Hartl, D.L.; Lozovskaya, E.R. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1995, 92, 8050–8054. [Google Scholar] [CrossRef]
- Vieira, J.; Vieira, C.P.; Hartl, D.L.; Lozovskaya, E.R. Factors contributing to the hybrid dysgenesis syndrome in Drosophila virilis. Genet. Res. 1998, 71, 109–117. [Google Scholar] [CrossRef]
- Blumenstiel, J.P. Whole genome sequencing in Drosophila virilis identifies Polyphemus, a recently activated Tc1-like transposon with a possible role in hybrid dysgenesis. Mob. DNA 2014, 5, 6. [Google Scholar] [CrossRef][Green Version]
- Erwin, A.A.; Galdos, M.A.; Wickersheim, M.L.; Harrison, C.C.; Marr, K.D.; Colicchio, J.M.; Blumenstiel, J.P. piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS Genet. 2015, 11, e1005332. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Hemmer, L.W.; Dias, G.B.; Smith, B.; Van Vaerenberghe, K.; Howard, A.; Bergman, C.; Blumenstiel, J.P. Hybrid dysgenesis in Drosophila virilis results in clusters of mitotic recombination and loss-of-heterozygosity but leaves meiotic recombination unaltered. Mob. DNA 2020, 11, 10. [Google Scholar] [CrossRef]
- Miller, D.E.; Staber, C.; Zeitlinger, J.; Hawley, R.S. Highly Contiguous Genome Assemblies of 15 Drosophila Species Generated Using Nanopore Sequencing. G3 Genes|Genomes|Genetics 2018, 8, 3131–3141. [Google Scholar] [CrossRef]
- Hubley, R. Dfam-consortium/RepeatAfterMe: RepeatAfterMe_V0.0.4. 2022. Available online: https://doi.org/10.5281/zenodo.7076442.
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Biessmann, H.; Mason, J.M.; Ferry, K.; D’Hulst, M.; Valgeirsdottir, K.; Traverse, K.L.; Pardue, M.-L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell 1990, 61, 663–673. [Google Scholar] [CrossRef]
- Casacuberta, E.; Pardue, M.-L. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc. Natl. Acad. Sci. USA 2003, 100, 3363–3368. [Google Scholar] [CrossRef]
- Pardue, M.-L.; DeBaryshe, P. Retrotransposons Provide an Evolutionarily Robust Non-Telomerase Mechanism to Maintain Telomeres. Annu. Rev. Genet. 2003, 37, 485–511. [Google Scholar] [CrossRef]
- Khurana, J.S.; Xu, J.; Weng, Z.; Theurkauf, W.E. Distinct Functions for the Drosophila piRNA Pathway in Genome Maintenance and Telomere Protection. PLoS Genet. 2010, 6, e1001246. [Google Scholar] [CrossRef]
- Savitsky, M.; Kwon, D.; Georgiev, P.; Kalmykova, A.; Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006, 20, 345–354. [Google Scholar] [CrossRef]
- Shpiz, S.; Kalmykova, A. Role of piRNAs in the Drosophila telomere homeostasis. Mob. Genet. Elements 2011, 1, 274–309. [Google Scholar] [CrossRef]
- Radion, E.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Lavrov, S.; Abramov, Y.; Komarov, P.A.; Glukhov, S.I.; Olovnikov, I.; Kalmykova, A. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin 2018, 11, 40. [Google Scholar] [CrossRef]
- Shpiz, S.; Olovnikov, I.; Sergeeva, A.; Lavrov, S.; Abramov, Y.; Savitsky, M.; Kalmykova, A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011, 39, 8703–8711. [Google Scholar] [CrossRef]
- Cui, M.; Bai, Y.; Li, K.; Rong, Y.S. Taming active transposons at Drosophila telomeres: The interconnection between HipHop’s roles in capping and transcriptional silencing. PLoS Genet. 2021, 17, e1009925. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Zurovcova, M.; Yao, J.G.; Lozovskaya, E.; Walter, M.F. A telomeric satellite in Drosophila virilis and its sibling species. Chromosoma 2000, 109, 372–380. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorador, A.P.; Dalikova, M.; Cerbin, S.; Stillman, C.M.; Zych, M.G.; Hawley, R.S.; Miller, D.E.; Ray, D.A.; Funikov, S.Y.; Evgen’ev, M.B.; et al. Paramutation-like Epigenetic Conversion by piRNA at the Telomere of Drosophila virilis. Biology 2022, 11, 1480. https://doi.org/10.3390/biology11101480
Dorador AP, Dalikova M, Cerbin S, Stillman CM, Zych MG, Hawley RS, Miller DE, Ray DA, Funikov SY, Evgen’ev MB, et al. Paramutation-like Epigenetic Conversion by piRNA at the Telomere of Drosophila virilis. Biology. 2022; 11(10):1480. https://doi.org/10.3390/biology11101480
Chicago/Turabian StyleDorador, Ana P., Martina Dalikova, Stefan Cerbin, Chris M. Stillman, Molly G. Zych, R. Scott Hawley, Danny E. Miller, David A. Ray, Sergei Y. Funikov, Michael B. Evgen’ev, and et al. 2022. "Paramutation-like Epigenetic Conversion by piRNA at the Telomere of Drosophila virilis" Biology 11, no. 10: 1480. https://doi.org/10.3390/biology11101480
APA StyleDorador, A. P., Dalikova, M., Cerbin, S., Stillman, C. M., Zych, M. G., Hawley, R. S., Miller, D. E., Ray, D. A., Funikov, S. Y., Evgen’ev, M. B., & Blumenstiel, J. P. (2022). Paramutation-like Epigenetic Conversion by piRNA at the Telomere of Drosophila virilis. Biology, 11(10), 1480. https://doi.org/10.3390/biology11101480





