Uncovering the Role of PhzC as DAHP Synthase in Shikimate Pathway of Pseudomonas chlororaphis HT66
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. DNA Manipulation and Transformation
2.3. Sequence and Protein Analysis of DAHP
2.4. Fermentation Process of P. chlororaphis and its Derivative Strains
2.5. Quantitative Real-Time PCR
2.6. Quantitative Assay for PCN Production
2.7. Statistical Analysis
3. Results
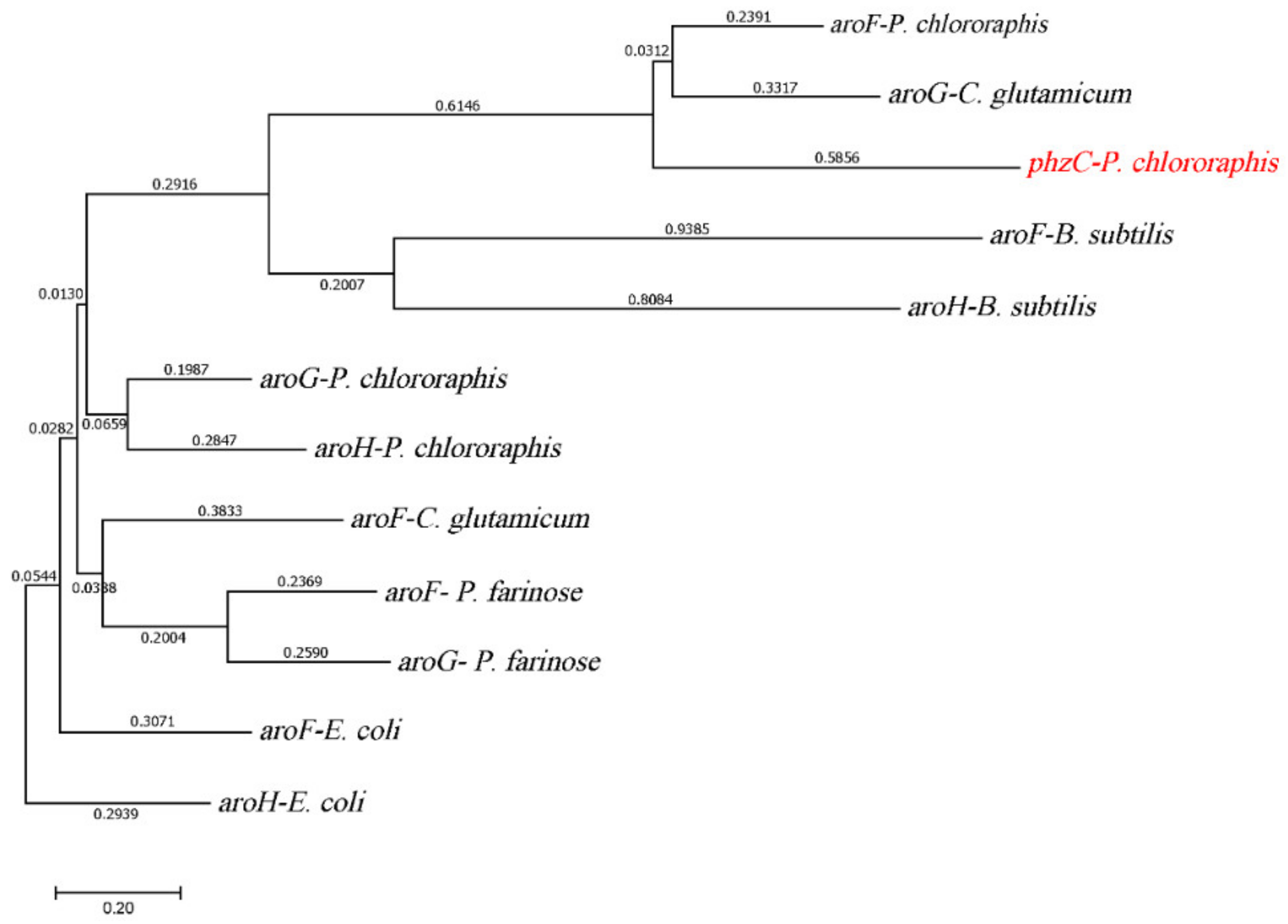
3.1. Uncovering DAHP Synthases in P. chlororaphis HT66
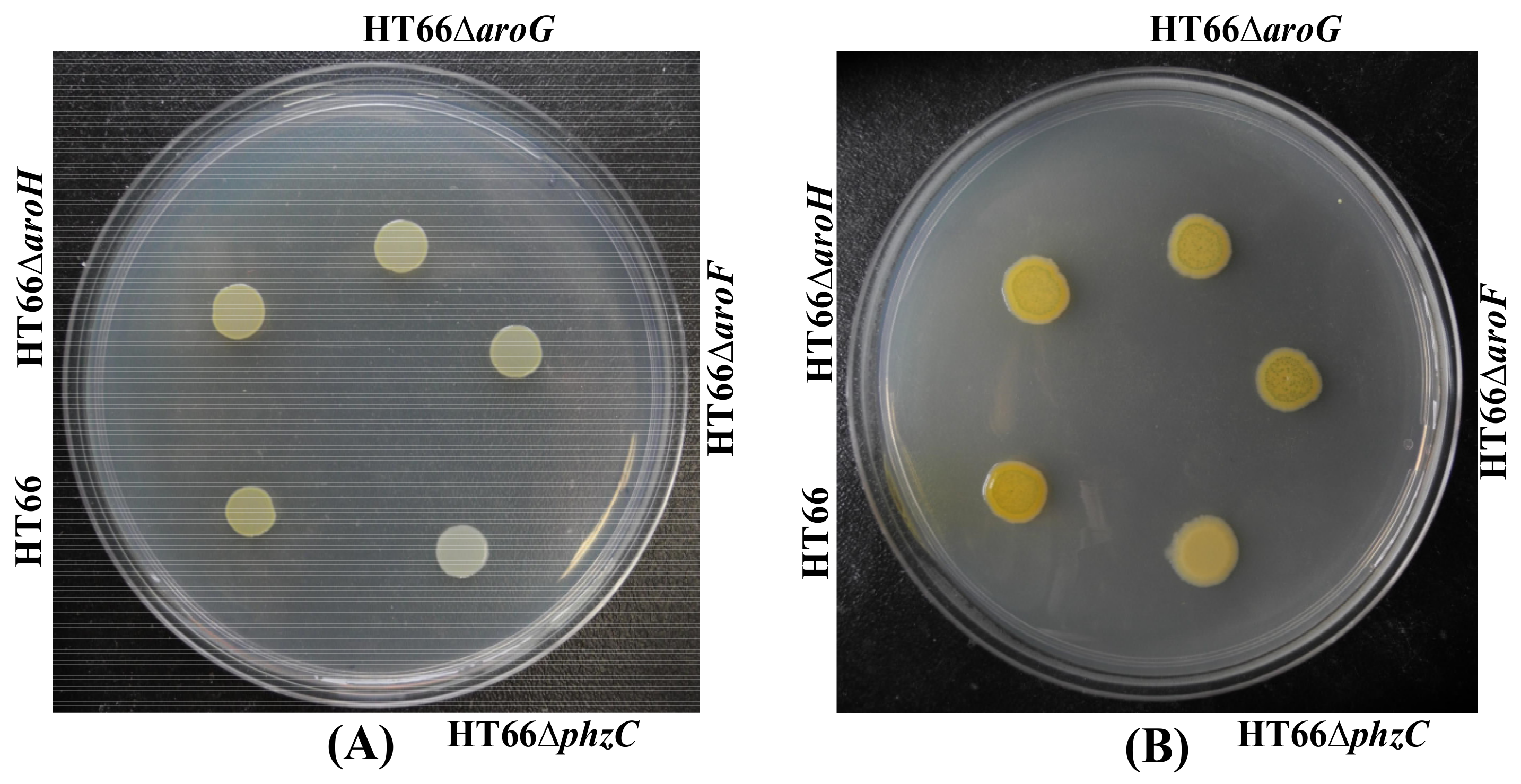
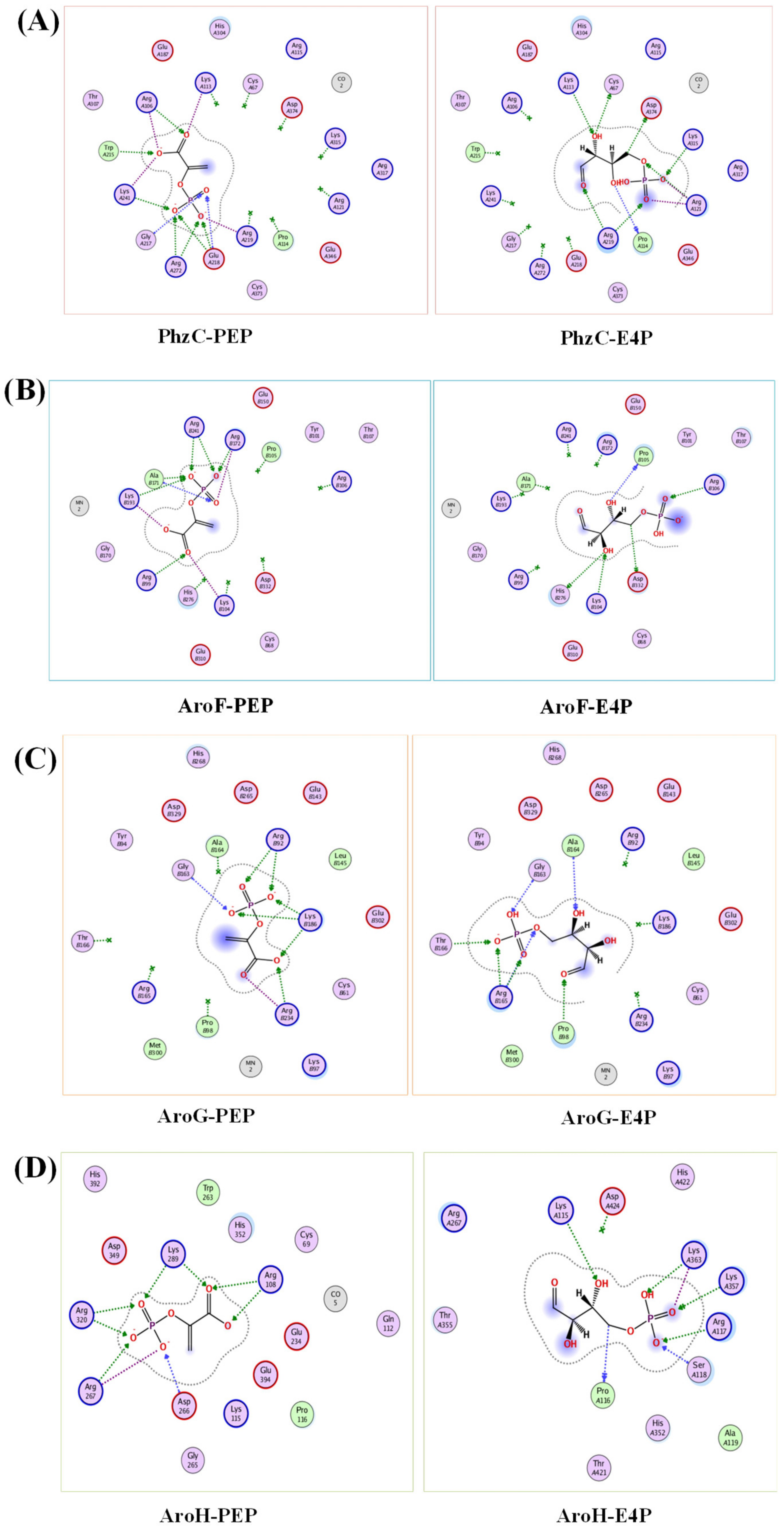
3.2. Mutation and Functional Characterization of DAHP Synthases
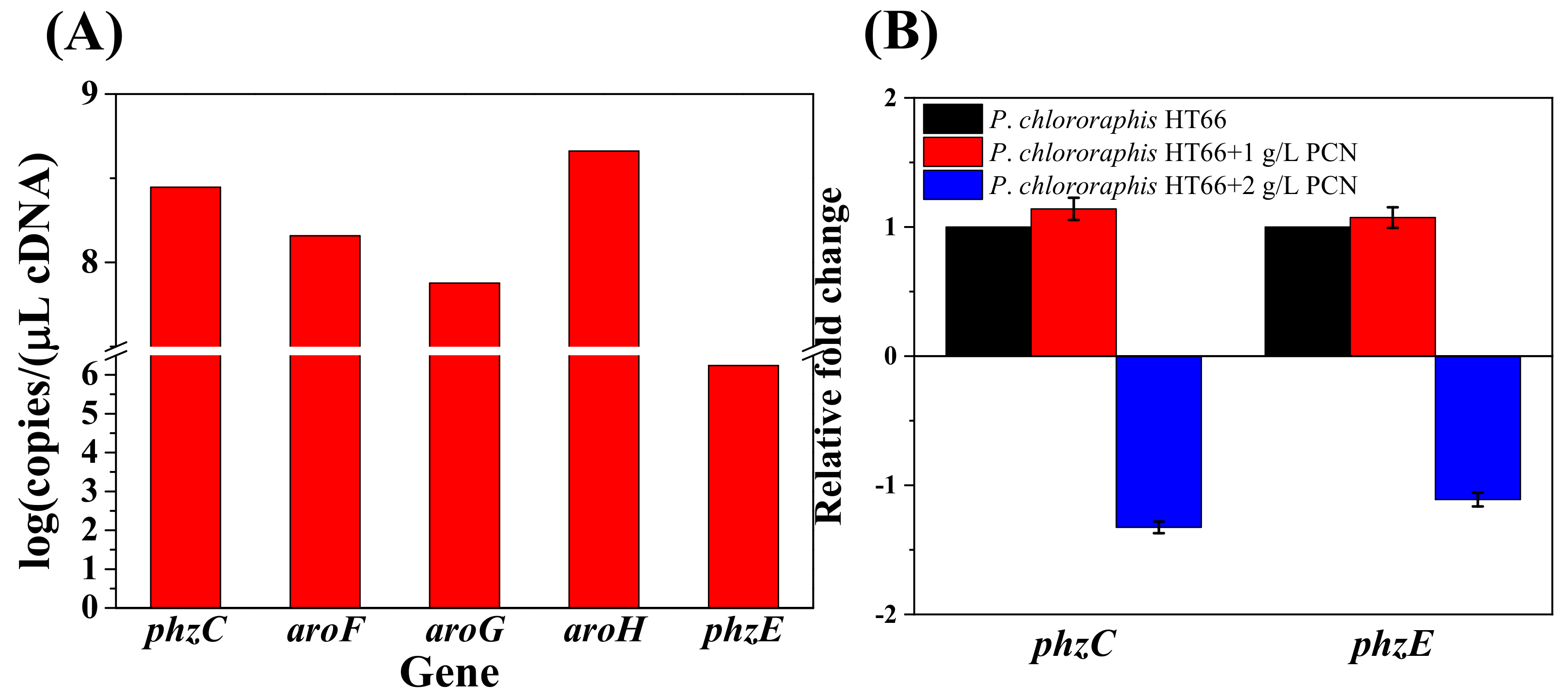
3.3. Gene Expression Level and Its Quantification
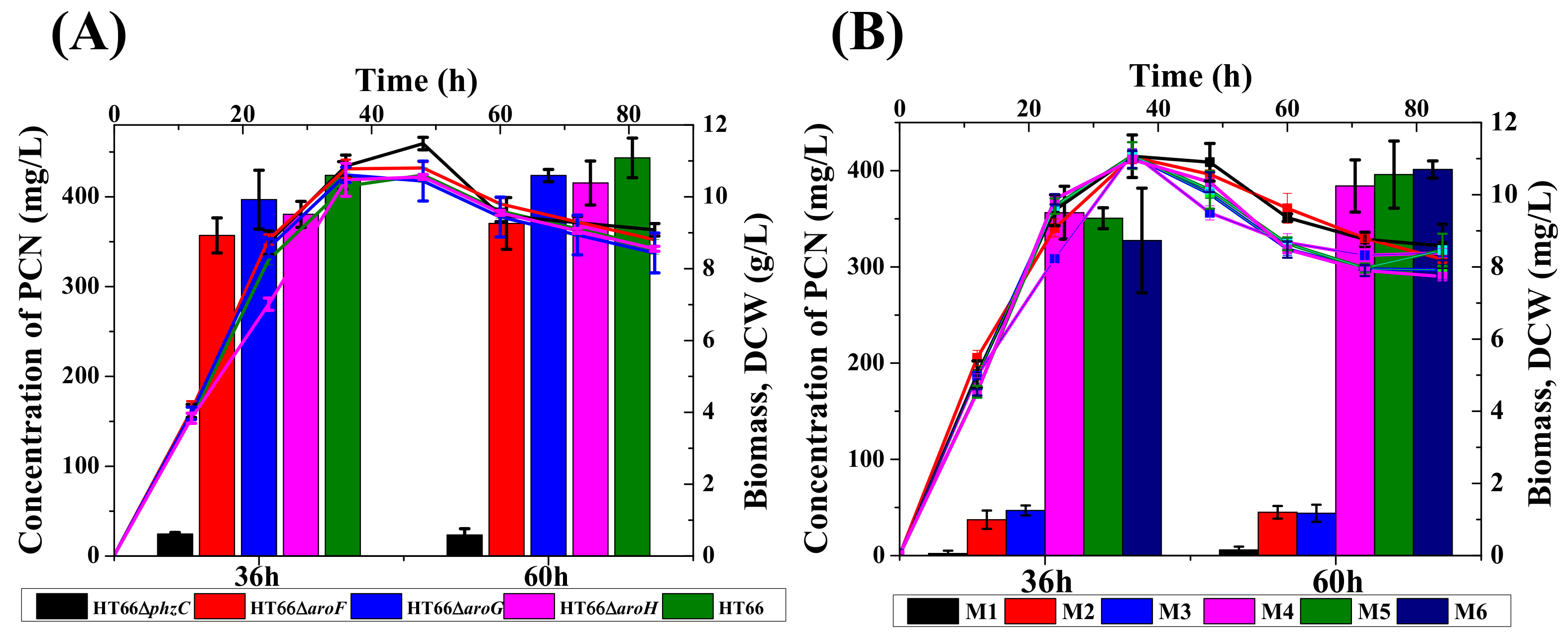
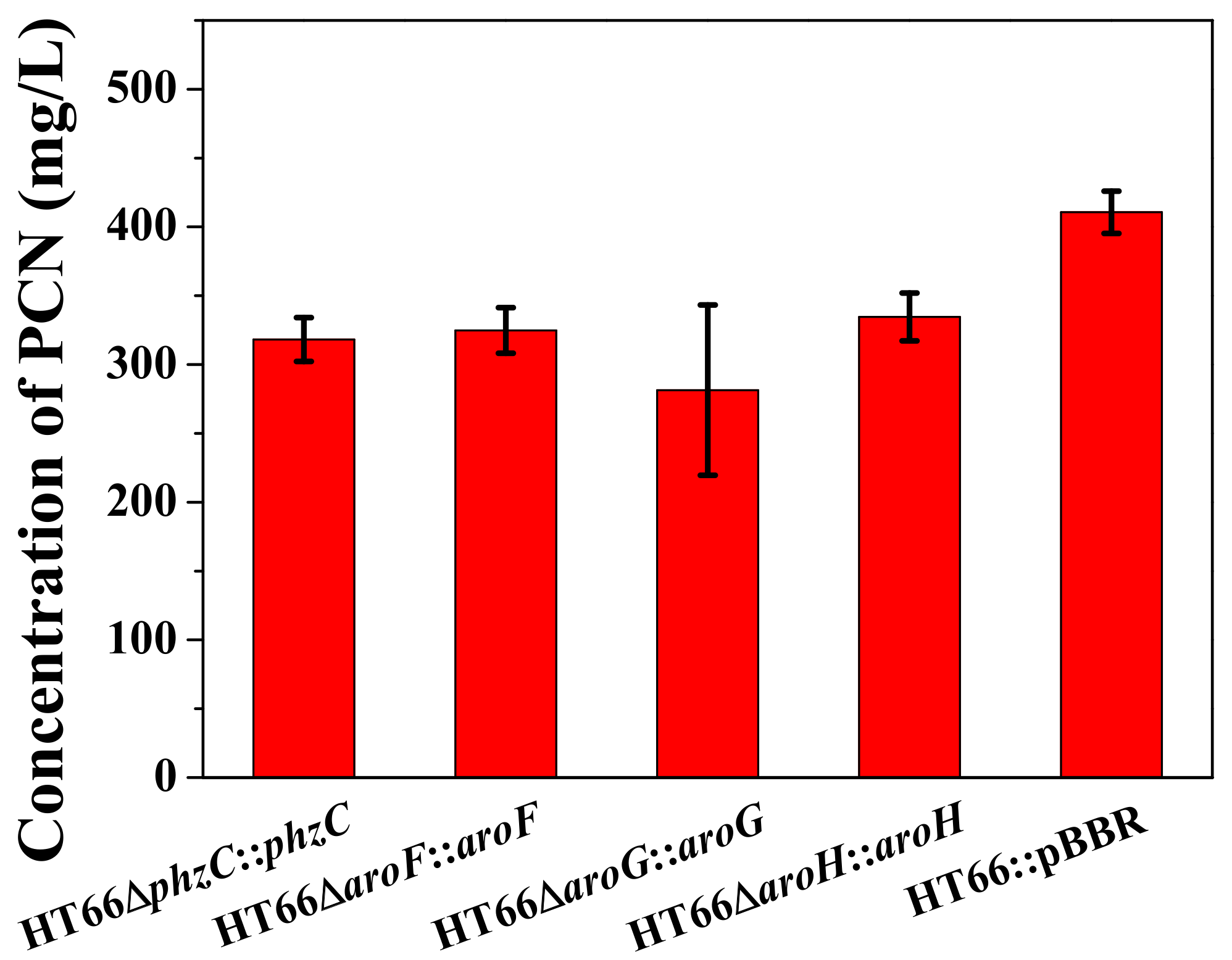
3.4. Effect of DAHP Synthase Gene Complementation in the Mutant Strains
3.5. Effect of DAHP Synthases on the Metabolism of Carbon and Nitrogen Sources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrmann, K.M.; Weaver, L.M. The Shikimate Pathway. Annu. Rev. Plant Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Averesch, N.J.H.; Krömer, J.O. Metabolic Engineering of the Shikimate Pathway for Production of Aromatics and Derived Compounds—Present and Future Strain Construction Strategies. Front. Bioeng. Biotechnol. 2018, 6, 32. [Google Scholar] [CrossRef]
- Floss, H.G. The Shikimate Pathway; Springer: Berlin/Heidelberg, Germany, 1979. [Google Scholar]
- Bentley, R.; Haslam, E. The Shikimate Pathway—A Metabolic Tree with Many Branche. Crit. Rev. Biochem. Mol. Biol. 1990, 25, 307–384. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef]
- Noda, S.; Shirai, T.; Oyama, S.; Kondo, A. Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 2016, 33, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bilal, M.; Zong, Y.; Hu, H.; Wang, W.; Zhang, X. Development of a Plasmid-Free Biosynthetic Pathway for Enhanced Muconic Acid Production in Pseudomonas chlororaphis HT66. ACS Synth. Biol. 2018, 7, 1131–1142. [Google Scholar] [CrossRef]
- Thykaer, J.; Nielsen, J.; Wohlleben, W.; Weber, T.; Gutknecht, M.; Lantz, A.E.; Stegmann, E. Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab. Eng. 2010, 12, 455–461. [Google Scholar] [CrossRef]
- Pontrelli, S.; Chiu, T.-Y.; Lan, E.I.; Chen, F.Y.-H.; Chang, P.; Liao, J.C. Escherichia coli as a host for metabolic engineering. Metab. Eng. 2018, 50, 16–46. [Google Scholar] [CrossRef]
- Contesini, F.J.; de Melo, R.R.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Bio-based production of chemicals, materials and fuels–Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 2012, 23, 631–640. [Google Scholar] [CrossRef]
- Thomas, C.M.; Haines, A.S.; Kosheleva, I.A.; Boronin, A. Pseudomonas: Model Organism, Pathogen, Cell Factory; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2008. [Google Scholar]
- Nielsen, J. Yeast cell factories on the horizon. Science 2015, 349, 1050–1051. [Google Scholar] [CrossRef]
- Kitade, Y.; Hashimoto, R.; Suda, M.; Hiraga, K.; Inui, M. Production of 4-Hydroxybenzoic Acid by an Aerobic Growth-Arrested Bioprocess Using Metabolically Engineered Corynebacterium glutamicum. Appl. Environ. Microbiol. 2018, 84, e02587-17. [Google Scholar] [CrossRef]
- Sun, X.; Lin, Y.; Huang, Q.; Yuan, Q.; Yan, Y. A Novel Muconic Acid Biosynthesis Approach by Shunting Tryptophan Biosynthesis via Anthranilate. Appl. Environ. Microbiol. 2013, 79, 4024–4030. [Google Scholar] [CrossRef]
- Shen, X.; Wang, J.; Wang, J.; Chen, Z.; Yuan, Q.; Yan, Y. High-level De novo biosynthesis of arbutin in engineered Escherichia coli. Metab. Eng. 2017, 42, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Noda, S.; Shirai, T.; Mori, Y.; Oyama, S.; Kondo, A. Engineering a synthetic pathway for maleate in Escherichia coli. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Kim, W.J.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Efficient Production of 2-Pyrone-4,6-dicarboxylic Acid from Glucose. ACS Synth. Biol. 2018, 7, 2296–2307. [Google Scholar] [CrossRef]
- Jin, K.; Zhou, L.; Jiang, H.; Sun, S.; Fang, Y.; Liu, J.; Zhang, X.; He, Y.-W. Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylic acid production. Metab. Eng. 2015, 32, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Mcdonald, M.; Mavrodi, D.V.; Thomashow, L.S.; Floss, H.G. Phenazine biosynthesis in Pseudomonas fluorescens: Branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J. Am. Chem. Soc. 2001, 123, 9459–9460. [Google Scholar] [CrossRef]
- Webby, C.J.; Baker, H.M.; Lott, J.S.; Baker, E.N.; Parker, E.J. The Structure of 3-Deoxy-D-arabino-heptulosonate 7-phosphate Synthase from Mycobacterium tuberculosis Reveals a Common Catalytic Scaffold and Ancestry for Type I and Type II Enzymes. J. Mol. Biol. 2005, 354, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.E.; Dunbar, B.; Hunter, L.S.; Nimmo, H.G.; Coggins, J.R. Evidence for a novel class of microbial 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase in Streptomyces coelicolor A3(2), Streptomyces rimosus and Neurospora crassa. Microbiology 1996, 142, 1973–1982. [Google Scholar] [CrossRef]
- Görlach, J.; Beck, A.; Henstrand, J.M.; Handa, A.K.; Herrmann, K.M.; Schmid, J.; Amrhein, N. Differential expression of tomato (Lycopersicon esculentum L.) genes encoding shikimate pathway isoenzymes. I. 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase. Plant Mol. Biol. 1993, 23, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Dong, X.N.; Ausubel, F.M.; Fink, G.R. Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc. Natl. Acad. Sci. USA 1991, 88, 8821–8825. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Herrmann, K.M. Cloning and Sequencing of a Second cDNA Encoding 3-Deoxy-d-arabino-Heptulosonate 7-Phosphate Synthase from Solanum tuberosum L. Plant Physiol. 1992, 100, 1075–1076. [Google Scholar] [CrossRef][Green Version]
- Biessy, A.; Filion, M. Phenazines in plant-beneficial Pseudomonas spp.: Biosynthesis, regulation, function and genomics. Environ. Microbiol. 2018, 20, 3905–3917. [Google Scholar] [CrossRef]
- Pierson, L.S., III; Gaffney, T.; Lam, S.; Gong, F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30-84. FEMS Microbiol. Lett. 1995, 134, 299–307. [Google Scholar] [CrossRef]
- Wang, S.; Cui, J.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. Pseudomonas spp. as cell factories (MCFs) for value-added products: From rational design to industrial applications. Crit. Rev. Biotechnol. 2020, 40, 1232–1249. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2011, 9, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Shumilin, I.A.; Bauerle, R.; Wu, J.; Woodard, R.W.; Kretsinger, R.H. Crystal Structure of the Reaction Complex of 3-Deoxy-d-arabino-heptulosonate-7-phosphate Synthase from Thermotoga maritima Refines the Catalytic Mechanism and Indicates a New Mechanism of Allosteric Regulation. J. Mol. Biol. 2004, 341, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ayilavarapu, S.; Doctor, A.; Lee, C.-T.; Tribble, G.D.; Chiu, Y.; Weltman, R.L.; Angelov, N. Altered human alveolar bone gene expression in type 2 diabetes—A cross-sectional study. J. Periodontal Res. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Haslam, E. Shikimic Acid: Metabolism and Metabolites; Wiley: Chichester, UK; New York, NY, USA, 1993. [Google Scholar]
- Jin, X.-J.; Peng, H.-S.; Hu, H.-B.; Huang, X.-Q.; Wang, W.; Zhang, X.-H. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci. Rep. 2016, 6, 27393. [Google Scholar] [CrossRef]
- Jiang, P.-H.; Shi, M.; Qian, Z.-K.; Li, N.-J.; Huang, W.-D. Effect of F209S Mutation of Escherichia coli AroG on Resistance to Phenylalanine Feedback Inhibition. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao Acta Biochim. Biophys. Sin. 2000, 32, 441–444. [Google Scholar]
- Lin, S.; Liang, R.; Meng, X.; Ouyang, H.; Yan, H.; Wang, Y.; Jones, G.S. Construction and expression of mutagenesis strain of aroG gene from Escherichia coli K-12. Int. J. Biol. Macromol. 2014, 68, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Ger, Y.-M.; Chen, S.-L.; Chiang, H.-J.; Shiuan, D. A Single Ser-180 Mutation Desensitizes Feedback Inhibition of the Phenylalanine-Sensitive 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate (DAHP) Synthetase in Escherichia coli. J. Biochem. 1994, 116, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Hu, C.; Shen, S.; Wang, W.; Jiang, P.; Huang, W. Requirement of the N-terminus for dimer formation of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate synthase AroG of Escherichia coli. J. Basic Microbiol. 2004, 44, 400–406. [Google Scholar] [CrossRef]
- Munack, S.; Roderer, K.; Ökvist, M.; Kamarauskaite, J.; Sasso, S.; van Eerde, A.; Kast, P.; Krengel, U. Remote Control by Inter-Enzyme Allostery: A Novel Paradigm for Regulation of the Shikimate Pathway. J. Mol. Biol. 2016, 428, 1237–1255. [Google Scholar] [CrossRef]
- Li, Q.-A.; Mavrodi, D.V.; Thomashow, L.S.; Rossle, M.; Blankenfeldt, W. Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE. J. Biol. Chem. 2011, 286, 18213–18221. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.F.; Calabrese, K.; Eisenstein, E.; Ladner, J.E. Structure and Mechanism of Pseudomonas aeruginosa PhzD, an Isochorismatase from the Phenazine Biosynthetic Pathway. Biochemistry 2003, 42, 5684–5693. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fu, C.; Liu, K.; Cui, J.; Hu, H.; Wang, W.; Zhang, X. Engineering a Synthetic Pathway for Gentisate in Pseudomonas Chlororaphis P3. Front. Bioeng. Biotechnol. 2021, 8, 622226. [Google Scholar] [CrossRef] [PubMed]



| Strains | Description | Source |
|---|---|---|
| S17-1 (λ pir) | E. coli res- pro mod+ integrated copy of RP4, mob+, used for incorporating constructs into P. chlororaphis | Lab stock |
| P. chlororaphis HT66 | P. chlororaphis wild-type, PCN, Apr, Spr | Lab stock |
| HT66ΔphzE | P. chlororaphis HT66 with phzE deleted | This study |
| HT66ΔphzC | P. chlororaphis HT66 with phzC deleted | This study |
| HT66ΔaroF | P. chlororaphis HT66 with aroF deleted | This study |
| HT66ΔaroG | P. chlororaphis HT66 with aroG deleted | This study |
| HT66ΔaroH | P. chlororaphis HT66 with aroH deleted | This study |
| HT66ΔphzCΔaroF | P. chlororaphis HT66 with phzC, aroF deleted | This study |
| HT66ΔphzCΔaroG | P. chlororaphis HT66 with phzC, aroG deleted | This study |
| HT66ΔphzCΔaroH | P. chlororaphis HT66 with phzC, aroH deleted | This study |
| HT66ΔaroFΔaroG | P. chlororaphis HT66 with aroF, aroG deleted | This study |
| HT66ΔaroHΔaroG | P. chlororaphis HT66 with aroH, aroG deleted | This study |
| HT66ΔaroFΔaroH | P. chlororaphis HT66 with aroF, aroH deleted | This study |
| HT66::pBBR | P. chlororaphis HT66 harboring pBBR | This study |
| HT66::phzC | P. chlororaphis HT66 harboring pBBR-Pphz-phzC | This study |
| HT66::aroF | P. chlororaphis HT66 harboring pBBR-Pphz-aroF | This study |
| HT66::aroG | P. chlororaphis HT66 harboring pBBR-Pphz-aroG | This study |
| HT66::aroH | P. chlororaphis HT66 harboring pBBR-Pphz-aroH | This study |
| HT66ΔphzC::phzC | P. chlororaphis HT66ΔphzC harboring pBBR-phzC | This study |
| HT66ΔaroF::aroF | P. chlororaphis HT66ΔaroF harboring pBBR-aroF | This study |
| HT66ΔaroG::aroG | P. chlororaphis HT66ΔaroG harboring pBBR-aroG | This study |
| HT66ΔaroH::aroH | P. chlororaphis HT66ΔaroH harboring pBBR-aroH | This study |
| Plasmids | Description | Source |
| pk18mobsacB | Broad-host-range gene replacement vector, Kmr | Lab stock |
| pk18-phzE | pk18mobsacB containing phzE upstream and phzE downstream, Kmr | This study |
| pk18-phzC | pk18mobsacB containing phzC upstream and phzC downstream, Kmr | This study |
| pk18-aroF | pk18mobsacB containing aroF upstream and aroF downstream, Kmr | This study |
| pk18-aroG | pk18mobsacB containing aroG upstream and aroG downstream, Kmr | This study |
| pk18-aroH | pk18mobsacB containing aroH upstream and aroH downstream, Kmr | This study |
| pBBR-Pphz-phzC | pBBR-MCS2 containing Pphz-phzC, Kmr | This study |
| pBBR-Pphz-aroF | pBBR-MCS2containing Pphz-aroF, Kmr | This study |
| pBBR-Pphz-aroG | pBBR-MCS2containing Pphz-aroG, Kmr | This study |
| pBBR-Pphz-aroH | pBBR-MCS2containing Pphz-aroH, Kmr | This study |
| Primer | Sequence (5′–3′) |
|---|---|
| For Gene Deletion | |
| phzC-1F | CATGATTACGAATTCACAACTAACCGCTAGCGACACCACT |
| phzC-1R | GATGCGATCACTCTCACGAGAGAATT |
| phzC-2F | TGCGCTTGAACTCAGGAGTCTTTGCCTGGAGTTTGTCGCCATGACCG |
| phzC-2R | GACTCTAGAGGATCCGGTGGAAATCAGTACCCCGACATG |
| aroF-1F | CATGATTACGAATTCAGTTCGATGGCCTCGACGTCTTC |
| aroF-1R | CATGGACTCGGGTGTTTTTTAAGGT |
| aroF-2F | ACCTTAAAAAACACCCGAGTCCATGACCCGTAGCGCTCGATCATCC |
| aroF-2R | GACTCTAGAGGATCCGAAGCAAGCGGCCTATTGCCT |
| aroG-1F | CATGATTACGAATTCACGGTTGCACACTATCAGCCTCG |
| aroG-1R | CGTGTTACTCGTCAGGTCACGGG |
| aroG-2F | CCCGTGACCTGACGAGTAACACGTCCCGTATCGCGGACACAAAA |
| aroG-2R | GACTCTAGAGGATCCGGTGCCAATGGTGCCTACTATTTGA |
| aroH-1F | CATGATTACGAATTCAAATCGCGACAGGATCAGTCCTG |
| aroH-1R | TTCCGCCCCTGTAGGAGCAG |
| aroH-2F | CTGCTCCTACAGGGGCGGAAATTCAAGGCTTCCTGGGCAGG |
| aroH-2R | GACTCTAGAGGATCCCGTGGCGAGTGTGTCATAAAACCT |
| For Gene Overexpression | |
| G-phzC-1F | TACCGGGCCCCCCCTCGAGTTTGAGCACCACTAAAGTTGAAAACAGG |
| G-phzC-1R | GGCGGCATCCTCCTTAGTTGGG |
| G-phzC-2F | CCCAACTAAGGAGGATGCCGCCATGGAAGACTTACTGAAACGGGTATTAAGTTG |
| G-phzC-2R | TGGCGGCCGCTCTAGATCAAAAGGAGGCAAGGGTTGAGGAG |
| G-aroF-1F | TACCGGGCCCCCCCTCGAGTTTGAGCACCACTAAAGTTGAAAACAGG |
| G-aroF-1R | GGCGGCATCCTCCTTAGTTGGG |
| G-aroF-2F | CCCAACTAAGGAGGATGCCGCCATGATGAGCCAACCCTGGAGCC |
| G-aroF-2R | TGGCGGCCGCTCTAGATCAACGCTTGACCTGTTTCAGGGTC |
| G-aroG-1F | TACCGGGCCCCCCCTCGAGTTTGAGCACCACTAAAGTTGAAAACAGG |
| G-aroG-1R | GGCGGCATCCTCCTTAGTTGGG |
| G-aroG-2F | CCCAACTAAGGAGGATGCCGCCATGGCTGATTTACCGATCAACGACC |
| G-aroG-2R | TGGCGGCCGCTCTAGATCAGGTACGAACCCGTTTTGGCA |
| G-aroH-1F | TACCGGGCCCCCCCTCGAGTTTGAGCACCACTAAAGTTGAAAACAGG |
| G-aroH-1R | GGCGGCATCCTCCTTAGTTGGG |
| G-aroH-2F | CCCAACTAAGGAGGATGCCGCCATGAACTCGTCCGTATCCGCTCTG |
| G-aroH-2R | TGGCGGCCGCTCTAGATCAGGCGGAAGCCGGAATGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Liu, D.; Bilal, M.; Wang, W.; Zhang, X. Uncovering the Role of PhzC as DAHP Synthase in Shikimate Pathway of Pseudomonas chlororaphis HT66. Biology 2022, 11, 86. https://doi.org/10.3390/biology11010086
Wang S, Liu D, Bilal M, Wang W, Zhang X. Uncovering the Role of PhzC as DAHP Synthase in Shikimate Pathway of Pseudomonas chlororaphis HT66. Biology. 2022; 11(1):86. https://doi.org/10.3390/biology11010086
Chicago/Turabian StyleWang, Songwei, Dongliang Liu, Muhammad Bilal, Wei Wang, and Xuehong Zhang. 2022. "Uncovering the Role of PhzC as DAHP Synthase in Shikimate Pathway of Pseudomonas chlororaphis HT66" Biology 11, no. 1: 86. https://doi.org/10.3390/biology11010086
APA StyleWang, S., Liu, D., Bilal, M., Wang, W., & Zhang, X. (2022). Uncovering the Role of PhzC as DAHP Synthase in Shikimate Pathway of Pseudomonas chlororaphis HT66. Biology, 11(1), 86. https://doi.org/10.3390/biology11010086







