Red Light Resets the Expression Pattern, Phase, and Period of the Circadian Clock in Plants: A Computational Approach
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
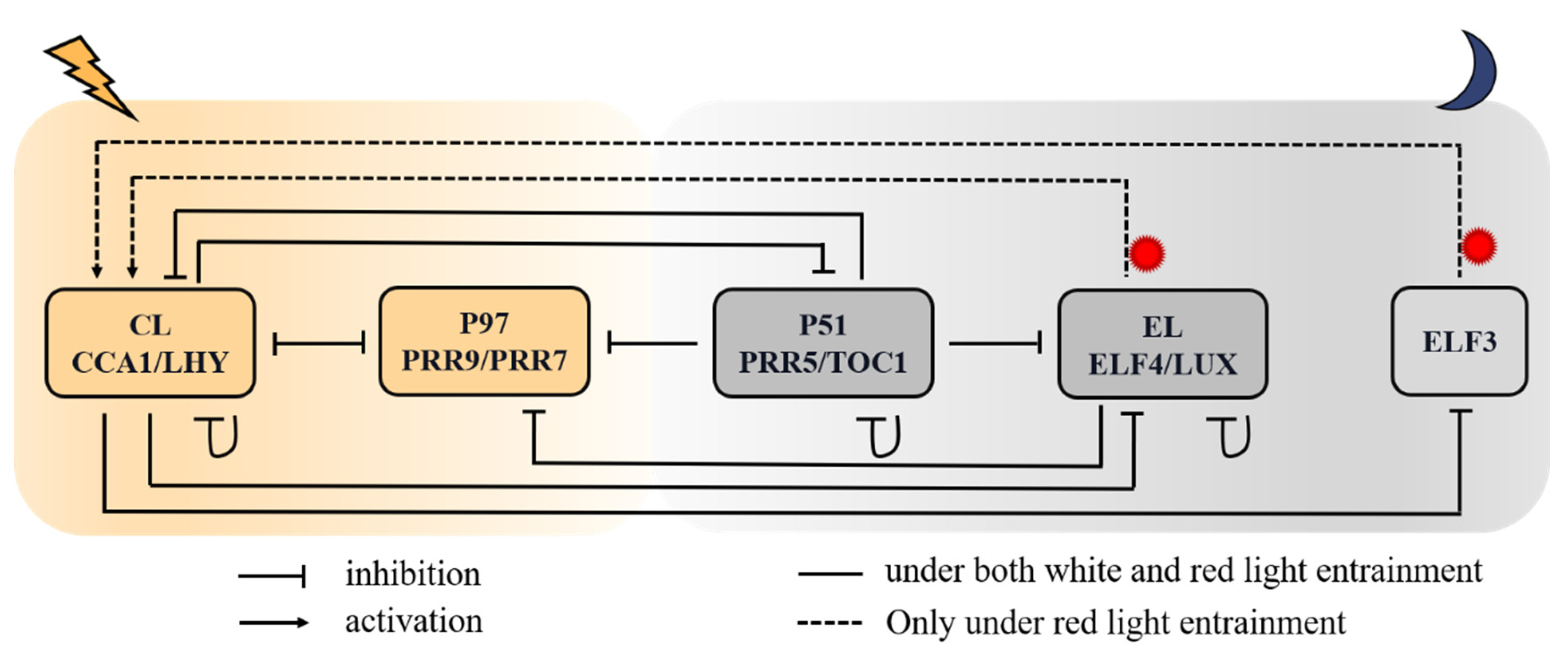
2.1. The Molecular Mechanism of Red-Light-Entrained Plant Circadian Clock
2.2. Formulation of Model Equations
2.3. Database and Simulation Tools
3. Results
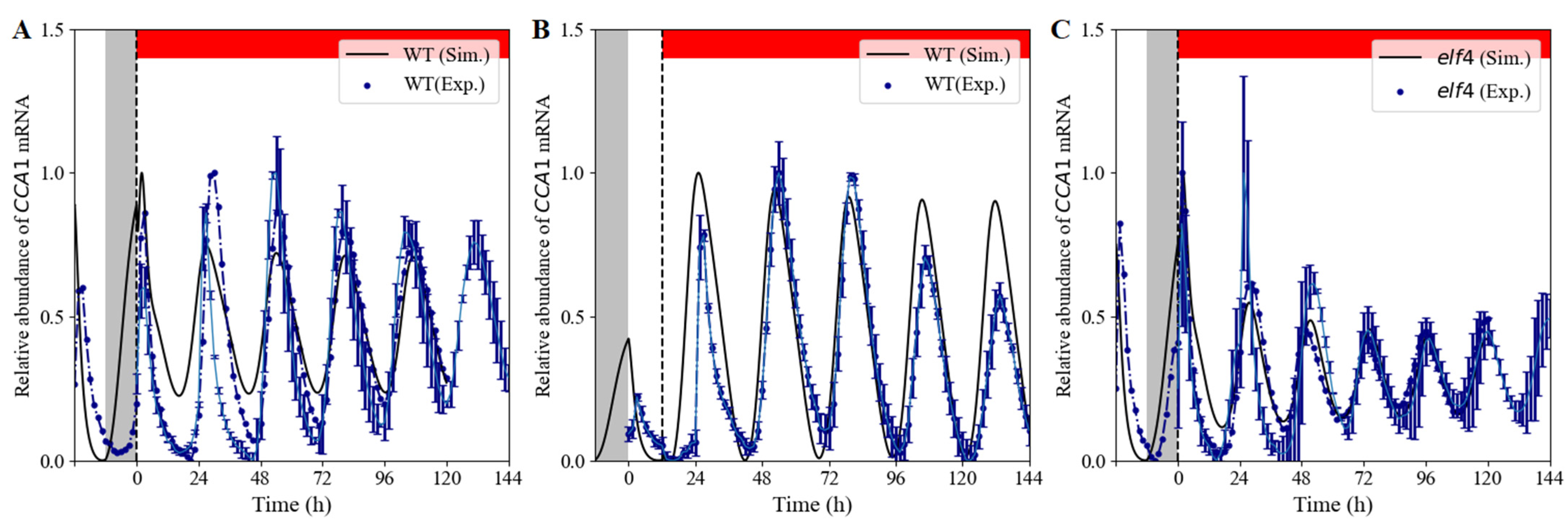
3.1. Model Predictions and Validation
3.1.1. Rhythmic Expression Patterns
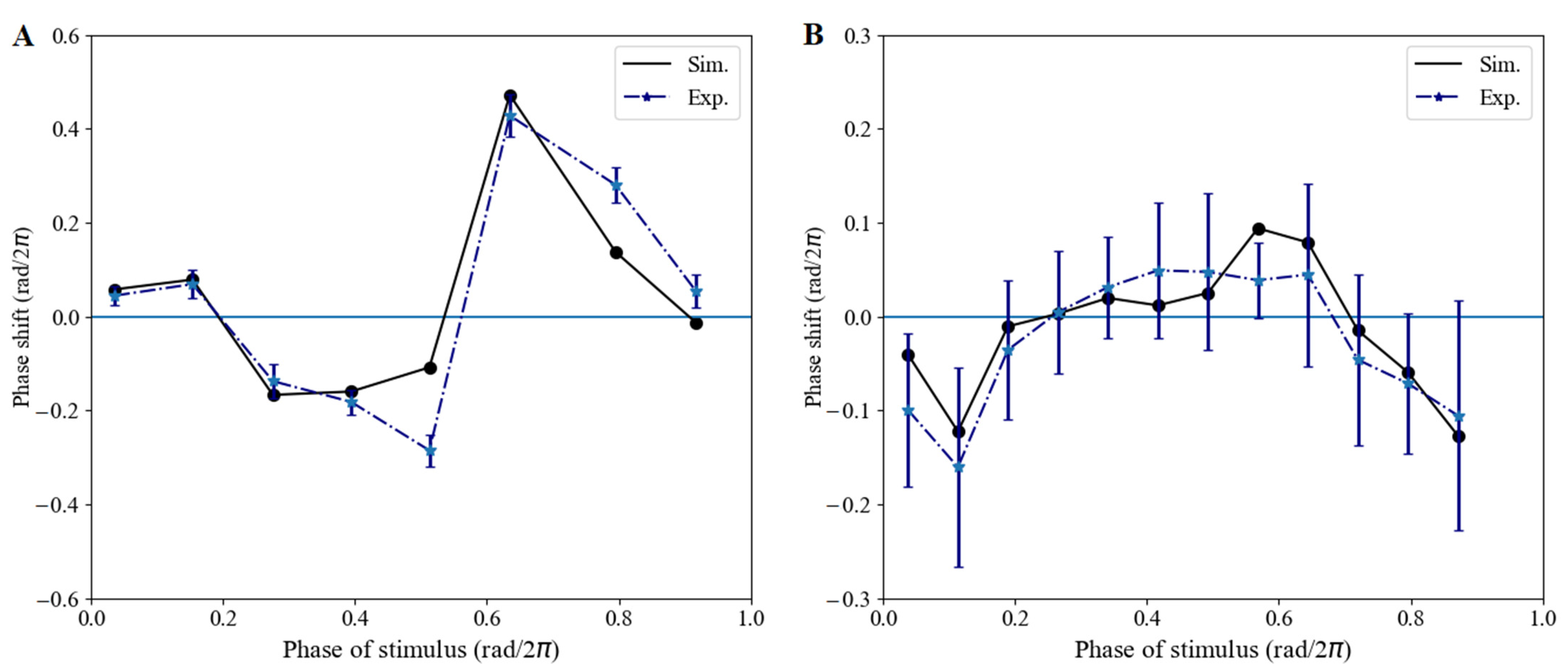
3.1.2. Phase Shifts Caused by Red-Light Pulses
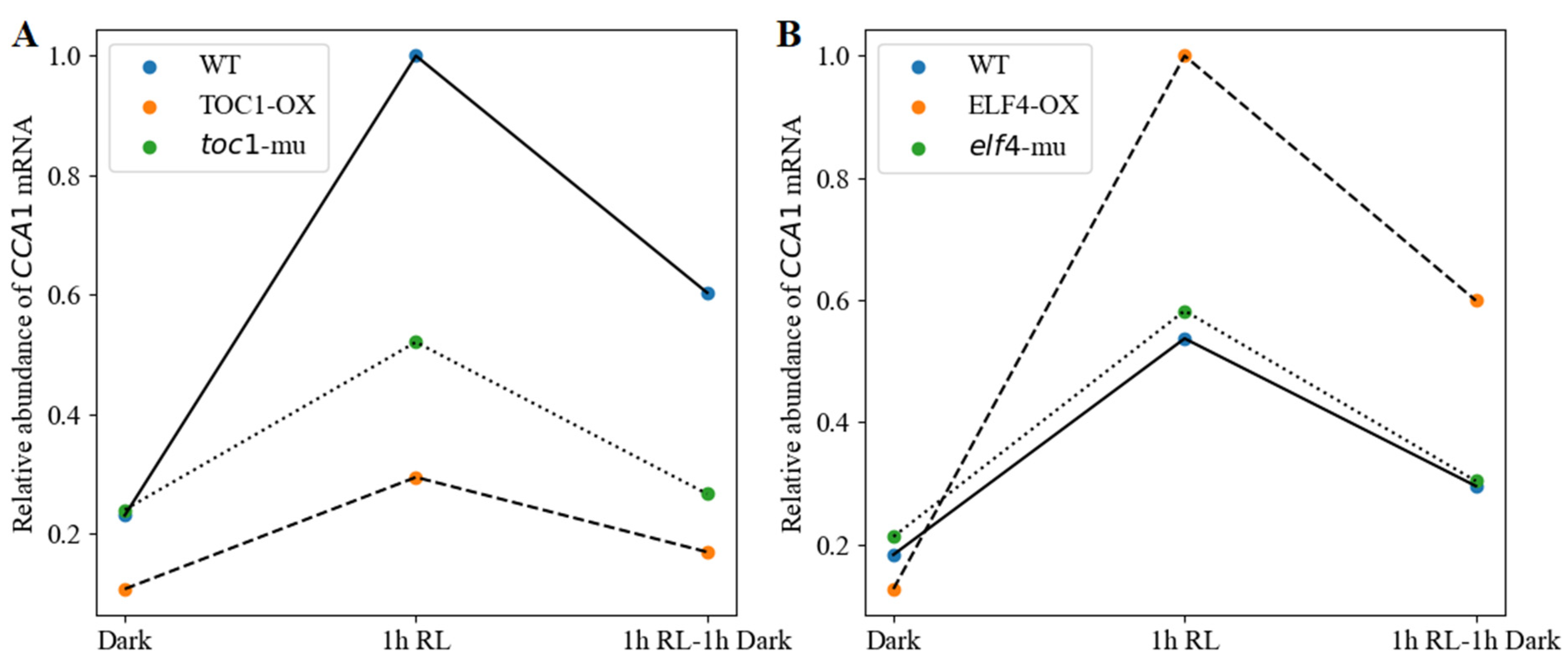
3.1.3. Red-Light Induction of CCA1 Expression with TOC1/ELF4 Overexpression and Mutation Lines
3.2. Red-Light Reset of the Expression Pattern, Period, and Phase of CL and P51 under Different Photoperiods
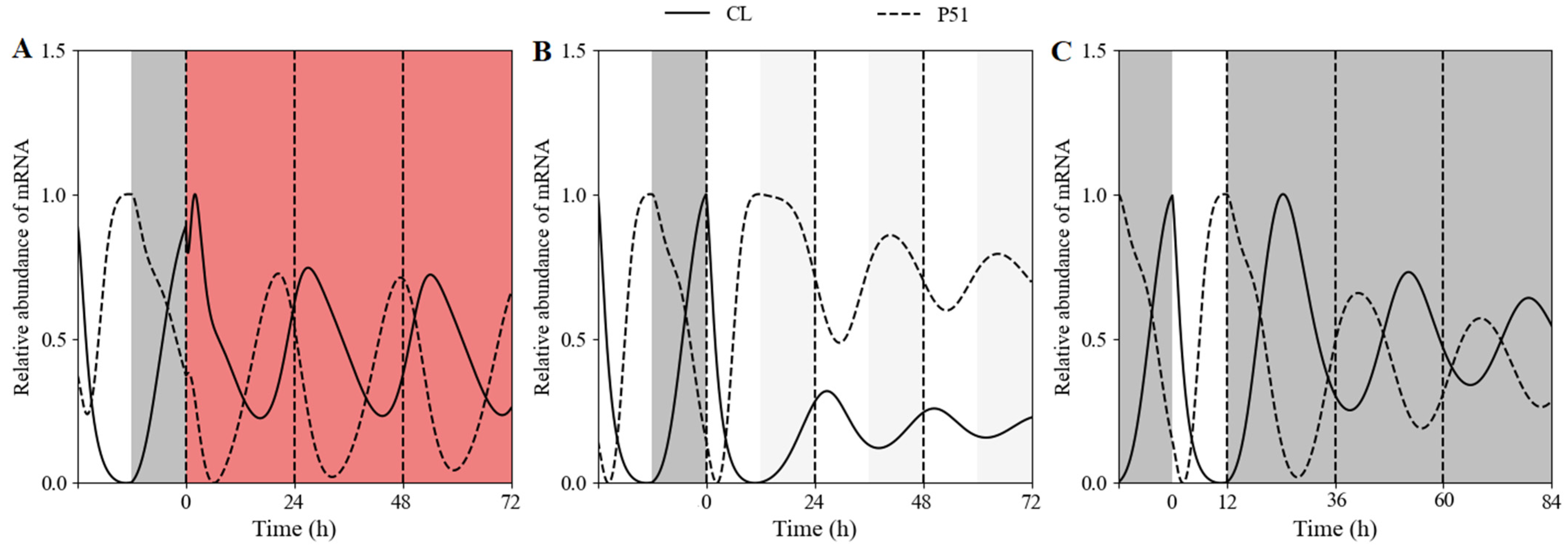
3.2.1. Clock Reset under Red-Light Free-Running Period in WT
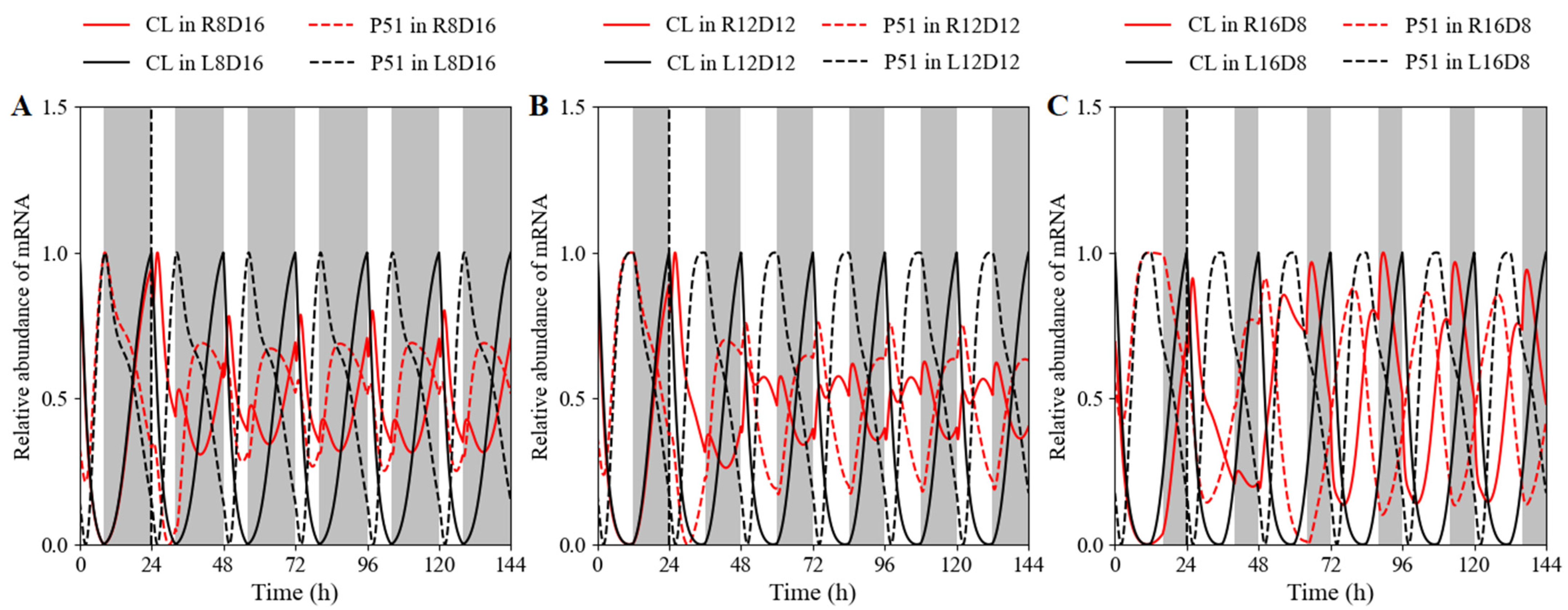
3.2.2. Clock Reset under Red-Light/Dark Cycles in WT
3.2.3. Clock Reset under Red-Light/Dark Cycles in Single or Double Mutant
3.2.4. Clock Reset under Extreme 24 h Red-Light Photoperiods
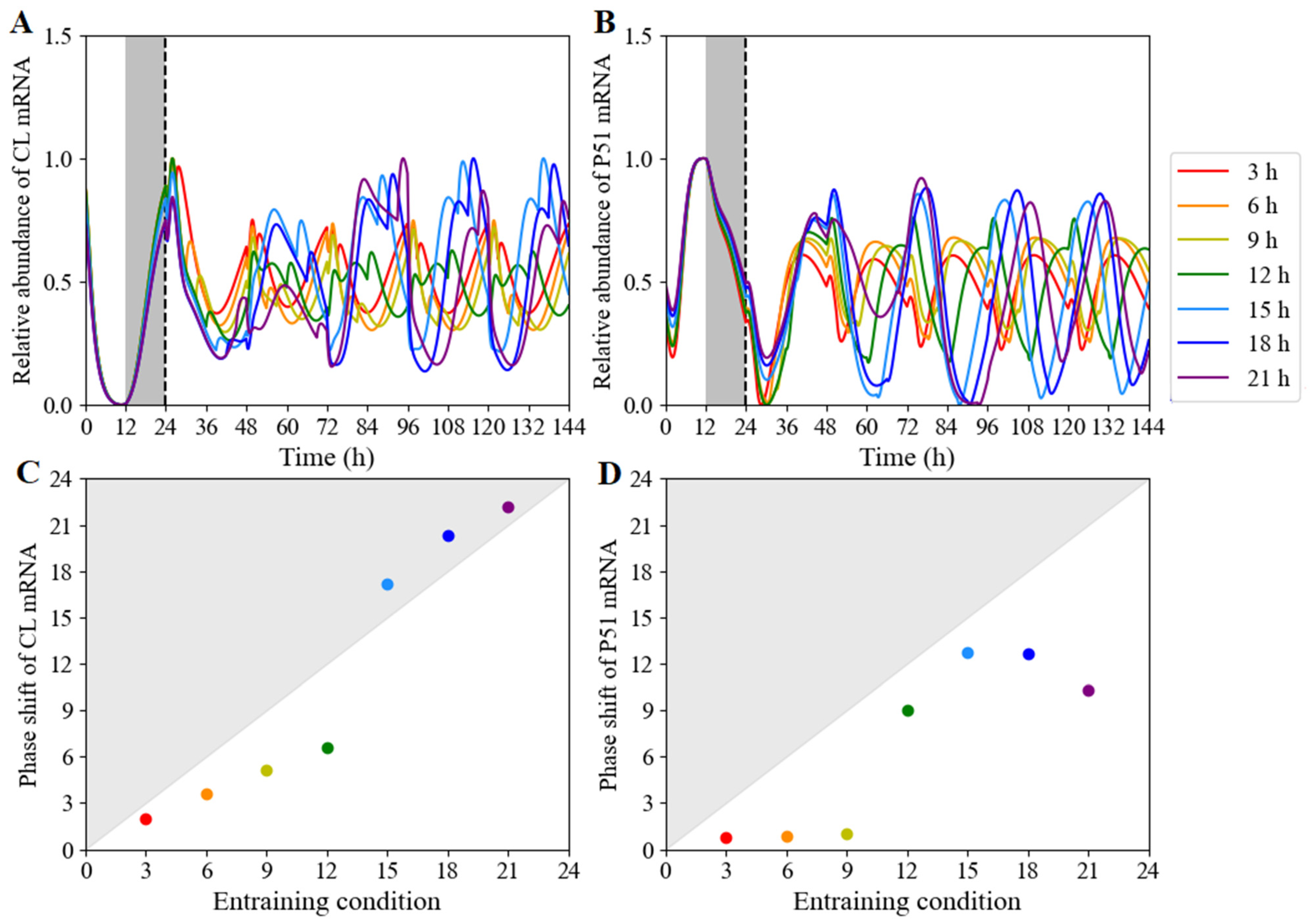
3.2.5. The Expression Pattern of CCA1 Determined by the Orders of Red-Light Input
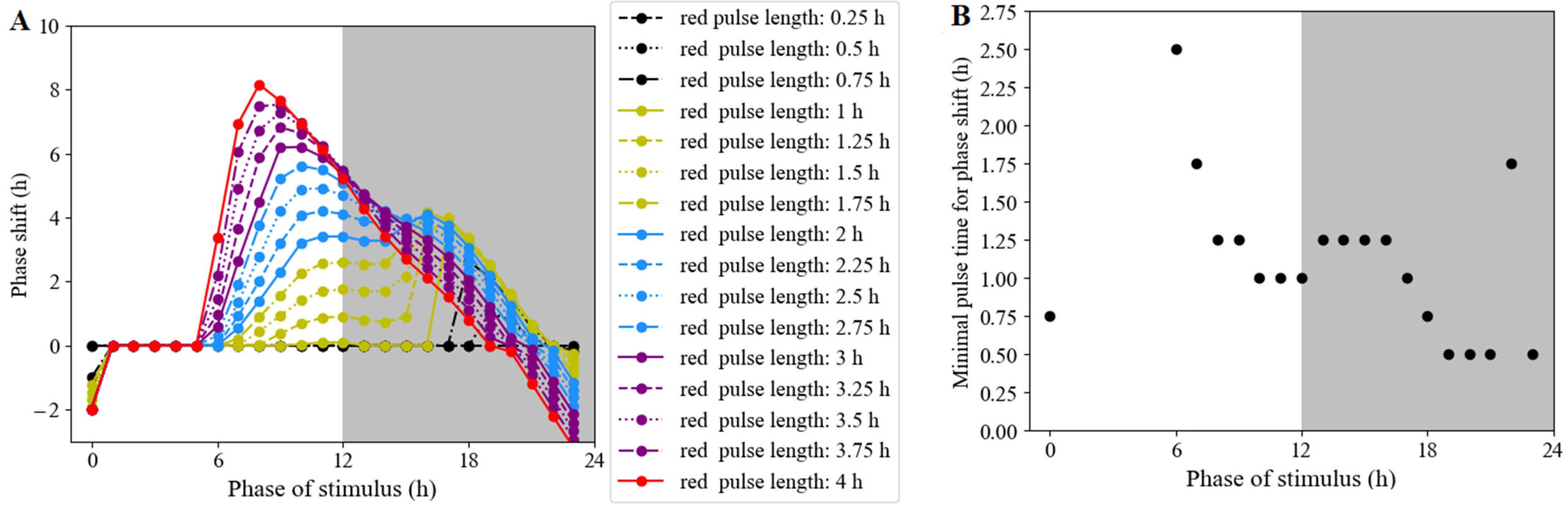
3.2.6. Phase Shifts Controlled by the Moments and Lengths of Red-Light Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Masuda, K.; Tokuda, I.T.; Nakamichi, N.; Fukuda, H. The singularity response reveals entrainment properties of the plant circadian clock. Nat. Commun. 2021, 12, 864. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Aas, O.; Jetmundsen, M.; Lee, Y.; Torre, S.; Fløistad, I.; Olsen, J. Day extension with far-red light enhances growth of subalpine fir (Abies Lasiocarpa (Hooker) Nuttall) seedlings. Forests 2018, 9, 175. [Google Scholar] [CrossRef]
- McClung, C.R. Wheels within wheels: New transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 2014, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Panda, S.; Liu, X.L.; Strayer, C.A.; Wagner, D.R.; Kay, S.A. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 2001, 13, 1305–1316. [Google Scholar] [CrossRef]
- Bordage, S.; Sullivan, S.; Laird, J.; Millar, A.J.; Nimmo, H.G. Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol. 2016, 212, 136–149. [Google Scholar] [CrossRef]
- Steed, G.; Ramirez, D.C.; Hannah, M.A.; Webb, A.A.R. Chronoculture, harnessing the circadian clock to improve crop yield and sustainability. Science 2021, 372, eabc9141. [Google Scholar] [CrossRef]
- Greenham, K.; Lou, P.; Puzey, J.R.; Kumar, G.; Arnevik, C.; Farid, H.; Willis, J.H.; McClung, C.R. Geographic variation of plant circadian clock function in natural and agricultural settings. J. Biol. Rhythm. 2017, 32, 26–34. [Google Scholar] [CrossRef]
- Creux, N.; Harmer, S. Circadian rhythms in plants. Cold Spring Harb. Perspect. Biol. 2019, 11, a034611. [Google Scholar] [CrossRef]
- Fogelmark, K.; Troein, C. Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput. Biol. 2014, 10, e1003705. [Google Scholar] [CrossRef]
- De Caluwé, J.; Xiao, Q.; Hermans, C.; Verbruggen, N.; Leloup, J.C.; Gonze, D. A compact model for the complex plant circadian clock. Front. Plant Sci. 2016, 7, 74. [Google Scholar] [CrossRef]
- Ye, J.H.; Lv, Y.Q.; Liu, S.R.; Jin, J.; Wang, Y.F.; Wei, C.L.; Zhao, S.Q. Effects of light intensity and spectral composition on the transcriptome profiles of leaves in shade grown tea plants (Camellia Sinensis L.) and regulatory network of flavonoid biosynthesis. Molecules 2021, 26, 5836. [Google Scholar] [CrossRef] [PubMed]
- Seif, M.; Aliniaeifard, S.; Arab, M.; Mehrjerdi, M.Z.; Shomali, A.; Fanourakis, D.; Li, T.; Woltering, E. Monochromatic red light during plant growth decreases the size and improves the functionality of stomata in chrysanthemum. Funct. Plant Biol. 2021, 48, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Amaki, W.; Watanabe, H. Effects of monochromatic light irradiation by led on the growth and anthocyanin contents in leaves of cabbage seedlings. Acta Hortic. 2011, 907, 179–184. [Google Scholar] [CrossRef]
- Materová, Z.; Sobotka, R.; Zdvihalová, B.; Oravec, M.; Nezval, J.; Karlický, V.; Vrábl, D.; Štroch, M.; Špunda, V. Monochromatic green light induces an aberrant accumulation of geranylgeranyled ghlorophylls in plants. Plant Physiol. Biochem. 2017, 116, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Asahina, M.; Tamaki, Y.; Sakamoto, T.; Shibata, K.; Nomura, T.; Yokota, T. Blue light-promoted rice leaf bending and unrolling are due to up-regulated brassinosteroid biosynthesis genes accompanied by accumulation of castasterone. Phytochemistry 2014, 104, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Shamsabad, M.R.M.; Esmaeilizadeh, M.; Roosta, H.R.; Dąbrowski, P.; Telesiński, A.; Kalaji, H.M. Supplemental light application can improve the growth and development of strawberry plants under salinity and alkalinity stress conditions. Sci. Rep. 2022, 12, 9272. [Google Scholar] [CrossRef]
- Nassarawa, S.S.; Belwal, T.; Javed, M.; Luo, Z. Influence of the red LEDs light irradiation on the quality and chemical attributes of postharvest table grape (Vitis Vinifera L.) during storage. Food Bioprocess Technol. 2022, 15, 1436–1447. [Google Scholar] [CrossRef]
- Dziwulska-Hunek, A.; Szymanek, M.; Stadnik, J. Impact of pre-sowing red light treatment of sweet corn seeds on the quality and quantity of yield. Agriculture 2020, 10, 165. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Song, S.; Su, W.; Hao, Y.; Liu, H. Supplementary red light results in the earlier ripening of tomato fruit depending on ethylene production. Environ. Exp. Bot. 2020, 175, 104044. [Google Scholar] [CrossRef]
- Lillo, C.; Appenroth, K.J. Light regulation of nitrate reductase in higher plants: Which photoreceptors are involved? Plant Biol. 2001, 3, 455–465. [Google Scholar] [CrossRef]
- Bian, Z.H.; Yang, Q.C.; Liu, W.K. Effects of light quality on the accumulation of phytochemicals in vegetables produced in controlled environments: A review: Effects of light on vegetable phytochemicals. J. Sci. Food Agric. 2015, 95, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.J.; Hubbard, K.E.; Hotta, C.T.; Dodd, A.N.; Webb, A.A.R. How plants tell the time. Biochem. J. 2006, 397, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Mazzella, M.A.; Whitelam, G.C.; Casal, J.J. Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J. Biol. Rhythms 2001, 16, 523–530. [Google Scholar] [CrossRef]
- Xu, X.; Xie, Q.; McClung, C.R. Robust circadian rhythms of gene expression in Brassica Rapa tissue culture. Plant Physiol. 2010, 153, 841–850. [Google Scholar] [CrossRef]
- Xiang, Y.; Sapir, T.; Rouillard, P.; Ferrand, M.; Jiménez-Gómez, J.M. Interaction between photoperiod and variation in circadian rhythms in tomato. BMC Plant Biol. 2022, 22, 187. [Google Scholar] [CrossRef]
- Wenden, B.; Kozma-Bognár, L.; Edwards, K.D.; Hall, A.J.W.; Locke, J.C.W.; Millar, A.J. Light inputs shape the Arabidopsis circadian system: Plant clocks with limited light input. Plant J. 2011, 66, 480–491. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and Temperature-entrainable Circadian Clock in Soybean Development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Oakenfull, R.J.; Davis, S.J. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017, 40, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.C.W.; Millar, A.J.; Turner, M.S. Modelling genetic networks with noisy and varied experimental data: The circadian clock in Arabidopsis thaliana. J. Theor. Biol. 2005, 234, 383–393. [Google Scholar] [CrossRef]
- Pokhilko, A.; Fernández, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef]
- Pokhilko, A.; Mas, P.; Millar, A.J. Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 2013, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, M.; Tokuda, I.T.; Locke, J.C.W. A spatial model of the plant circadian clock reveals design pinciples for coordinated timing. Mol. Syst. Biol. 2022, 18, e10140. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.H.; Huang, M.Y.; Huang, W.D.; Hsu, M.H.; Yang, Z.W.; Yang, C.M. The effects of red, blue, and white light-emitting diodes on the growth, development, and edible quality of hydroponically grown Lettuce (Lactuca Sativa L. Var. Capitata). Sci. Hortic. 2013, 150, 86–91. [Google Scholar] [CrossRef]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef]
- Yeom, M.; Kim, H.; Lim, J.; Shin, A.Y.; Hong, S.; Kim, J.I.; Nam, H.G. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol. Plant 2014, 7, 1701–1704. [Google Scholar] [CrossRef]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-Binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.J.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef]
- Kikis, E.A.; Khanna, R.; Quail, P.H. ELF4 is a phytochrome-regulated component of a negative−feedback loop involving the central oscillator components CCA1 and LHY: ELF4 and Rc induction of the circadian clock. Plant J. 2005, 44, 300–313. [Google Scholar] [CrossRef]
- Liu, X.L.; Covington, M.F.; Fankhauser, C.; Chory, J.; Wagner, D.R. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 2001, 13, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; López-Salmerón, V.; Davière, J.-M.; Prat, S. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Anna, B.B.; Grzegorz, B.; Marek, K.; Piotr, G.; Marcin, F. Exposure to high-intensity light systemically induces micro-transcriptomic changes in Arabidopsis thaliana roots. Int. J. Mol. Sci. 2019, 20, 5131. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Whitelam, G.C. Light signals, phytochromes and cross-talk with other environmental cues. J. Exp. Bot. 2003, 55, 271–276. [Google Scholar] [CrossRef]
- Liu, X.Y.; Xue, C.; Kong, L.; Xu, Z.G.; Hua, J. Interactive effects of light quality and temperature on Arabidopsis growth and immunity. Plant Cell Physiol. 2020, 61, 933–941. [Google Scholar] [CrossRef]
- Pay, M.L.; Kim, D.W.; Somers, D.E.; Kim, J.K.; Foo, M. Modelling of plant circadian clock for characterizing hypocotyl growth under different light quality conditions. In Silico. Plants 2022, 4, diac001. [Google Scholar] [CrossRef]
- Nimmo, H.G.; Laird, J.; Bindbeutel, R.; Nusinow, D.A. The evening complex is central to the difference between the circadian clocks of Arabidopsis thaliana shoots and roots. Physiol Plant. 2020, 169, 442–451. [Google Scholar] [CrossRef]
- Ohara, T.; Fukuda, H.; Tokuda, I.T. An extended mathematical model for reproducing the phase response of Arabidopsis thaliana under various light conditions. J. Theor. Biol. 2015, 382, 337–344. [Google Scholar] [CrossRef]
- Más, P.; Alabadí, D.; Yanovsky, M.J.; Oyama, T.; Kay, S.A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 2003, 15, 223–236. [Google Scholar] [CrossRef]
- Ito, S.; Niwa, Y.; Nakamichi, N.; Kawamura, H.; Yamashino, T.; Mizuno, T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled Circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 201–213. [Google Scholar] [CrossRef]
- Salomé, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 2005, 17, 791–803. [Google Scholar] [CrossRef] [PubMed]
- McWatters, H.G.; Kolmos, E.; Hall, A.; Doyle, M.R.; Amasino, R.M.; Gyula, P.; Nagy, F.; Millar, A.J.; Davis, S.J. ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 2007, 144, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.U.; Boikoglou, E.; Herrero, E.; Hallstein, M.; Davis, A.M.; James, G.V.; Nagy, F.; Davis, S.J. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 2014, 3, e02206. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.D.; Akman, O.E.; Knox, K.; Lumsden, P.J.; Thomson, A.W.; Brown, P.E.; Pokhilko, A.; Kozma-Bognar, L.; Nagy, F.; Rand, D.A.; et al. Quantitative analysis of regulatory flexibility under changing environmental conditions. Mol. Syst. Biol. 2010, 6, 424. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef]
- Pagliarini, S.; Korobeinikov, A. A mathematical model of marine bacteriophage evolution. R. Soc. Open Sci. 2018, 5, 171661. [Google Scholar] [CrossRef]
- Niwa, Y.; Matsuo, T.; Onai, K.; Kato, D.; Tachikawa, M.; Ishiura, M. Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2013, 110, 13666–13671. [Google Scholar] [CrossRef]
- Schmal, C.; Myung, J.; Herzel, H.; Bordyugov, G. A theoretical study on seasonality. Front. Neurol. 2015, 6, 94. [Google Scholar] [CrossRef]
- Lopez, L.; Fasano, C.; Perrella, G.; Facella, P. Cryptochromes and the circadian clock: The story of a very complex relationship in a spinning world. Genes 2021, 12, 672. [Google Scholar] [CrossRef]
- Hogewoning, S.W.; Trouwborst, G.; Maljaars, H.; Poorter, H.; van Ieperen, W.; Harbinson, J. Blue light dose-responses of leaf photosynthesis, morphology, and chemical composition of cucumis sativus grown under different combinations of red and blue light. J. Exp. Bot. 2010, 61, 3107–3117. [Google Scholar] [CrossRef]
- Nimmo, H.G. Entrainment of Arabidopsis roots to the light:dark cycle by light piping: Light piping entrains roots. Plant Cell Environ. 2018, 41, 1742–1748. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, M.L.; Pokhilko, A.; Fernández, A.P.; Halliday, K.J.; Millar, A.J.; Hillston, J. Stochastic properties of the plant circadian clock. J. R. Soc. Interface 2012, 9, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, M.L.; Akman, O.E.; van Ooijen, G. Stochastic models of cellular circadian rhythms in plants help to understand the impact of noise on robustness and clock structure. Front. Plant Sci. 2014, 5, 564. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Kubota, A.; Kwon, M.S.; Covington, M.F.; Lee, N.; Taagen, E.R.; Cintrón, D.L.; Hwang, D.Y.; Akiyama, R.; Hodge, S.K.; et al. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat. Plants 2018, 4, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Balcerowicz, M. PHYTOCHROME-INTERACTING FACTORS at the interface of light and temperature signalling. Physiol. Plant. 2020, 169, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Hiltbrunner, A. Phytochromes: Methods and Protocols, 1st ed.; Springer: New York, NY, USA, 2019; pp. 179–190. [Google Scholar]
- Hoops, S.; Hontecillas, R.; Abedi, V.; Leber, A.; Philipson, C.; Carbo, A.; Bassaganya-Riera, J. Ordinary Differential Equations (ODEs) Based Modeling. In Computational Immunology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 63–78. [Google Scholar]
- Leloup, J.C.; Goldbeter, A. Modeling the mammalian circadian clock: Sensitivity analysis and multiplicity of oscillatory mechanisms. J. Theor. Biol. 2004, 230, 541–562. [Google Scholar] [CrossRef]
- MacGregor, D.R.; Gould, P.; Foreman, J.; Griffiths, J.; Bird, S.; Page, R.; Stewart, K.; Steel, G.; Young, J.; Paszkiewicz, K.; et al. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic MRNA export in Arabidopsis. Plant Cell 2013, 25, 4391–4404. [Google Scholar] [CrossRef]
- Zhang, R.; Gonze, D.; Hou, X.; You, X.; Goldbeter, A. A computational model for the cold response pathway in plants. Front. Physiol. 2020, 11, 591073. [Google Scholar] [CrossRef]
- Hairer, E.; Lubich, C.; Wanner, G. Geometric Numerical Integration: Structure-Preserving Algorithms for Ordinary Differential Equations, 2nd ed.; Springer Series in Computational Mathematics; Springer: Berlin, Germany; New York, NY, USA, 2006; pp. 27–50. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, T.; Shui, Y.; Wu, Y.; Hou, X.; You, X. Red Light Resets the Expression Pattern, Phase, and Period of the Circadian Clock in Plants: A Computational Approach. Biology 2022, 11, 1479. https://doi.org/10.3390/biology11101479
Huang T, Shui Y, Wu Y, Hou X, You X. Red Light Resets the Expression Pattern, Phase, and Period of the Circadian Clock in Plants: A Computational Approach. Biology. 2022; 11(10):1479. https://doi.org/10.3390/biology11101479
Chicago/Turabian StyleHuang, Ting, Yao Shui, Yue Wu, Xilin Hou, and Xiong You. 2022. "Red Light Resets the Expression Pattern, Phase, and Period of the Circadian Clock in Plants: A Computational Approach" Biology 11, no. 10: 1479. https://doi.org/10.3390/biology11101479
APA StyleHuang, T., Shui, Y., Wu, Y., Hou, X., & You, X. (2022). Red Light Resets the Expression Pattern, Phase, and Period of the Circadian Clock in Plants: A Computational Approach. Biology, 11(10), 1479. https://doi.org/10.3390/biology11101479






