Simple Summary
An efficient immobilized bacterial consortium of Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus, and Staphylococcus xylosus was used to degrade MO under high salinity conditions. The biodegradation efficiency was confirmed by UV−visible spectrophotometric analysis, FTIR, and the assaying of degradative enzymes. The cytotoxicity of such metabolic byproducts was evaluated on two human normal cell lines. In addition, the immobilized consortium showed the synergistic interaction between immobilized consortium and indigenous microorganism for degrading MO in artificially contaminated agricultural and industrial effluents.
Abstract
Methyl orange (MO) is categorized among the recalcitrant and refractory xenobiotics, representing a significant burden in the ecosystem. To clean-up the surrounding environment, advances in microbial degradation have been made. The main objective of this study was to investigate the extent to which an autochthonous consortium immobilized in alginate beads can promote an efficient biodegradation of MO. By employing response surface methodology (RSM), a parametric model explained the interaction of immobilized consortium (Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus and Staphylococcus xylosus) to assimilate 200 mg/L of MO in the presence of 40 g/L of NaCl within 120 h. Physicochemical analysis, including UV-Vis spectroscopy and FTIR, and monitoring of the degrading enzymes (azoreductase, DCIP reductase, NADH reductase, laccase, LiP, MnP, nitrate reductase and tyrosinase) were used to evaluate MO degradation. In addition, the toxicity of MO-degradation products was investigated by means of phytotoxicity and cytotoxicity. Chlorella vulgaris retained its photosynthetic performance (>78%), as shown by the contents of chlorophyll-a, chlorophyll-b and carotenoids. The viability of normal lung and kidney cell lines was recorded to be 90.63% and 99.23%, respectively, upon exposure to MO-metabolic outcomes. These results reflect the non-toxicity of treated samples, implying their utilization in ferti-irrigation applications and industrial cooling systems. Moreover, the immobilized consortium was employed in the bioremediation of MO from artificially contaminated agricultural and industrial effluents, in augmented and non-augmented systems. Bacterial consortium remediated MO by 155 and 128.5 mg/L in augmented systems of agricultural and industrial effluents, respectively, within 144 h, revealing its mutual synergistic interaction with both indigenous microbiotas despite differences in their chemical, physical and microbial contents. These promising results encourage the application of immobilized consortium in bioaugmentation studies using different resources.
1. Introduction
The genetic diversity and wealthy nature of microbial metabolism are considered the paramount agents in its governing several vital medical, pharmaceutical, agricultural, environmental and industrial applications. At the environmental level, microbial metabolism was recruited in resolving numerous environmental issues. Through bioremediation, either bacterial [1] or mycoremediation [2], several man-made complicated xenobiotic compounds are degraded to simpler inorganic constituents or even mineralized into water and carbon dioxide. Such a naturally occurring phenomenon offers many advantages over physical and chemical methods, including eco-friendliness, cost-effectiveness and less sludge generation or incomplete degraded byproducts, which represent additional environmental burdens [3,4]. However, the effective implementation of biodegradation processes depends mainly on the behavior of microbes toward xenobiotic compounds via the secretion of adequate enzymatic batteries or non-enzymatic redox-active molecules. Actually, the employing of advanced trials in molecular, bioinformatics, biotechnological and even basic microbiological applications, either segregated or in alliance, aid in accelerating the biodegradation process efficiently. Interestingly, genetic transformation, bio-stimulation, bio-augmentation, mixed cultures and immobilization are among the attractive techniques pertaining to the proficient microbial degradation of recalcitrant substances [5,6]. Bioaugmentation is the addition of exogenous microorganisms, characterized by their biodegradation capacities, to indigenous or in situ microbial populations for improving the treatment process of pollutants [7]. The success in this task is mostly based on the proliferative and competitional proficiency of the introduced microbes [8]. However, the comprehensive understanding of addition/maintenance techniques, and knowledge of molecular biology and the capabilities of commercial products, are prerequisites [9]. Additionally, adjusting and monitoring some parameters such as temperature, pH, dissolved oxygen, nutrition availability, toxicity and microbial pressures during the treatment process contributes to its success; otherwise, it fails, as revealed in [10].
Notably, the employment of statistical approaches to the design of experiments (DOEs) such as central composite design (CCD) in the remediation process, either biologically or chemically, has recently become more common [11,12,13]. Through these chemometric means, the optimum conditions of multiple independent variables can be defined. Importantly, the interaction effect of each examined variable on the response would be determined without exerting more effort, with more time saved and the process being more cost-effective [14,15]. Interestingly, the utilization of CCD in chemical processes such as wastewater flocculation and coagulation, protein extraction from wastewater and dye degradation was revealed [16]
Remarkably, the comparison of the biodegradative performance of monocultures and mixed cultures always favors the mixed cultures, as evidenced by Ahmadi et al. [17] and Phan et al. [18]. Microbial consortia demonstrate stability in the cleavage of synthetic contaminants; the metabolic burden of synthetic pollutant degradation is divided across multiple microbes, and each microbe in the consortium participates with its own mechanism in the degradation process [19]. Such combinational action of all the consortium’s microbial partners speeds up the biodegradation rate [20]. Additionally, the microbes in their mixed system seem to be resistant to abiotic stressors. A number of recent studies listed the enhanced impact of different microbial consortiums in the removal of a wide array of pollutants, including heavy metals [21], organochlorine pesticide [22], organophosphate pesticides [23], azo dye [24] and organic hydrocarbons [25,26]. On the other hand, the use of microbial biomass in the form of an immobilized state is advantageous as compared to the freely suspended state [27]. This could be explained by the protection of the immobilized cells from the toxic impact of pollutants or their byproducts, and their stability for reuse. Thus, the immobilization technique is considered as a powerful tool that can be utilized towards various applications in an economic manner [28,29]. The entrapment approach is used in the overwhelming majority of immobilization techniques, especially in alginate beads. It is a natural polymer, and is cheap, nontoxic, biocompatible to microbial cells, readily available, highly viscous and exhibits good stability. Furthermore, its gel nature furnishes suitable porosity, allowing the continuous diffusion of substrates and byproducts [30]. In broader terms, the utilization of alginate gel immobilized with microbial biomass was recorded in different applications, such as alcoholic wine fermentation [31], dye decolorization [32], crack remediation [33] waste oil degradation [34] and heavy metal removal [24,35].
The combination of such approaches in the degradation of recalcitrant substrates generated from anthropogenic activities, such as azo dye, seems to be promising [36,37]. Actually, azo dye represents about almost 80% of synthetic dyes that enter water bodies from textile discharge [38], causing environmental injuries such as reductions in dissolved oxygen concentrations, the blocking of sunlight penetration, and the blocking of photosynthesis [39,40,41]. These negative effects are compounded by its adverse impact on human health, including respiratory problems, diarrhea, vomiting and nausea [42]. The continuous presence of azo dyes in ecosystems further leads to poisoning, carcinogenicity, teratogenicity and mutagenicity [43]. It is worth mentioning that textile discharge is characterized by its salty nature, as stated by Mirbolooki et al. [44]. In fact, higher concentrations of NaCl are added during the staining process to neutralize the surface charge of negatively-charged fibers; hence, the fixation and adsorption of anionic dyes can be improved. By these means, the leakage of considerable quantities of salt along with dyestuff takes place, which varies from 1 to 10 g/L, causing further encumbrance.
On this basis, the current study focused on the biodegradation of Methyl orange (MO), as one of the most extensively used hazardous anionic azo dyes [45]. Methyl orange is an azo dye that has been sulphonated [46]. In laboratories, it is also used as a pH indicator. It is brightly colored and highly soluble in water, making it difficult to remediate using traditional treatment methods (physicochemical methods) [47]. Its toxic nature and resistance to degradation necessitate the discovery of more azo dye-degrading microorganisms. A statistical experiment design was employed to study the interactive effect of some variables on the degradation performance of the immobilized microbial consortium. Moreover, the activity of some oxidoreductive enzymes, which are responsible for biodegradation, were quantitatively detected during the process. In addition, UV-Visible spectra and FTIR analysis were employed to confirm the breakdown and detoxification of MO. Furthermore, the acute toxicity assay, which encompassed a phytotoxicity assessment, was conducted, with the results being evaluated against those obtained for algae. Additionally, the cytotoxicity of the degraded products, as compared to the MO dye, was determined on human and animal cell lines. Additionally, a bioaugmentation study of the consortium immobilized in alginate beads was conducted on real effluents that were artificially contaminated with MO in comparison to free consortium.
2. Materials and Methods
2.1. Screening, Selection and Growth Condition Analysis of MO-Degrading Microorganisms
In this article, strains from the in-house culture collection isolated from various environmental sources (sediments and fresh water) and industrial effluents were examined for degradation of MO. The coordinates of the screening sites were as follows: Naba Alhamra, Wadi El Natrun (morphotectonic depression at the northeast corner of the Western Desert between the longitudes of 30°00′ and 30°30′ E and the latitudes of 30°15′ and 30°40′ N, Al-Beheira governorate, Egypt), El-Mahmoudeya Canal (45-mile-long (72 km) sub-canal derived from the Nile River and influx into the Mediterranean Sea, latitude 31°11′2″ N and longitude 30°31′27″ E, Alexandria governorate, Egypt) and Jeddah’s Second Industrial City neighborhood (latitude 21°48′ N and longitude 39°19′ E, Saudi Arabia). The isolated strains were directly screened for MO degradation using a plate test [48]. Using mineral salt medium (MSM) (g/L) (1.27 K2HPO4; 0.42 MgSO4·7H2O; 0.42 NaNO3; 0.29 KCl; 2 NaCl; 0.85 KH2PO4, pH 7.0) supplemented with 50 mg/L of MO, constant microbial lawns were spotted separately on the surface of the solidified agar plates and incubated for 72 h at 30 °C. A discoloration zone was observed around some colonies, which demonstrated their preliminary ability for degradation. The colonies with the most discolored areas, measured from the colony’s edge, were selected to construct the consortium.
2.2. Antagonism Assay for Detecting the Compatibility of the Consortium Strains
The bacterial compatibility was evaluated for consortia as described by [49]. Briefly, about 50 μL of each bacterial suspension (about 106 (CFU) mL−1) was spotted onto nutrient agar plates (g/L) (2 yeast extract; 5 peptone; 5 sodium chloride; 15 agar, pH 7.0) with 1 cm spaces between the spots. The inoculated plates were then incubated at 30 °C in a static condition for 72 h. The overlapping of the spots implied that there was compatibility without symptoms of inhibition being shown [50].
2.3. Inoculum Preparation and Consortium Immobilization in Alginate Microspheres
A pure culture of each bacterial strain was inoculated in NB and incubated at 30 °C in an orbital shaker (150 rpm) (MCL-D21 Rotary shaker) until the OD600 reached 0.6 to 0.8. The cultures were then collected aseptically and centrifuged at 10,000 rpm for 10 min. At equal cell densities, the harvested pellets were resuspended in distilled water and homogeneously mixed. The consortium was immobilized by entrapment in a 3% sodium alginate solution following the method described by Eltarahony et al. [51]. A quantity of 50 mL of bacterial biomass was mixed thoroughly with a slurry of 3% sodium alginate (50 mL) in a 250 mL Erlenmeyer flask. The homogeneous mixture was dropped into 100 mL of 2% chilled sterile CaCl2 (w/v) using a syringe, thereby forming gel beads (~4 mm diameter). The immobilized consortium beads were kept in CaCl2 (2%) for 1 h. at 30 °C to complete the hardening process. Thereafter, the collected beads were washed several times using sterile distilled water to remove any remains before freeze-drying for 8 h at 36 °C [52,53].
2.4. Characterization of Immobilized Beads by Scanning Electron Microscopy (SEM)
SEM (JSM 6360LA, JEOL, Akishima-shi, Japan) was used to examine the morphology of the beads. Some beads (before and after bacterial immobilization) were dehydrated at 30 °C using a vacuum stove until constant weight was achieved, and hydrogels were freeze-dried for 8 h at −20 °C and 0.022 mmHg using a Rificor S.H lyophilizer. Dehydrated and lyophilized beads were cryo-fractured in liquid nitrogen before being mounted on bronze stubs and metallized with an argon plasma metallizer (PELCO 91000 sputter coater) [53].
2.5. Biodegradation Test of Immobilized Beads
MO degradation for immobilized consortium was screened initially in a 100 mL MSM broth (mentioned previously) supplemented with 100 mg/L MO and 7 immobilized beads containing 106 CFU/mL. All tests were performed in triplicates and samples were incubated for 7 days at 30 °C in a rotary shaker set to 150 rpm (MaxQ 6000, Thermo Fisher, Waltham, MA, USA). At time intervals of 6 h throughout the incubation period, the MO residues were measured using a spectrophotometer at wavelength of 465 nm [54,55].
2.6. MO Degradation by Consortium Immobilized in Alginate Beads Using Central Composite Design (CCD)
In this step, the biodegradation performance was examined simultaneously and statistically in relation to certain variables [56]. The statistical modeling of the degradation solution was constructed based on 5 experimental levels, −α, −1, 0, +1, and +α, for four independent variables (degradation time, inoculated consortium, MO and NaCl concentrations) in a 31-trail matrix. The experimental levels of the examined independent variables are illustrated in Table 1.

Table 1.
The concentrations of examined variables (incubation time, inoculum size, MO conc. and NaCl) at each level in the CCD experiment.
The statistical calculation that describes the relationship between the coded and actual values is represented in Equation (1):
where Xi is the coded value of the ith variable, Ui is the actual value of the ith variable, Ui0 is the actual value of the ith variable at the center point and ΔUi is the step change of the variable. The relationship between the examined independent variables and the response (MO detoxification) was calculated using the equation of second-degree polynomial (Equation (2)):
where: Y is the predicted response; X1, X2 and X3 are input variables that influence the response variable Y; β0 is the intercept; β1, β2 and β3 are linear coefficients; β11, β22 and β33 are squared or quadratic coefficients, and β12, β13 and β23 are interaction coefficients.
Xi = Ui − Ui0/ΔUi
Y = β0 + β1X1 + β2X2 + β3X3 + β11X11+ β22X22 + β33X33 + β12X1X2 + β13X1X3 + β23X2X3
2.7. Statistical Analysis
The experimental designs, the regression analysis for determining the analysis of variance (ANOVA), three-dimensional surface plots (3D) and two-dimensional contour plots (2D) were established using the Minitab 14.0 statistical software (Minitab Inc., State College, PA, USA).
2.8. Decolorization Assay of MO
For each CCD-trial, MO decolorization was assessed by determining the change in absorbance of clear supernatants at the absorption maxima (λ max) of 465 nm with a UV–Vis spectrophotometer (Labomed model, Inc., Los Angeles, CA, USA). Abiotic control or un-inoculated medium was prepared in the CCD matrix for each trial. The experiments were performed in triplicate and the average was considered. The color removal efficiency was calculated as the percentage ratio based on the following equation [49,57]:
2.8.1. Extraction and Analysis of Degradation By-Products
Both MO and its degraded metabolites were subjected to further analysis to confirm the accomplishment of the biodegradation process. The immobilized consortium was inoculated in MSM broth containing 200 mg/L of MO and incubated at 30 °C in a 150 rpm orbital shaker for 5 days. Simultaneously, a control experiment was conducted in exactly the same manner as outlined previously but without consortium inoculum. At the end of the incubation time, a 50 mL quantity of the MO (control dye without degradation) and the degraded sample was centrifuged at 10,000 rpm for 10 min. The obtained supernatants were extracted using ethyl acetate in equal proportion followed by evaporation by rotary evaporator (IKA Rotary evaporator RV 8 V-C). The generated extract was suspended in water (5 mL) for the further analysis.
2.8.2. UV−Visible Spectrophotometric Analysis
A UV–Visible spectrophotometer (Labomed model, Inc., USA) was used to assess biodegradation at the zero time point and after the completion of the process. A clear supernatant was scanned from 200 to 800 nm within the absorption spectrum.
2.8.3. FTIR
The FTIR spectra of MO and its degraded metabolites were determined using the Shimadzu FTIR-8400S (Kyoto, Japan). Dehydrated and lyophilized samples were triturated into fine powder, combined with KBr (1.0 percent w/w) and crushed into transparent discs in a hydraulic press. Spectra were captured at 4 cm−1 resolution over a range of 4000–400 cm −1 with an accumulation of 64 scans and using dried air as a background [58].
2.9. Enzymes Activity Assays
The degradation performance of the bacterial consortium was examined at flask level in MSM broth supplemented with 200 mg/L of MO (i.e., the highest concentration was degraded efficiently by the consortium). About 106 CFU mL−1 was utilized as an inoculum and incubated as mentioned previously. The activities of the enzymes, including manganese peroxidase (MnP), nitrate reductase (NR), tyrosinase, lignin peroxidase (LiP), NADH-DCIP reductase, azoreductase and laccase, were quantified during the biodegradation process at a constant interval time (12 h). The crude enzymes were prepared by cell lysis using the physical disruption method (Ultrasonic Disruptor UD-200, Tommy, Tokyo, Japan) for 2 min intervals based on a 40-amplitude output at 4 °C. The obtained slurry was subsequently centrifuged for 5 min at 4 °C and 15,000 rpm, and the clear solution was used as a crude enzyme. The enzyme assays were determined as reported by Glenn and Gold [59], Redinbaugh and Campbell [60], Shanmugam et al. [61], Sambasiva Rao et al. [62], Zhao et al. [63], El-Fakharany et al. [64], Rekik et al. [65], and Sari and Simarani [66,67] Negative controls (enzymes denatured by boiling) were run in parallel.
2.10. Protein Content Determination
The protein content in all tested samples was determined by reading the OD values of samples at 280 nm or using the method outlined in [68] with BSA as a standard protein.
2.11. Acute Toxicity Assessments
2.11.1. Phytotoxicity Bioassays with Chlorella vulgaris
The toxic effects of MO (200 mg/L) before degradation and its byproduct (after degradation) were compared with control (without any treatment). In three Erlenmeyer flasks (500 mL), the C. vulgaris cells were inoculated into 100 mL of Bold’s basal media (BBM) medium alone and in combination with 200 mg/L of either MO or the degradation byproducts. All flasks were incubated for 4 days at 25 °C under illumination [69]. Following the completion of the incubation period, a fixed volume of algal cultures was centrifuged at 3000 rpm for 20 min. The obtained biomass was subjected to pigment fractionation in accordance with Shebany et al. [70]. Pigments were extracted in hot methanol (70 °C) for 10 min. Centrifugation (Sorvall RC-3C Plus refrigerated floor model) was used to remove cell debris, and the clear supernatant, which contained the pigments, was aspirated and diluted to a specific volume. At 452.5, 644 and 663 nm, the extinction coefficient was measured using a spectrophotometer against a methanol blank. Taking dilution into account, the pigment fractions’ contents (µg/mL algal suspension) were calculated using the following equations [70]:
2.11.2. Determination of Cytotoxicity against Normal Lung and Kidney Cell Lines
A human normal lung fibroblast cell line (Wi-38) and a normal adult African green monkey kidney cell line (Vero) were used to investigate the toxicity of MO dye before and after bioremediation according to the tetrazolium salt (MTT) assay [71]. The MTT assay was performed to detect the viability of the treated cells as compared to healthy untreated cells. Only viable active metabolic cells have the ability to reduce yellow soluble tetrazolium salt to insoluble purple formazan by means of their mitochondrial dehydrogenase enzyme. Human Wi-38 and mammalian Vero cells were maintained in Dulbecco’s Modified Eagle (DMEM) medium (Lonza, Bend, OR, USA) containing 10% fetal bovine serum. These cell lines were sub-cultured for 2 weeks before an assay was conducted using trypsin EDTA (Lonza, USA). Their viability and counting were detected by trypan blue staining and with the use of a hemocytometer. Wi-38 and Vero cells were seeded in a 96-well culture plate at 1 × 104 cells per well and incubated at 37 °C in a 5% CO2 incubator. After 24 h, for the cell attachment, the cells were exposed to MO (200 mg/L) and its degraded byproducts. Following 72 h of incubation in a 5% CO2 incubator, 20 µL of MTT solution (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) (5 mg/mL) was added to each well before incubation at 37 °C for 4 h in a 5% CO2 incubator. MTT (Sigma, St. Louis, MO, USA) solution was removed and the insoluble blue formazan crystals trapped in cells were solubilized with 150 µL of 100% DMSO at 37 °C for 10 min. The absorbance of each well was measured with a microplate reader (BMG LabTech, Ortenberg, Germany) at 570 nm to estimate cell viability.
2.12. Bioaugmentation of Free and Immobilized Cells in Real Waste Effluents
The biodegradative potential of immobilized consortium beads in real wastewater containing various pollutant loads was determined in this step. In comparison to free non-immobilized consortium, the immobilized beads were employed as a modular bioaugmentation tool for remediating two effluents that were artificially inoculated with MO (200 mg/L) in two forms, namely sterilized (non-augmented) and non-sterilized (augmented) states. The industrial effluent was collected, in December 2020, from Discharge Line Industrial Zone, Borg Al Arab, Alexandria, Egypt, while the agricultural effluent was collected from Bahig Canal, Borg Elarab, Alexandria, Egypt, in October 2020. The quality criteria of the samples were identified and their quality was measured according to these standards. In parallel, non-immobilized alginate beads were run to detect the absorption percentage. For bench-scale bioremediation trials, fixed numbers of immobilized alginate beads, of equivalent size to the free consortium, and non-immobilized alginate beads were inoculated in 1 L flasks containing 600 mL of industrial and agricultural effluents in both augmented and non-augmented states. The incubation conditions were performed as recorded previously. At each time interval (24 h), 2 mL quantities of samples were drawn, centrifuged and measured spectrophotometrically at 465 nm. The biodegradation percentage was calculated using Equation (3) [72].
3. Results
3.1. Selection of MO-Degrading Microorganisms and Their Compatibility
The potential of 39 bacterial strains to degrade MO was examined; four of them were selected based on the highest values of the clearance zone that surrounded their colonies. The selected strains were: Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus and Staphylococcus xylosus. Their 16S-rDNA genes were identified and deposited in GenBank under accession numbers MK551748, MN631047, MT940225 and MT940226, respectively. Such strains were isolated from different ecosystems that contained different pollutant loads. They exhibited characteristic biochemical and physiological capabilities and some of them were utilized previously in the bioremediation of heavy metals [58]. As noticed, the selected strains are affiliated to different bacterial classes encompassing alphaproteobacteria, gammaproteobacterial and firmicutes, which imply different behaviors in terms of the handling of substrates. However, their combination in the consortium depends mainly on the absence of interspecies antagonism, which was clearly demonstrated in the compatibility experiment. Such corporative interactions facilitate their application, as noted by Fuentes et al. [73]. On highly nutritive media such as nutrient broth, MO was easily and entirely degraded in time range of 20–28 h by four selected strains as monocultures; upon combination into a mixed culture, removal was achieved within 18 h (data not shown). Nonetheless, the reason behind utilizing MSM in the present study is based on its capacity to resemble reality and mimic the effluents that are full of stressors and have a lack of nutrients. Herein, MO was assimilated by the examined consortium and 30% of it was eliminated during 7 days of incubation at the basal MSM, where it consumed MO as a carbon source and attacked its structure to obtain enough energy for survival and proliferation. Individually, in terms of discoloration, Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus and Staphylococcus xylosus showed 7, 9, 13 and 16% color removal, respectively, in MSM within the exact conditions, reflecting the enhancement of degradation by the action of combinatorial configuration. The porous structure of alginate microsphere that granted the migration of the dye’s molecules from the solution into the bulk structure of microspheres is shown in Figure 1A. Moreover, successful entrapment of the oval shaped bacterial consortium cells onto the porous alginate microsphere matrix was indicated via SEM (Figure 1B). It is evident from this figure that the porous structure of alginate enables the interior migration of MO dye’s molecules onto the alginate microspheres, which allows them to interact with bacterial strains, thereby facilitating efficient dye degradation.
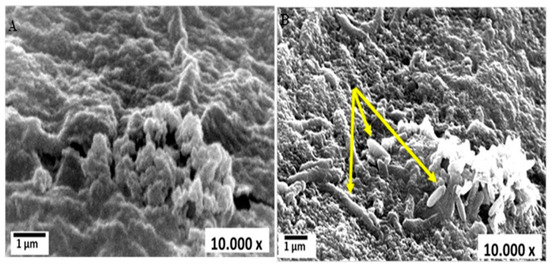
Figure 1.
Scanning electron microscope (SEM) micrograph of blank alginate microsphere before and after bacterial cell immobilization: (A) blank alginate microsphere; (B) immobilized bacterial consortium in alginate microsphere.
3.2. MO Degradation by Consortium Immobilized in Alginate Beads Using Central Composite Design (CCD)
To study the degradation behavior of immobilized consortium with different inoculated bacterial loads, in response to different ranges of MO and NaCl concentrations, over different incubation time periods, CCD was employed in 31 experimental runs. To our knowledge, this has rarely been investigated to date. The design matrix, which was composed of 16 factorial points (cubic points), 8 axial or star points (points with an axial distance to the center of (a = ±2)), and 7 replicates of center points, is represented, along with experimental, predicted responses and the studentized residual results, in Table 2. As observed, the highest percentage of MO degradation, at 46.38%, was recorded for trial 2 (axial point); however, the minimum removal percentage (0.27%) was noted for trial 6 (axial point), reflecting the interaction effect of different parameters at varied levels on response.

Table 2.
Central composite design (CCD) matrix of MO degradation using immobilized consortium influenced by incubation time, inoculum size, MO and NaCl concentrations along with the predicted responses and residuals.
3.3. Multiple Regression Analysis and ANOVA
The multiple regression analysis (Table 3) and ANOVA (Table 4) methods were employed to analyze the CCD results. By these means, the individual model coefficients, the significance of the regression model, the lack-of fit, the coefficient values of each examined parameter, their probability p-values, and their Linear interactions and quadratic effects were estimated. In addition, the coefficients of determination R2 and adj-R2 were also applied to evaluate the efficiency of the polynomial regression model. These values were identified to be 0.969 and 0.942, respectively, revealing that 96.9% of the variation in MO degradation could be explained by the investigated variables and that only 3.1% of the variation could not be elucidated by these variables. As documented in [74], regression models with high R2 values (0.9–1) are deemed to have the strongest and most positive correlations. Higher R2 values also indicate that a model is good and, more specifically, that it can explain the variation in experimental data as compared to the predicted data. Furthermore, the adjusted-R2 (adj-R2) value adjusts the R2 value based on the sample size and the number of variables in the model. The adj-R2 value should be in considerable agreement with the R2 value (≤2%) [75]. In this study, the excellent agreement between R2 and adj-R2 values verified the aptness of the model. On the other hand, the lack of fit test, which describes the variation in the data around the fitted model, revealed a value of 0.093, as inferred using the ANOVA method (Table 4). Broadly, an insignificant lack-of-fit indicates a good model [56]. Moreover, the model exhibited evident correlation between the observed (experimental) and the predicted values, as inferred from the studentized residual (Table 2). A small residual value (≤ ±2) is accepted. It estimates the error between points and detects outliers. Clearly, the residual values of our model did not deviate from accepted limits and fall on a straight line with only one outlier; this implies that the errors fell within a normal distribution, as demonstrated in the normal probability plots of the residuals (Figure 2). Thus, the model used in this study is optimal for predicting MO detoxification within the range of experimental parameters.

Table 3.
Estimated effect, regression coefficients and corresponding T and p values for determining the effects of different variables on MO degradation by means of immobilized consortium using CCD.

Table 4.
Analysis of variance (ANOVA) for MO degradation by means immobilized consortium obtained using CCD.
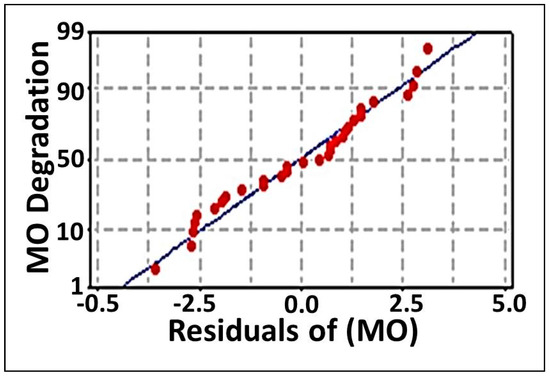
Figure 2.
The normal probability plot of the residuals for MO degradation by immobilized consortium determined by the second-order polynomial equation.
It is noteworthy that the p-value is an important tool for assessing the consequences and significance of the model and also each independent variable in the design. Commonly, a low probability ‘p’ value (prob > F <0.05) denotes high significance of the corresponding coefficient [56]. In this case, it was found that the ANOVA results, which are tabulated in Table 4, illustrate that the model is highly appropriate and adequate, as evidenced by the very low probability value (0.000). Subsequently, all these data indicated that the model is significant, adequate and well-fitted to the experimental data. Additionally, the linear coefficients of incubation time, inoculum size, and MO and NaCl concentrations were significant for MO degradation with probability values of 0.0, 0.001, 0.0 and 0.016, respectively. However, the quadratic coefficient of the MO concentration, in contrast to the quadratic coefficients of the other variables, was only significant at a level of p = 0.0. Generally, the linear effects of the tested parameters had more pronounced effects in terms of enhancing MO degradation than the quadratic and interaction effects, as summarized in Table 4. Additionally, the sign of each coefficient determined either a negative or a positive effect of the corresponding variable on the MO detoxification; specifically, the negative coefficient values of MO concentration (linear effect) implied their negative impact on MO removal at higher levels. However, the positive coefficients of incubation time, inoculum size and NaCl (linear effect) reflected their positive contribution to MO degradation as their values were increased up to certain limits. Similarly, the influence of the interaction between incubation time and NaCl concentration exerted a positive effect on MO degradation (Table 3). Finally, MO degradation as a response could be expressed in the terms of a second-order polynomial equation Equation (7) as following:
MO degradation (%) = 8.64 + 3.36 incubation time + 2.19 inoculum size − 9.44 MO concentration + 1.39 NaCl concentration + 0.05 (incubation time)2 –0.6 (inoculum size)2 + 4.0 (MO concentration)2 – 0.53 (NaCl concentration)2 – 0.82 incubation time × inoculum size – 1.08 incubation time × MO concentration + 1.34 incubation time × NaCl concentration – 2.52 inoculum size × MO concentration–0.396 Inoculum size × NaCl concentration – 0.399 MO concentration MO concentration
3.4. Graphical Interpretation of the Response Surface Model
In order to illustrate the relationships between the response and the interactions between the tested variables, three-dimensional (3D) surface plots and two-dimensional (2D) contour plots were generated (Figure 3). The MO degradation was plotted on the Z-axis versus and the two examined parameters were plotted on X and Y axes, while other parameters were kept fixed at their midpoints. The simultaneous effect of incubation time and MO concentration on MO detoxification, at constant level of NaCl concentration and inoculum size, was studied, and the results are presented in Figure 3A,B. It can be seen that more than 40% removal was obtained at the lowest concentration of MO (200 mg/L) under 5 days incubation (the highest incubation time). However, as elucidated in Figure 3C,D, the obvious increase in MO detoxification was conjugated with a mutual increase in both the incubation time and the NaCl concentration, reflecting a synergistic interaction effect. Moreover, an antagonistic relationship between inoculum size and MO concentration was observed, as shown in Figure 3E,F. The highest concentrations of MO resulted in lower MO detoxification within a low range of MO (200–250 mg/L), and high bacterial inoculum sizes (10–20%) assisted in significantly elevating the removal percentage of MO, which reached 50% and above. The same antagonistic trend was also clearly observed in the efficacy of MO degradation as a function of the MO and NaCl concentrations. The maximum degradation of MO (more than 40%) could be achieved at a wide range of concentrations of NaCl (5–40 g) and at an even higher range of MO concentrations (300–400 mg/L) (Figure 3G,H).
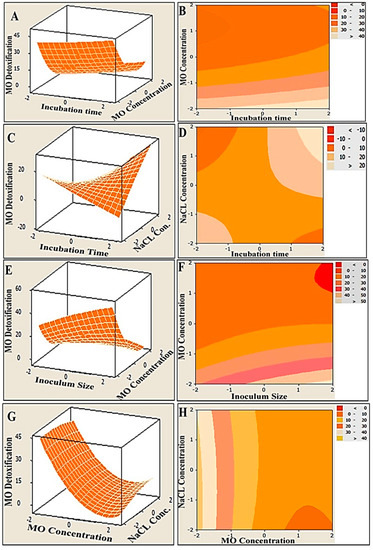
Figure 3.
Three-dimensional surface and two-dimensional contour plots of MO degradation by immobilized consortium, showing the interactive impact of two variables at constant zero levels of the other variables. Both plots were created using MINITAB-14 software. (A,B) are the effect of incubation time and MO concentration on MO detoxification, (C,D) show obvious increase in MO detoxification with a mutual increase in both incubation time and NaCl concentration, (E,F) show antagonistic relationship between inoculum size and MO concentration, (G,H) show the maximum degradation of MO at a wide range of concentrations of NaCl and high range of MO concentrations.
To predict the maximum MO degradation under the examined conditions, the reduced regression model was solved using the optimizer tool in MINITAB 14.0, which calculates individual desirability using a desirability function. Such functions define the sufficient combination of variables to gain the maximum response. The result ranges from zero (less than the accepted limit) to one (the accepted limit) [56]. Therefore, the predicted optimal levels of the process variables were as follows: incubation time—5 days; inoculum size—20%; MO concentration—200 mg/L; and NaCl concentration—40 g/L.
3.5. Analysis of Degradation By-Products
3.5.1. UV−Visible Spectrophotometric Analysis
As demonstrated in Figure 4A, the changes in the UV–vis spectra (from 200 to 800 nm) of MO were observed upon consortium treatment. The spectrum profile of MO (control) showed the main absorption peak to be at 465 nm, due to the azo bond, which was almost completely vanished in the supernatant of the treated solution. The dissipation of the peak at 465 nm indicates that these consortia were successful in removing MO. Thus, the decolorization of MO by these consortia may be attributed largely to biodegradation. Our results are in agreement with those reported by Akansha et al. [76] and Haque et al. [49].
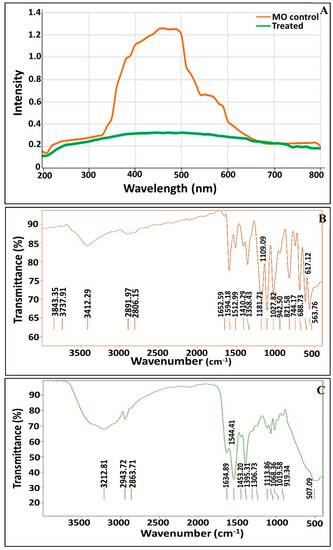
Figure 4.
UV-Vis spectroscopy profile of MO before and after treatment by immobilized consortium (A); FTIR spectra of MO before degradation (B) and after degradation by immobilized consortium (C).
3.5.2. FTIR Analysis
In order to affirm the complete decolorization and demethylation of anionic MO dye, a comparative investigation of the FTIR spectrum of the supernatant in the presence of dye was conducted before and after the degradation process (Figure 4B,C). Generally, the spectrum of decolorized MO after the completion of the decolorization process displayed significant phenomena such as the disappearance of some characteristic peaks as compared to the control spectrum, the appearance of new bands, and also shifts in existing bands. Figure 4B illustrates the different absorbance peaks assigned to the MO dye molecules, including a peak at 2892 cm−1 for asymmetric CH3 stretching vibrations, peaks at 1512 and 1410 cm−1 for C=C-H in the bend of the C-H plane. Moreover, absorbance peaks were recorded at 1109, 1027, 942 and 821 cm−1 for ring vibrations, and a peak was found at 744 cm−1 for the disubstituted benzene ring. Remarkably, their presence denoted the aromatic nature of the dye. The peak at 1652 cm−1 for the –N=N- stretch and the peaks at 1358 and 1181 cm−1 for -C-N revealed the azo nature of the dye. The peaks at 688, 617 and 563 cm−1 for the -C-S- stretching vibrations and the S=O stretching vibrations were attributed to the sulfonic nature of the dye [77,78]. The FTIR spectra of MO after the treatment process was found to display a peak at 3212 cm−1 for the N-H bend and a peak at 2943 cm−1 for the asymmetric -CH3 stretching vibrations (Figure 4C). Notably, the absence of peaks at 1594, 1512, 1410, 942, 821, 744, 688 and 617 cm−1 reflected the breakdown of aromatic C=C. Additionally, the absence of a peak at 1652 implied the cleavage of the azo bond. Our results are consistent with those reported in [49,76]. However, the appearance of a new band at 1247 cm−1 implied the generation of aromatic amines with -C-N vibrations, as highlighted by Masarbo et al. [79] and Baena-Baldiris et al. [80].
3.6. Enzymes Activity Assays
A huge number of microbial enzymes have been involved in the decolorization and biodegradation of synthetic dyes. These bioremediation-related microbial enzymes include laccases, oxido-reductases and hydrolases. In the current study, eight biodegradative enzymes produced by the bacterial consortium were assayed in relation to the decolorization of MO as a model of anionic azo dyes. The optimum decolorization time was observed after 96 h of incubation with MO. Bacterial cell lysates exhibited more potent intracellular activities than extracellular supernatant for all tested enzymes. The decolorization of azo dyes such as MO mainly occurred as a result of the biotransformation process, which was mediated by reductive and oxidative enzymes [81]. The results for all tested enzymes (nitrate reductase, azoreductase, DCIP reductase, NADH reductase, lignin peroxidase (LiP), manganese peroxidase (MnP), laccase and tyrosinase) are summarized in Figure 5. The induction of nitrate reductase, azoreductase, DCIP reductase and NADH reductase was estimated to be 21.61 ± 1.96, 23.05 ± 1.74, 306.97 ± 4.78 and 135.985 ± 2.79 IU/min/mg protein, respectively, after incubation periods of 4 days (Figure 5).
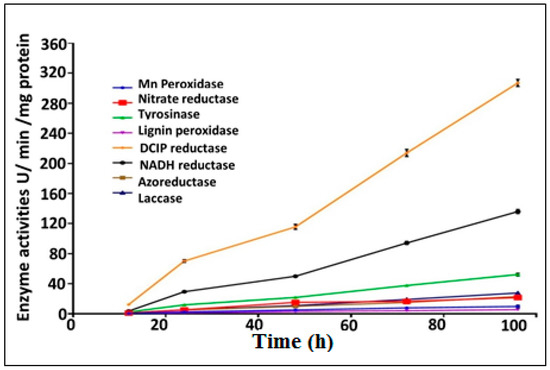
Figure 5.
Quantitative bio-induction of eight different degrading enzymes during the decolorization of MO dye by immobilized consortium.
The expression of LiP and MnP was found to be 5.53 ± 0.64 and 9.76 ± 1.69 IU/min/mg protein, respectively. Moreover, the expression of laccase was found to be increased dramatically with the increasing of the incubation time, and was estimated to be 1.45 ± 0.43, 5.68 ± 0.43, 10.72 ± 0.57, 19.60 ± 0.91 and 27.63 ± 1.24 IU/min/mg protein after incubation times of 0.5, 1.0, 2.0, 3.0 and 4.0 days, respectively. The expression of tyrosinase was also estimated to be maximally induced after 4.0 days of incubation with enzymatic activity of 52.72 ± 1.06 IU/min/mg protein. Further significance is suggested by the fact that the enzymatic activity of all tested enzymes was found to be increased by the used bacterial consortium in a time-dependent manner throughout the MO decolorization process.
3.7. Acute Toxicity Assessment
3.7.1. Phytotoxicity Bioassays with C. vulgaris
In medium derived from CCD containing 200 mg/L of MO (control experiment), chlorophyll-a was reduced before treatment with bacterial consortium. The maximum decrease in chlorophyll-a was 62.16% compared to the control (C. vulgaris 100%); however, the decrease in chlorophyll-a after treatment with consortium was 14.88% as compared to the control (C. vulgaris 100%) (Figure 6A). The decrease in chlorophyll-b was 51.28% before treatment with consortium and 21.49% after treatment with consortium relative to the control (Figure 6B). On the other hand, the carotenoid content was sharply increased before treatment with consortium (218.73%) and sharply decreased after treatment with consortium (22%) (Figure 6C).
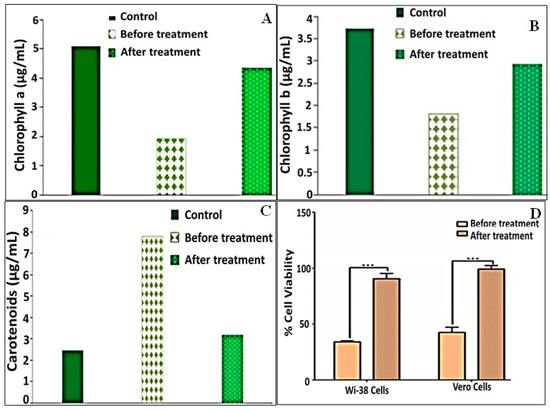
Figure 6.
Phytotoxicity of methyl orange (200 mg/L) and remediated byproducts against C. vulgaris content of (A) chlorophyll-a, (B) chlorophyll-b and (C) carotenoids. (D) Cytotoxicity against normal (human Wi-38 and mammalian Vero) cell lines in terms of cell viability.
3.7.2. Cytotoxicity against Normal Human and Animal Cell Lines
To assess the cytotoxicity of MO and its degradation metabolites on human and animal cells, they were incubated with human normal lung (Wi-38) and monkey kidney (Vero) cells. Then, the viability of these treated cells was determined by means of an MTT assay and by estimation, relative to the untreated healthy cells, based on the formation of insoluble formazan products by active cells. MO dye (200 mg/L) lowered the viability of Wi-38 and Vero cells to 34.10% and 42.57%, respectively. However, upon treatment by immobilized consortium, the remediated solution maintained the viability of both normal cells at 90.63% and 99.23%, respectively (Figure 6D). Accordingly, the results revealed that the treatment with immobilized consortium exhibited most significant potency in terms of reducing the toxicity of MO in both normal cell lines in comparison to dye alone (p < 0.0), which reflected the success of MO degradation using the examined consortium.
3.8. Bioaugmentation of Free and Immobilized Cells in Real Waste Effluents
The performance of immobilized consortium in comparison to free consortium was examined in real effluent samples, collected from discharge lines of industrial and agricultural zones in Alexandria, Egypt, either solely or in synchronization with indigenous microbiota (i.e., augmentation). The chemical, physical and biological criteria of the examined wastewater were tested and their results are tabulated in Table 5. Some general observations were recorded and listed. Commencing with the non-immobilized alginate beads, which were run in parallel as a control to examine the absorption, an uptake percentage ranging from 10.82 to 12.88% was recorded. Furthermore, the biodegradation process was time dependent; specifically, higher biodegradation was implemented gradually as a function of the time, as shown in Figure 7. In this process, the immobilized consortium degraded 75.5 and 61.5 mg/L within 72 h in the augmented system of agricultural and industrial effluent, respectively; however, at 144 h., 155 and 128.5 mg/L were remediated by immobilized consortium in the augmented system of agricultural and industrial effluent, respectively. Meanwhile, more efficient biodegradation of MO was observed in both augmented systems, either free or immobilized, as compared to the non-bioaugmented systems. Increments in MO removal of about 13.53 and 8.64% in the augmented systems of the agricultural and industrial effluents, respectively, were attained with the use of immobilized consortium. However, for the freely-added consortium, 8.59 and 5.43% increments in MO elimination were recorded in the agricultural and industrial effluents’ augmented systems, respectively. Notably, the immobilized consortium exhibited higher biodegradation potential than free consortium, with 77.59 and 62.38% removal in the bioaugmented system of agricultural effluent; for comparison, 69 and 53.74% removal were attained in the non-bioaugmented system. Regarding the industrial effluent, 64.78 and 51.38% removal was obtained with immobilized and free augmented consortium, respectively; in the non-bioaugmented system, the immobilized and free consortium eliminated 59.35 and 37.85% of MO, respectively. Broadly, the MO bioremediation efficiency in agricultural wastewater displayed better results than those recorded for industrial effluent (Figure 7).

Table 5.
Quality analysis of agricultural and industrial effluents.
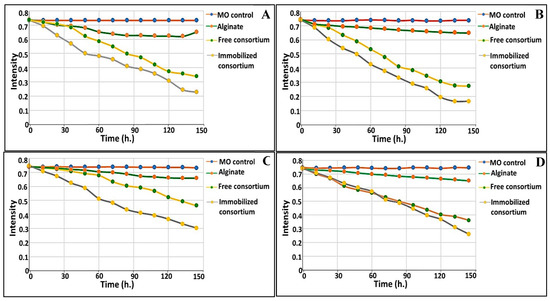
Figure 7.
MO bioremediation from agricultural and industrial effluents using microbial consortium either freely suspended or immobilized in both augmented and non-augmented states: (A) non-augmented state of agricultural effluent; (B) augmented state of agricultural effluent; (C) non-augmented state of industrial effluent; (D) augmented state of industrial effluent.
4. Discussion
The discharge of dye effluents into water bodies is becoming a real threat. During the dying process, about 10–70% unfixed dye is disposed, causing substantial ecological deteriorations such as damaging the photosynthesis process, changing the water quality and bringing about adverse aesthetic influences. These issues are further compounded by the gradual accumulation of dye in the tissues of aquatic creatures, which subsequently causes serious issues for human health through the food chain [82]. In the present study, the immobilized microbial consortium was employed to bioremediate MO in a real simulation model. The process began with the accurate selection of microbes that possess biodegradative capabilities. The selected strains Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus and Staphylococcus xylosus exhibited promising biodegradation traits within time range of 20–28 h., either solely or combined in a consortium, achieving complete degradation in 18 h with the use of NB medium. Interestingly, the presence of compatibility and mutually beneficial corporation in the mixed co-culture system accelerates the bioremediation process, as mentioned by Weihua Tang et al. [83].
Notably, numerous dyestuff degradation studies focused on the testing of degradation and decolorization using highly nutritive microbiological ingredients [49,69,76,84]. In fact, peptone and yeast extract were widely used for this purpose as carbon sources, nitrogen sources or both. Both trigger the generation of NADH, which acts as redox mediator during the metabolization process, and thus, aids in energy production, survival and multiplication of microorganisms. Nonetheless, their presence may cause only decolorization with incomplete decomposition. In fact, the microorganisms first utilize easily metabolized rich nutrients, rather than assimilating recalcitrant compounds as dye, in order to survive and proliferate [49]. Additionally, the effluents are not always full of easily metabolized nutrients such as complex microbiological media. However, effluents may be poorly nutritive and supplemented with various impurities such as surfactants, pesticides and biocides, which are relatively often accompanied by high salinity [79]. This directed us to examine the assimilation behavior of consortium toward MO in MSM that lacked complex nutrients, in a way that mimicked what happens in reality. In this case, the bacterial consortium was imposed to decompose MO into simpler structures for growth and multiplication. Remarkably, the combinatory state of microbial species could permit full degradation. In fact, individual bacterial species could utilize MO incompletely and its intermediate byproduct could be consumed by another bacterial species in such mixed-culture consortiums; ultimately, full decomposition of the dye was achieved without the accumulation of hazard residues [49]. Different studies supported our finding regarding MO assimilation [85]. However, the utilization of freely suspended biomass for the purpose of degradation of toxic materials, especially in situ, would restrict its efficacy. This fact could be attributed to the low mechanical strength of microbial cells at this state, as well as their susceptibility to harsh environmental conditions and toxic byproducts, which would alternate their metabolic activities. To circumvent such limitations, the immobilization approach was employed. Such an approach provides stability and protection for microbial cells, and ensures viability over a prolonged period. Importantly, it also permits in situ applications and recovery/reuse of microbial cells [24,86]. Thus, the entrapment of the consortium in ionotropic hydrogels such as alginates would lead to the promotion of the bioremediation process. The natural origin of alginate, its biocompatibility, low toxicity, easy gelling operation and inexpensiveness encourage its applications in biotechnological, medical and environmental usages. Furthermore, the research presented in [87] reinforces this point of view. In this study, it was shown that the entrapment of cells in a suitable matrix would increase the retention time of the cells in wastewater; consequently, the treatment process would be simplified.
It is plausible to study the effects of different parameters on MO’s degradation performance. Several levels of various parameters were tested simultaneously in a systematic, cost-effective, time-saving approach, which was attained via CCD. In fact, the conventional single-dimensional “one variable at time (OVAT)” strategy is a time consuming, laborious practice due to large number of required experiments and often fails to explain the interaction effects among the multivariable parameters [56,75]. Through the statistical design of the experiment (CCD), different levels of dye concentrations, NaCl concentrations, and inoculated sizes were studied in 31 trials within a time range of 1–5 days. The statistical model revealed that a 20% inoculation of the biomass with the immobilized consortium effectively eliminated 200 mg/L of MO in the presence of 40 g/L of NaCl within 5 days of incubation. Interestingly, our immobilized consortium was characterized by its ability to remove MO under high salinity. On the other hand, Weihua Tang et al. highlighted the importance of the salt tolerance property in the dye decolorization process, especially from wastewater [83]. He and coworkers referred to the removal efficiency of the fungus-algae consortium to decolorize 58.24% of dye from simulated wastewater in the presence of 20 g/L of NaCl. In addition, Akansha et al. recorded the decline in MO decolorization by Bacillus stratosphericus SCA1007 in the salinity range of 4–7%, attributing this case to cell plasmolysis [76]. Moreover, Haque et al. pointed out the ability of biofilm consortium to detoxify 68.34% of MO in a highly nutritive peptone medium yeast extract (YEP) containing 10% NaCl [49]. Thus, it could be deduced that our consortium is promising in terms salt-tolerant azo degradation and that it has significant potential to be applied in high salinity environments.
Remarkably, the existence of adequate inoculum sizes can enhance dye-treatment operations. As noted by Sari and Simarani [66], the performance of the L. fusiformis strain W1B6 in azo dye decolorization increased with the elevation of the inoculum size from 5% to 10%. Furthermore, Haque et al. stated that the azo dye molecules obstructed the active azoreductase sites of lower biomass ratios of inoculated consortium, reflecting the significance of microbial biomass availability in terms of improving dye bioremediation via the donation of more azoreductase active sites [49]. In the same context, the decolorization performance of azo dye was inversely proportional to the initial dye concentration [83]. At higher dye concentrations, the decolorization performance declined due to the saturation of degradative enzymes’ active sites with dye molecules, the inactivation of the transport system and the overcrowding of dye byproducts, which ultimately led to the inhibition of microbial metabolism. In agreement with our results, Haque et al. documented that the MO removal efficiency and also its growth rate were significantly diminished by increasing the MO concentration from 100 to 400 mg/L [49]. The exact observation was recorded elsewhere [66,88]. On the other hand, the biodegradation is directly proportional to the incubation time. In fact, MO removal was ameliorated within the examined time course, which was in line with the results obtained by Haque et al. [49], who found that MO removal increased from 59.34 to 79.21% with the increasing of the incubation time from 36 to 48 h. Broadly, dyeing wastewater always contains high salt and dye concentrations of 3–10% and 10–200 mg/L, respectively [49,89]. Therefore, for environmental applications, the ability of microorganisms to withstand high concentrations of dye and salinity is an indispensable property in terms of guaranteeing successful bioremediation.
Based on results of the enzyme assay regarding the synchronization of the UV-Vis spectra and the FTIR of the biodegraded solution, the mechanism of the degradation process could be speculated upon. The absence of both orange color from remediated solution and also the absorption maxima at 465 nm indicated complete cleavage of the N=N bond, as highlighted by Costa et al. [12]. Such results were affirmed by FTIR via the absence of the azo bond at 1652 cm−1, which signified the initiation of the biodegradation process by the action of azoreductase, which mediated the reductive disputing of the N=N bond. However, the disappearance of some peaks unveiled the breakdown of the benzene ring by the dint of DCIP reductase and NADH reductase, which were considered to be key enzymes for biodegradation in the current study. Nonetheless, other oxidative enzymes, such as LiP, MnP and lacasse, played important roles in the accomplishment of the MO bioremediation process. They mediated the oxidation of aromatic and inorganic compounds and their mineralization into simpler structures such as 1, 4-benzene diamine and N, N-dimethyl, as inferred from the appearance of new peaks in FTIR. Interestingly, several mechanisms conducted MO degradation enzymatically, either through non-specific free radical mechanisms, as was the case for laccases, or through free-radical production that oxidizes azo dyes, as was observed for LiP, MnP and peroxidases. Furthermore, the non-enzymatic route of degradation involves low molecular weight redox mediators and biogenic reductants that mediate chemical reduction [90]. Our results agreed with those reported by Haque et al. [49] who documented the induction of azoreductase, NADH-DCIP reductase and laccase by their biofilm consortia for MO decolorization in a similar time frame. Additionally, Ayed et al. [91] found that the microbial consortium of Sphingomonas paucimobilis, Bacillus cereusATCC14579 and Bacillus cereusATCC11778 decolorized MO (>84%) within 48 h under optimal conditions. They revealed a significant increase in the expression of azoreductase, LiP and laccase. On the other hand, Sheela and Sadasivam noticed an increase in laccase level expression caused by Bacillus cereus SKB12 during the biodegradation of textile dyes, while the level of expression of the NADH-DCIP reductase and MnP enzymes was found to be poor [92]. Furthermore, Thanavel et al. revealed that textile dye degradation was successfully implemented by a strain of Aeromonas hydrophila SK16, which showed a significant expression of degradative enzymes such as tyrosinase, LiP, riboflavin reductase, azoreductase and laccase with the increasing of their levels during the degradation process [93]. However, the decolorization and cleavage of azo dye did not ensure the safety and nontoxicity of metabolic end products; thus, a toxicity study should accompany these results. Generally, phytotoxicity was examined through the seed germination of plants such as Vigna radiata [76], Triticum aestivum and Sorghum vulgare [49,79], and Phaseolus mungo and Sorghum vulgare [94]. Herein, the phytotoxicity assay was utilized to assess the impact of the original dyes’ and metabolites’ outcomes on the growth and photosynthesis performance of C. vulgaris. As it is a photosynthetic unicellular microalgae, it is characterized by its rapid growth and its sensitivity to toxic compounds; hence, it could provide a quick insinuation of the effect of bioremediated wastewater on photosynthesis and could possibly be applied in the subsequent irrigation of plants [95]. The phytotoxicity results revealed the toxic effect of MO (200 mg/L) and the inhibition of the photosynthesis process as compared to the control, which were deduced from the diminishing of chlorophyll-a and chlorophyll-b and the increase in carotenoid content. However, the impact of bioremediated water on the algae photosynthesis process was much lower than that observed in the original MO (200 mg/L) due to the lowering of the chlorophyll-a and chlorophyll-b contents to 14.88 and 21.49% and the lowering of the carotenoid content to 22% The discrepancy in the behaviors of the chlorophyll and the carotenoids in response to either MO or its metabolic byproducts could be attributed to the antioxidant activity displayed by carotenoids under stress conditions in order to tolerate and adapt to hazardous compounds [96,97].This is consistent with the findings of Liao et al. [98], who documented that the EC50 of azo dye, Reactive Black B (RBB), was 48 mg/L against the growth of the freshwater algal C. vulgaris, assigned this result to the malfunctioning of chloroplasts. On the other hand, the cytotoxicity of MO and its bioremediated byproduct against the human cell lines Wi-38 and Vero cells ensured the safety and reduced toxicity of MO-bioremediated water via the maintenance of the viability of both normal cells at 90.63% and 99.23%, respectively. Broadly, our toxicity results were promising, suggesting the safe employment of MO-bioremediated wastewater, through the use of the immobilized consortium under study, for ferti-irrigation purposes to overcome water crisis issues.
To emphasize the effectiveness of the consortium under study and compare its efficiency in different effluents, it was employed in the bioremediation of real agricultural and industrial wastewater supplemented artificially with 200 mg/L of MO, using augmented and non-augmented systems, in both free and immobilized states. As noticed, the water quality parameters were analyzed and indicated some common differences between both effluents, which would aid in explaining and perceiving the overall performance of the bioremediation process. The agricultural effluents yielded higher values in terms of total count, biological oxygen demand (B.O.D), sulfate, carbonate, calcium, total nitrogen and phosphate contents as compared to the industrial effluents. That could be ascribed to the presence of fertilizers, nitrogenous/phosphorous nutrients, soil stabilizers, herbicides and pesticide-containing residues. On the other hand, the industrial effluents yielded higher amounts of oil, phenolic compounds, chemical oxygen demand (C.O.D) and several heavy metals. By applying our microbial consortium either as free or immobilized in these real effluents, a difference in bioremediation performance was observed according to the method of application, namely the augmented and the non-augmented system, and also the nature of effluents. As stated by Qu et al. [99] and Zommere et al. [86], the success or failure of a bioaugmentation process depends substantially on the activity and persistence of the introduced consortium in addition to its competitive interaction with indigenous microbiota in the contaminated area. Despite simplicity and success at the laboratory scale, several studies reported failures in the bioaugmentation operation owing to the previously mentioned reasons [86,100]. In the current research, our mixed-culture consortium achieved the biodegradation and noticeable removal of MO in augmented systems of both examined effluents, reflecting its corporative interaction with native indigenous microorganisms without any competitive inhibition despite the variations in the natures and contents of the two different wastewaters. However, the observed degradative capability of the consortium in the non-augmented systems of both wastewaters revealed its ability to withstand harsh circumstances (inhibitory substances such as heavy metals, phenols, oil, nitrogen/phosphorous content, osmotic stress, etc.). Furthermore, the higher removal capacity of the immobilized state compared to the freely suspended state could be attributed to the presence of microbial cells in the highly diffusive protective matrix, which provided better contact with MO due to its absorption capability, without direct contact with other toxic pollutants and in a protective manner. This result concurred with that given in [24]. Meanwhile, the bioremoval potency of the immobilized consortium in agricultural wastewater was higher than that observed in industrial effluent. This could be assigned to the presence of more stressor substances such as heavy metals, oils and phenols. Such contents could have a negative influence on the degradative performance of the consortium. In addition, the presence of a higher microbial count in agricultural effluent, as a result of the higher contents of nutritive substances, could assist in achieving improved degradation of MO. Generally, the mutual and synergistic interaction among the introduced consortium and indigenous microorganisms enhanced the decolorization and degradation of MO in this bioaugmentation operation. It is plausible to mention that the prolongation of the incubation time would result in a progressive enhancement in the bioremediation process, which is consistent with the findings of Jublee Jasmine and Suparna Mukherji [101] and Poi et al. [100]. The promising results of the bioaugmentation operation with our characteristic immobilized consortium encourage us to study the dynamics of bioaugmentation processes that are specifically designed for a wider variety of bioaugmented systems, in order to meet industrial requirements and comply with environmental legislation.
5. Conclusions
The current study aimed to construct an efficient bacterial consortium of Raoultella planticola, Ochrobactrum thiophenivorans, Bacillus flexus and Staphylococcus xylosus to degrade MO. The consortium exhibited considerable ability to assimilate MO as a carbon source in mineral salt media with stronger efficiency than bacteria alone. The degradation behavior of the immobilized consortium using CCD indicated its efficacy in degrading 200 mg/L of MO under high salinity (40 g/L of NaCl) within 5 days of incubation. The degradation potency was emphasized by the expression of degradative enzymes such as azoreductase, DCIP reductase, NADH reductase, laccase, LiP, MnP and tyrosinase. The UV−Visible spectrophotometric analysis and FTIR examination of degradation outcomes revealed the breakdown of the azo bond and the aromatic groups. The toxicity study of such metabolic byproducts was assessed by determining their influence on the photosynthetic performance of C. vulgaris and the viability percentage of human cell lines (Wi-38 and Vero cells). Moreover, the immobilized consortium, in comparison to freely suspended culture, was employed in the bioremediation of MO, from artificially contaminated agricultural and industrial effluents, in augmented and non-augmented systems. The results highlighted the mutually corporative interaction between immobilized consortium and indigenous microorganisms in the biodegradation of MO in both examined effluents, reflecting the adaptation capacity of our consortium to effluents of various kinds and from various sources.
Author Contributions
A.I.: conceptualization, methodology, validation, formal analysis, investigation, software, resources, writing—original draft preparation, editing, supervision and visualization; E.M.E.-F.: software, methodology, validation, formal analysis, data curation, writing—review and editing, and visualization; M.M.A.-S.: software, methodology, validation, formal analysis, data curation, writing—review and editing, and visualization; M.F.E.: software, methodology, validation, formal analysis, data curation, writing—review and editing, and visualization; M.E.: conceptualization, methodology, validation, formal analysis, investigation, resources, writing—original draft preparation, editing, supervision and visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
References
- Asad, S.; Amoozegar, M.A.; Pourbabaee, A.; Sarbolouki, M.; Dastgheib, S. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour. Technol. 2007, 98, 2082–2088. [Google Scholar] [CrossRef]
- El-Sayed, G.O.; Hazaa, M.M.; El-Komy, A.M. Biotreatment of water polluted with methyl orange dye by using different forms of yeast. J. Basic Environ. Sci. 2018, 5, 217–221. [Google Scholar]
- Mishra, S.; Nayak, J.K.; Maiti, A. Bacteria-mediated bio-degradation of reactive azo dyes coupled with bio-energy generation from model wastewater. Clean Technol. Environ. Policy 2020, 22, 651–667. [Google Scholar] [CrossRef]
- Sinha, S.; Chattopadhyay, P.; Pan, I.; Chatterjee, S.; Chanda, P.; Bandyopadhyay, D.; Das, K.; Sen, S. Microbial transformation of xenobiotics for environmental bioremediation. Afr. J. Biotechnol. 2011, 8, 6016–6027. [Google Scholar] [CrossRef]
- Fernandez, M.; Paisio, C.E.; Perotti, R.; Pereira, P.P.; Agostini, E.; González, P.S. Laboratory and field microcosms as useful experimental systems to study the bioaugmentation treatment of tannery effluents. J. Environ. Manag. 2019, 234, 503–511. [Google Scholar] [CrossRef]
- Gaur, N.; Narasimhulu, K.; PydiSetty, Y. Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J. Clean. Prod. 2018, 198, 1602–1631. [Google Scholar] [CrossRef]
- Sayed, K.; Baloo, L.; Sharma, N. Bioremediation of Total Petroleum Hydrocarbons (TPH) by Bioaugmentation and Biostimulation in Water with Floating Oil Spill Containment Booms as Bioreactor Basin. Int. J. Environ. Res. Public Health 2021, 18, 2226. [Google Scholar] [CrossRef] [PubMed]
- Magan, N.; Fragoeiro, S.; Bastos, A.C. Environmental Factors and Bioremediation of Xenobiotics Using White Rot Fungi. Mycobiology 2010, 38, 238–248. [Google Scholar] [CrossRef]
- Herrero, M.; Stuckey, D. Bioaugmentation and its application in wastewater treatment: A review. Chemosphere 2015, 140, 119–128. [Google Scholar] [CrossRef]
- Raper, E.; Stephenson, T.; Anderson, D.; Fisher, R.; Soares, A. Industrial wastewater treatment through bioaugmentation. Process. Saf. Environ. Prot. 2018, 118, 178–187. [Google Scholar] [CrossRef]
- Basturk, I.; Varank, G.; Murat-Hocaoglu, S.; Yazici-Guvenc, S.; Oktem-Olgun, E.E.; Canli, O. Characterization and Treatment of Medical Laboratory Wastewater by Ozonation: Optimization of Toxicity Removal by Central Composite Design. Ozone Sci. Eng. 2021, 43, 213–227. [Google Scholar] [CrossRef]
- Costa, A.F.S.; Albuquerque, C.D.C.; Salgueiro, A.A.; Sarubbo, L.A. Color removal from industrial dyeing and laundry effluent by microbial consortium and coagulant agents. Process. Saf. Environ. Prot. 2018, 118, 203–210. [Google Scholar] [CrossRef]
- Asfaram, A.; Ghaedi, M.; Dashtian, K.; Ghezelbash, G.R. Preparation and Characterization of Mn0.4Zn0.6Fe2O4 Nanoparticles Supported on Dead Cells of Yarrowia lipolytica as a Novel and Efficient Adsorbent/Biosorbent Composite for the Removal of Azo Food Dyes: Central Composite Design Optimization Study. ACS Sustain. Chem. Eng. 2018, 6, 4549–4563. [Google Scholar] [CrossRef]
- Machrouhi, A.; Alilou, H.; Farnane, M.; El Hamidi, S.; Sadiq, M.; Abdennouri, M.; Tounsadi, H.; Barka, N. Statistical optimization of activated carbon from Thapsia transtagana stems and dyes removal efficiency using central composite design. J. Sci. Adv. Mater. Devices 2019, 4, 544–553. [Google Scholar] [CrossRef]
- Azargohar, R.; Dalai, A. Production of activated carbon from Luscar char: Experimental and modeling studies. Microporous Mesoporous Mater. 2005, 85, 219–225. [Google Scholar] [CrossRef]
- Ghelich, R.; Jahannama, M.R.; Abdizadeh, H.; Torknik, F.S.; Vaezi, M.R. Central composite design (CCD)-Response surface methodology (RSM) of effective electrospinning parameters on PVP-B-Hf hybrid nanofibrous composites for synthesis of HfB2-based composite nanofibers. Compos. Part B Eng. 2019, 166, 527–541. [Google Scholar] [CrossRef]
- Ahmadi, M.; Jorfi, S.; Kujlu, R.; Ghafari, S.; Soltani, R.D.C.; Haghighifard, N.J. A novel salt-tolerant bacterial consortium for biodegradation of saline and recalcitrant petrochemical wastewater. J. Environ. Manag. 2017, 191, 198–208. [Google Scholar] [CrossRef]
- Phan, D.C.; Goodwin, D.G.; Frank, B.P.; Bouwer, E.J.; Fairbrother, D.H. Biodegradability of carbon nanotube/polymer nanocomposites under aerobic mixed culture conditions. Sci. Total Environ. 2018, 639, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Gangola, S.; Bhandari, G.; Zhang, W.; Maithani, D.; Mishra, S.; Chen, S. New insights into the degradation of synthetic pollutants in contaminated environments. Chemosphere 2021, 268, 128827. [Google Scholar] [CrossRef]
- Sarkar, P.; Dey, A. 4-Nitrophenol biodegradation by an isolated and characterized microbial consortium and statistical optimization of physicochemical parameters by Taguchi Methodology. J. Environ. Chem. Eng. 2020, 8, 104347. [Google Scholar] [CrossRef]
- Muñoz, R.; Alvarez, M.T.; Muñoz, A.; Terrazas, E.; Guieysse, B.; Mattiasson, B. Sequential removal of heavy metals ions and organic pollutants using an algal-bacterial consortium. Chemosphere 2006, 63, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Abbes, C.; Ben Mouhoub, R.; Ben Hassine, S.; Landoulsi, A. Enhancement of mixture pollutant biodegradation efficiency using a bacterial consortium under static magnetic field. PLoS ONE 2019, 14, e0208431. [Google Scholar] [CrossRef]
- Abdel-Razek, M.A.; Abozeid, A.M.; Eltholth, M.M.; Abouelenien, F.A.; El-Midany, S.A.; Moustafa, N.Y.; Mohamed, R.A. others Bioremediation of a pesticide and selected heavy metals in wastewater from various sources using a consortium of microalgae and cyanobacteria. Slov. Vet. 2019, 56, 61–73. [Google Scholar]
- Allam, N. Bioremediation Efficiency of Heavy Metals and Azo Dyes by Individual or Consortium Bacterial Species Either as Free orImmobilized Cells: A Comparative Study. Egypt. J. Bot. 2017, 57, 555–564. [Google Scholar] [CrossRef]
- Pizarro-Tobias, P.; Niqui, J.L.; Roca, A.; Solano, J.; Fernández, M.; Bastida, F.; García, C.; Ramos, J.L. Field trial on removal of petroleum-hydrocarbon pollutants using a microbial consortium for bioremediation and rhizoremediation. Environ. Microbiol. Rep. 2014, 7, 85–94. [Google Scholar] [CrossRef]
- Nozari, M.; Samaei, M.R.; Dehghani, M.; Ebrahimi, A.A. Bioremediation of Alkane Hydrocarbons Using Bacterial Consortium From Soil. Health Scope 2018, 7, e12524. [Google Scholar] [CrossRef]
- Hameed, B.B.; Ismail, Z.Z. Decolorization, biodegradation and detoxification of reactive red azo dye using non-adapted immobilized mixed cells. Biochem. Eng. J. 2018, 137, 71–77. [Google Scholar] [CrossRef]
- Ismail, Z.Z.; Khudhair, H.A. Recycling of immobilized cells for aerobic biodegradation of phenol in a fluidized bed bioreactor. Syst. Cybern. Inform. 2015, 13, 81–86. [Google Scholar]
- Maniyam, M.N.; Yaacob, N.S.; Azman, H.H.; Ab Ghaffar, N.A.; Abdullah, H. Immobilized cells of Rhodococcus strain UCC 0004 as source of green biocatalyst for decolourization and biodegradation of methyl orange. Biocatal. Agric. Biotechnol. 2018, 16, 569–578. [Google Scholar] [CrossRef]
- Dogan, N.M.; Sensoy, T.; Doganli, G.A.; Bozbeyoglu, N.N.; Arar, D.; Akdogan, H.A.; Canpolat, M. Immobilization of Lycinibacillus fusiformis B26 cells in different matrices for use in turquoise blue HFG decolourization. Arch. Environ. Prot. 2016, 42, 92–99. [Google Scholar] [CrossRef][Green Version]
- García, J.M.; Garcia-Martinez, T.; Mauricio, J.C.G.; Moreno, J. Yeast Immobilization Systems for Alcoholic Wine Fermentations: Actual Trends and Future Perspectives. Front. Microbiol. 2018, 9, 241. [Google Scholar] [CrossRef]
- Akpor, O.B. Research Article Dye Decolouration by Immobilized and Free Bacterial Cells at Different Glucose Concentration. 2018. Res. J. Environ. Sci. 2018, 12, 33–40. [Google Scholar] [CrossRef][Green Version]
- Susilowati, P.E.; A Rajiani, N.; Hermawan, H.; Zaeni, A.; Sudiana, I.N. The use Immobilized Bacteria-Alginate-Chitin for crack remediation. IOP Conf. Ser. Earth Environ. Sci. 2019, 299, 012010. [Google Scholar] [CrossRef]
- Jeon, Y.; Bissessur, A.; Singh, P. Novel immobilization techniques of Acinetobacter (V2) and Paenibacillus (D9) bacterial strains for waste oil degradation. Biotechnol. Biotechnol. Equip. 2019, 33, 911–920. [Google Scholar] [CrossRef]
- Staroń, P.; Chwastowski, J. Raphia-Microorganism Composite Biosorbent for Lead Ion Removal from Aqueous Solutions. Materials 2021, 14, 7482. [Google Scholar] [CrossRef] [PubMed]
- Vaiano, V.; Sacco, O.; Libralato, G.; Lofrano, G.; Siciliano, A.; Carraturo, F.; Guida, M.; Carotenuto, M. Degradation of anionic azo dyes in aqueous solution using a continuous flow photocatalytic packed-bed reactor: Influence of water matrix and toxicity evaluation. J. Environ. Chem. Eng. 2020, 8, 104549. [Google Scholar] [CrossRef]
- Mapukata, S.; Kobayashi, N.; Kimura, M.; Nyokong, T. Asymmetrical and symmetrical zinc phthalocyanine-cobalt ferrite conjugates embedded in electrospun fibers for dual photocatalytic degradation of azo dyes: Methyl Orange and Orange G. J. Photochem. Photobiol. A Chem. 2019, 379, 112–122. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Liu, J.; Wang, Y.-X.; He, C.-S.; Zhang, L.-S.; Mu, Y.; Li, W.-H. Bioelectrochemical decolorization of a reactive diazo dye: Kinetics, optimization with a response surface methodology, and proposed degradation pathway. Bioelectrochemistry 2019, 128, 9–16. [Google Scholar] [CrossRef]
- Kolekar, Y.M.; Nemade, H.N.; Markad, V.L.; Adav, S.S.; Patole, M.S.; Kodam, K.M. Decolorization and biodegradation of azo dye, reactive blue 59 by aerobic granules. Bioresour. Technol. 2012, 104, 818–822. [Google Scholar] [CrossRef]
- Al-Baldawi, I.A.; Abdullah, S.R.S.; Almansoory, A.F.; Ismail, N.I.; Abu Hasan, H.; Anuar, N. Role of Salvinia molesta in biodecolorization of methyl orange dye from water. Sci. Rep. 2020, 10, 13980. [Google Scholar] [CrossRef]
- Croce, R.; Cinà, F.; Lombardo, A.; Crispeyn, G.; Cappelli, C.I.; Vian, M.; Maiorana, S.; Benfenati, E.; Baderna, D. Aquatic toxicity of several textile dye formulations: Acute and chronic assays with Daphnia magna and Raphidocelis subcapitata. Ecotoxicol. Environ. Saf. 2017, 144, 79–87. [Google Scholar] [CrossRef]
- Manzar, M.S.; Waheed, A.; Qazi, I.W.; Blaisi, N.I.; Ullah, N. Synthesis of a novel epibromohydrin modified crosslinked polyamine resin for highly efficient removal of methyl orange and eriochrome black T. J. Taiwan Inst. Chem. Eng. 2019, 97, 424–432. [Google Scholar] [CrossRef]
- Prabhakar, Y.; Gupta, A.; Kaushik, A. Enhanced decolorization of reactive violet dye 1 by halo-alkaliphilic Nesterenkonia strain: Process optimization, short acclimatization and reusability analysis in batch cycles. Process. Saf. Environ. Prot. 2019, 131, 116–126. [Google Scholar] [CrossRef]
- Mirbolooki, H.; Amirnezhad, R.; Pendashteh, A.R. Treatment of high saline textile wastewater by activated sludge microorganisms. J. Appl. Res. Technol. 2017, 15, 167–172. [Google Scholar] [CrossRef]
- Benkhaya, S.; M’Rabet, S.; El Harfi, A. Classifications, properties, recent synthesis and applications of azo dyes. Heliyon 2020, 6, e03271. [Google Scholar] [CrossRef] [PubMed]
- Masarbo, R.S. Decolorization of Sulfonated Azo Dye Methyl Orange by Bacterial Species. Int. J. Curr. Res. Acad. Rev. 2016, 4, 172–178. [Google Scholar] [CrossRef][Green Version]
- Chen, S.; Zhang, J.; Zhang, C.; Yue, Q.; Li, Y.; Li, C. Equilibrium and kinetic studies of methyl orange and methyl violet adsorption on activated carbon derived from Phragmites australis. Desalination 2010, 252, 149–156. [Google Scholar] [CrossRef]
- Zabłocka-Godlewska, E.; Przystaś, W.; Grabińska-Sota, E. Dye Decolourisation Using Two Klebsiella Strains. Water Air Soil Pollut. 2015, 226, 2249. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Haque, A.; Mosharaf, K.; Marcus, P.K. Decolorization, degradation and detoxification of carcinogenic sulfonated azo dye methyl orange by newly developed biofilm consortia. Saudi J. Biol. Sci. 2021, 28, 793–804. [Google Scholar] [CrossRef]
- Rahman, M.M.; Khan, A.A.; Ali, M.E. others Screening of antagonistic bacteria against bacterial wilt of tomato, eggplant and potato in Bangladesh. Int. J. Agric. Biol. 2013, 15, 973–975. [Google Scholar]
- Eltarahony, M.; Zaki, S.; Kheiralla, Z.; Abd-El-Haleem, D. NAP enzyme recruitment in simultaneous bioremediation and nanoparticles synthesis. Biotechnol. Rep. 2018, 18, e00257. [Google Scholar] [CrossRef] [PubMed]
- Devi, P.; Wahidullah, S.; Sheikh, F.; Pereira, R.; Narkhede, N.; Amonkar, D.; Tilvi, S.; Meena, R.M. Biotransformation and Detoxification of Xylidine Orange Dye Using Immobilized Cells of Marine-Derived Lysinibacillus sphaericus D3. Mar. Drugs 2017, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.C.; López, O.V.; Ciolino, A.E.; Morata, V.I.; Villar, M.A.; Ninago, M.D. Immobilization of enological pectinase in calcium alginate hydrogels: A potential biocatalyst for winemaking. Biocatal. Agric. Biotechnol. 2019, 18, 101091. [Google Scholar] [CrossRef]
- Fang, L.; Xu, Y.; Xu, L.; Shi, T.; Ma, X.; Wu, X.; Li, Q.X.; Hua, R. Enhanced biodegradation of organophosphorus insecticides in industrial wastewater via immobilized Cupriavidus nantongensis X1T. Sci. Total Environ. 2020, 755, 142505. [Google Scholar] [CrossRef]
- Ruan, B.; Wu, P.; Chen, M.; Lai, X.; Chen, L.; Yu, L.; Gong, B.; Kang, C.; Dang, Z.; Shi, Z.; et al. Immobilization of Sphingomonas sp. GY2B in polyvinyl alcohol–alginate–kaolin beads for efficient degradation of phenol against unfavorable environmental factors. Ecotoxicol. Environ. Saf. 2018, 162, 103–111. [Google Scholar] [CrossRef]
- Abu-Elreesh, G.; El-Shall, H.; Eltarahony, M.; Abdelhaleem, D. Conversion of Cost-Effective agricultural wastes into valued oil using the fungus Curvularia Sp: Isolation, Optimization and Statistical analysis. Biosci. Res. 2019, 16, 3006–3024. [Google Scholar]
- Chen, K.-C.; Wu, J.-Y.; Liou, D.-J.; Hwang, S.-C.J. Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 2003, 101, 57–68. [Google Scholar] [CrossRef]
- Eltarahony, M.; Ibrahim, A.; El-Shall, H.; Ibrahim, E.; Althobaiti, F.; Fayad, E. Antibacterial, Antifungal and Antibiofilm Activities of Silver Nanoparticles Supported by Crude Bioactive Metabolites of Bionanofactories Isolated from Lake Mariout. Molecules 2021, 26, 3027. [Google Scholar] [CrossRef] [PubMed]
- Glenn, J.K.; Gold, M.H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 1985, 242, 329–341. [Google Scholar] [CrossRef]
- Redinbaugh, M.G.; Campbell, W. Quaternary structure and composition of squash NADH:nitrate reductase. J. Biol. Chem. 1985, 260, 3380–3385. [Google Scholar] [CrossRef]
- Shanmugam, V.; Kumari, M.; Yadav, K.D. n-Propanol as a substrate for assaying the ligninperoxidase activity of Phanerochaete chrysoporium. Indian J. Biochem. Biophys. 1999, 36. [Google Scholar]
- Sambasiva Rao, K.R.S.; Tripathy, N.K.; Srinivasa Rao, D.; Prakasham, R.S. Production, characterization, catalytic and inhibitory activities of tyrosinase. Res. J. Biotechnol. 2013, 8, 1. [Google Scholar]
- Zhao, M.; Sun, P.-F.; Du, L.-N.; Wang, G.; Jia, X.-M.; Zhao, Y.-H. Biodegradation of methyl red by Bacillus sp. strain UN2: Decolorization capacity, metabolites characterization, and enzyme analysis. Environ. Sci. Pollut. Res. 2014, 21, 6136–6145. [Google Scholar] [CrossRef] [PubMed]
- El Fakharany, E.; Hassan, M.A.; Taha, T.H. Production and Application of Extracellular Laccase Produced by Fusarium oxysporum EMT. Int. J. Agric. Biol. 2016, 18, 939–947. [Google Scholar] [CrossRef]
- Rekik, H.; Jaouadi, N.Z.; Bouacem, K.; Zenati, B.; Kourdali, S.; Badis, A.; Annane, R.; Bouanane-Darenfed, A.; Bejar, S.; Jaouadi, B. Physical and enzymatic properties of a new manganese peroxidase from the white-rot fungus Trametes pubescens strain i8 for lignin biodegradation and textile-dyes biodecolorization. Int. J. Biol. Macromol. 2019, 125, 514–525. [Google Scholar] [CrossRef]
- Sari, I.P.; Simarani, K. Decolorization of selected azo dye by Lysinibacillus fusiformis W1B6: Biodegradation optimization, isotherm, and kinetic study biosorption mechanism. Adsorpt. Sci. Technol. 2019, 37, 492–508. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Geng, A. Expression of a novel manganese peroxidase from Cerrena unicolor BBP6 in Pichia pastoris and its application in dye decolorization and PAH degradation. Biochem. Eng. J. 2020, 153, 107402. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zaki, S.A.; Eltarahony, M.; Abd-El-Haleem, D.A. Disinfection of water and wastewater by biosynthesized magnetite and zerovalent iron nanoparticles via NAP-NAR enzymes of Proteus mirabilis 10B. Environ. Sci. Pollut. Res. 2019, 26, 23661–23678. [Google Scholar] [CrossRef]
- Shebany, Y.M.; El-Dawy, E.G.; Gherbawy, Y.A. Potential of Different Fungi Species in Biodegradation Field of Phenolic Compounds. Annu. Res. Rev. Biol. 2019, 1–9. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Bharti, V.; Vikrant, K.; Goswami, M.; Tiwari, H.; Sonwani, R.K.; Lee, J.; Tsang, D.C.; Kim, K.-H.; Saeed, M.; Kumar, S.; et al. Biodegradation of methylene blue dye in a batch and continuous mode using biochar as packing media. Environ. Res. 2019, 171, 356–364. [Google Scholar] [CrossRef]
- Fuentes, M.S.; Briceño, G.E.; Saez, J.M.; Benimeli, C.; Diez, M.C.; Amoroso, M.J. Enhanced Removal of a Pesticides Mixture by Single Cultures and Consortia of Free and Immobilized Streptomyces Strains. BioMed Res. Int. 2013, 2013, 392573. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; Rabei, N.H.; El-Malkey, S.E. Eco-friendly approach for biosorption of Pb2+ and carcinogenic Congo red dye from binary solution onto sustainable Ulva lactuca biomass. Sci. Rep. 2020, 10, 16021. [Google Scholar] [CrossRef]
- Eltarahony, M.; Zaki, S.; Abd-El-Haleem, D. Aerobic and anaerobic removal of lead and mercury via calcium carbonate precipitation mediated by statistically optimized nitrate reductases. Sci. Rep. 2020, 10, 4029. [Google Scholar] [CrossRef]
- Akansha, K.; Chakraborty, D.; Sachan, S.G. Decolorization and degradation of methyl orange by Bacillus stratosphericus SCABiocatal. Agric. Biotechnol. 2019, 18, 101044. [Google Scholar] [CrossRef]
- Kalyani, D.C.; Telke, A.A.; Govindwar, S.P.; Jadhav, J.P. Biodegradation and Detoxification of Reactive Textile Dye by IsolatedPseudomonassp. SUK1. Water Environ. Res. 2009, 81, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, P.S.; Joseph, L.; Dhanya, A. Photocatalytic degradation of textile dyes by hydrogel supported titanium dioxide nanoparticles. J. Environ. Eng. Ecol. Sci. 2013, 2. [Google Scholar]
- Masarbo, R.S.; Ismailsab, M.; Monisha, T.R.; Nayak, A.S.; Karegoudar, T.B. Enhanced decolorization of sulfonated azo dye methyl orange by single and mixed bacterial strains AK1, AK2 and VKY1. Bioremediat. J. 2018, 22, 136–146. [Google Scholar] [CrossRef]
- Baena-Baldiris, D.; Montes-Robledo, A.; Baldiris-Avila, R. Franconibacter sp., 1MS: A New Strain in Decolorization and Degradation of Azo Dyes Ponceau S Red and Methyl Orange. ACS Omega 2020, 5, 28146–28157. [Google Scholar] [CrossRef]
- Raghukumar, C.; Chandramohan, D.; Michel, F.C.; Redd, C.A. Degradation of lignin and decolorization of paper mill bleach plant effluent (BPE) by marine fungi. Biotechnol. Lett. 1996, 18, 105–106. [Google Scholar] [CrossRef]
- Sghaier, I.; Ouertani, R.; Mahjoubi, M.; El-Hidri, D.; Hassen, W.; Chamkhi, A.; Chouchane, H.; Jaouani, A.; Cherif, A.; Neifar, M. Application of a mixture design to optimize textile azo-dye decolorization using a bacterial consortium. Biom. Biostat. Int. J. 2019, 8, 58–63. [Google Scholar] [CrossRef]
- Tang, W.; Xu, X.; Ye, B.-C.; Cao, P.; Ali, A. Decolorization and degradation analysis of Disperse Red 3B by a consortium of the fungus Aspergillus sp. XJ-2 and the microalgae Chlorella sorokiniana XJK. RSC Adv. 2019, 9, 14558–14566. [Google Scholar] [CrossRef]
- Contreras, M.; Grande-Tovar, C.D.; Vallejo, W.; Chaves-López, C. Bio-Removal of Methylene Blue from Aqueous Solution by Galactomyces geotrichum KL20A. Water 2019, 11, 282. [Google Scholar] [CrossRef]
- Shah, M.P.; Patel, K.A.; Darji, A.M. Microbial Degradation and Decolorization of Methyl Orange Dye by an Application of Pseudomonas Spp. ETL-1982. Int. J. Environ. Bioremediation Biodegrad. 2013, 1, 26–36. [Google Scholar] [CrossRef]
- Zommere, Ž.; Nikolajeva, V. others Immobilization of bacterial association in alginate beads for bioremediation of oil-contaminated lands. Environ. Exp. Biol. 2017, 15, 105–111. [Google Scholar]
- Giese, E.C. Mining Applications of Immobilized Microbial Cells in an Alginate Matrix: An Overview. Rev. Int. Contam. Ambient. 2020, 36, 775–787. [Google Scholar] [CrossRef]
- Das, A.; Mishra, S. Removal of textile dye reactive green-19 using bacterial consortium: Process optimization using response surface methodology and kinetics study. J. Environ. Chem. Eng. 2017, 5, 612–627. [Google Scholar] [CrossRef]
- Chenthamara, D.; Subramaniam, S.; Ramakrishnan, S.G.; Krishnaswamy, S.; Essa, M.M.; Lin, F.H.; Qoronfleh, M.W. Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 2019, 23, 20. [Google Scholar] [CrossRef]
- Cui, D.; Zhang, H.; He, R.; Hao, Z. The Comparative Study on the Rapid Decolorization of Azo, Anthraquinone and Triphenylmethane Dyes by Anaerobic Sludge. Int. J. Environ. Res. Public Health 2016, 13, 1053. [Google Scholar] [CrossRef]
- Ayed, L.; Chaieb, K.; Cheref, A.; Bakhrouf, A. Biodegradation of triphenylmethane dye Malachite Green by Sphingomonas paucimobilis. World J. Microbiol. Biotechnol. 2009, 25, 705–711. [Google Scholar] [CrossRef]
- Sheela, T.; Sadasivam, S.K. Dye degradation potential and its degradative enzymes synthesis of Bacillus cereus SKB12 isolated from a textile industrial effluent. J. Appl. Biol. Biotechnol. 2020, 8, 42–46. [Google Scholar]
- Thanavel, M.; Kadam, S.K.; Biradar, S.P.; Govindwar, S.P.; Jeon, B.-H.; Sadasivam, S.K. Combined biological and advanced oxidation process for decolorization of textile dyes. SN Appl. Sci. 2018, 1, 97. [Google Scholar] [CrossRef]
- Bedekar, P.A.; Kshirsagar, S.D.; Gholave, A.R.; Govindwar, S.P. Degradation and detoxification of methylene blue dye adsorbed on water hyacinth in semi continuous anaerobic–aerobic bioreactors by novel microbial consortium-SB. RSC Adv. 2015, 5, 99228–99239. [Google Scholar] [CrossRef]
- Oukarroum, A.; Zaidi, W.; Samadani, M.; Dewez, D. Toxicity of Nickel Oxide Nanoparticles on a Freshwater Green Algal Strain ofChlorella vulgaris. BioMed Res. Int. 2017, 2017, 9528180. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W. Antioxidants in Photosynthesis and Human Nutrition. Science 2002, 298, 2149–2153. [Google Scholar] [CrossRef] [PubMed]
- Novoveská, L.; Ross, M.E.; Stanley, M.S.; Pradelles, R.; Wasiolek, V.; Sassi, J.-F. Microalgal Carotenoids: A Review of Production, Current Markets, Regulations, and Future Direction. Mar. Drugs 2019, 17, 640. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-S.; Hung, C.-H.; Chao, S.-L. Decolorization of azo dye reactive black B by Bacillus cereus strain HJ-1. Chemosphere 2013, 90, 2109–2114. [Google Scholar] [CrossRef]
- Qu, Y.; Zhou, J.; Wang, J.; Song, Z.; Xing, L.; Fu, X. Bioaugmentation of Bromoamine Acid Degradation with Sphingomonas xenophaga QYY and DNA Fingerprint Analysis of Augmented Systems. Biodegradation 2006, 17, 83–91. [Google Scholar] [CrossRef]
- Poi, G.; Shahsavari, E.; Aburto-Medina, A. Large Scale Bioaugmentation of Municipal Waste Water Contaminated with Petroleum Hydrocarbons. JSM Environ. Sci. Ecol. 2016, 4, 1036. [Google Scholar]
- Jasmine, J.; Mukherji, S. Evaluation of bioaugmentation and biostimulation effects on the treatment of refinery oily sludge using 2n full factorial design. Environ. Sci. Process. Impacts 2014, 16, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).