Development of a Quantitative PCR Assay for Four Salmon Species Inhabiting the Yangyangnamdae River Using Environmental DNA
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Species and Molecular Assay Development
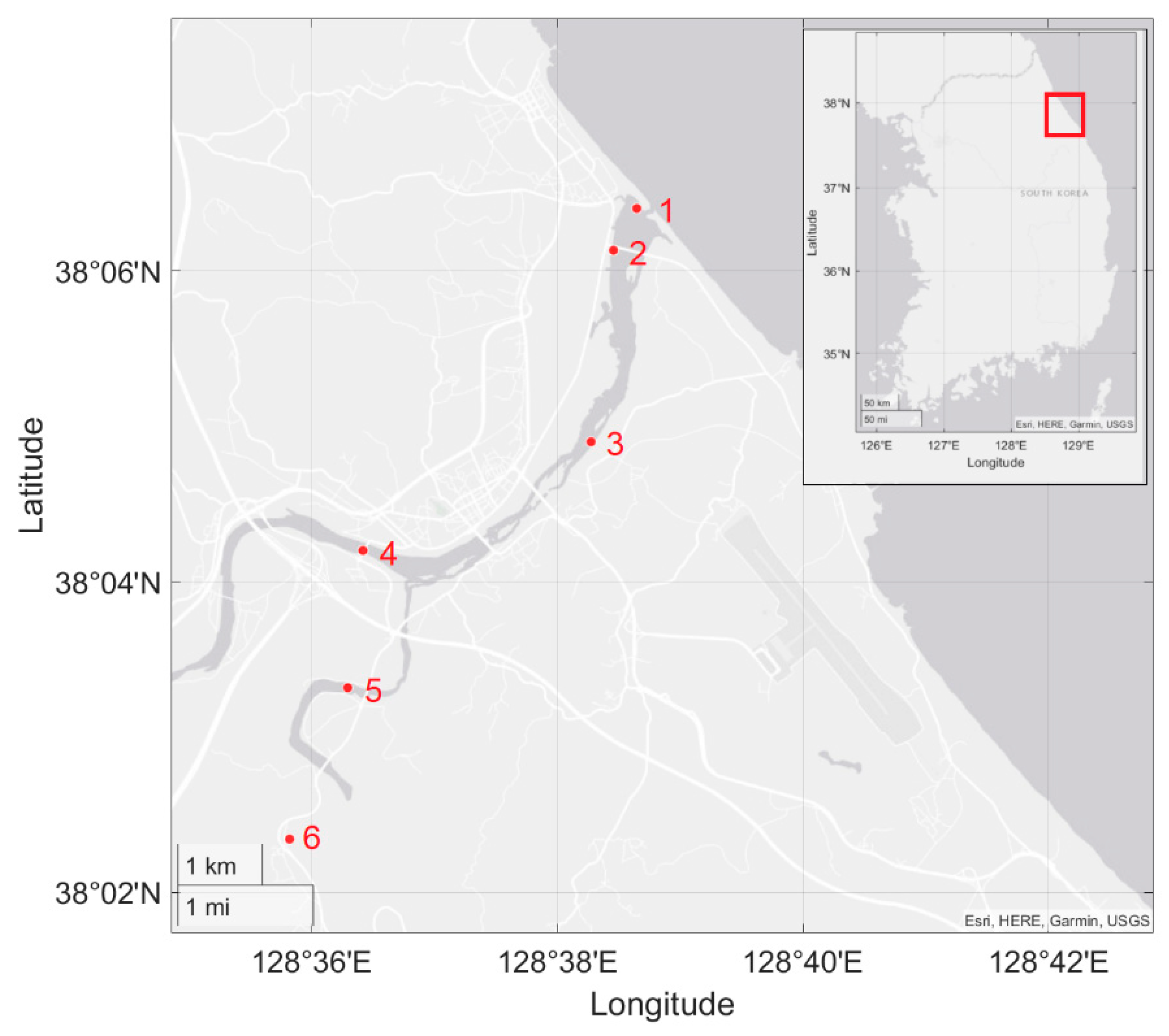
2.2. Water Sample Collection and Environmental DNA Extraction
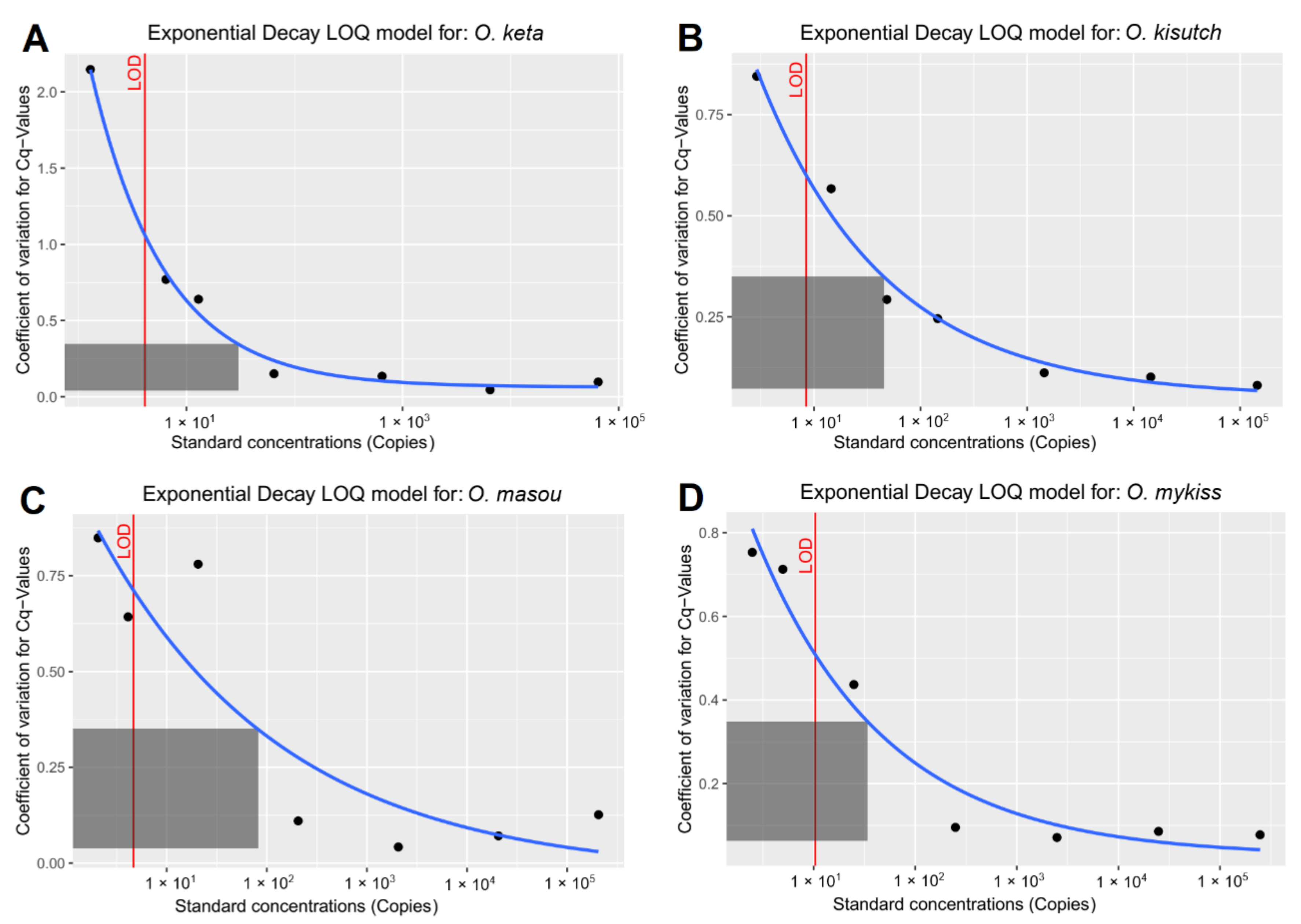
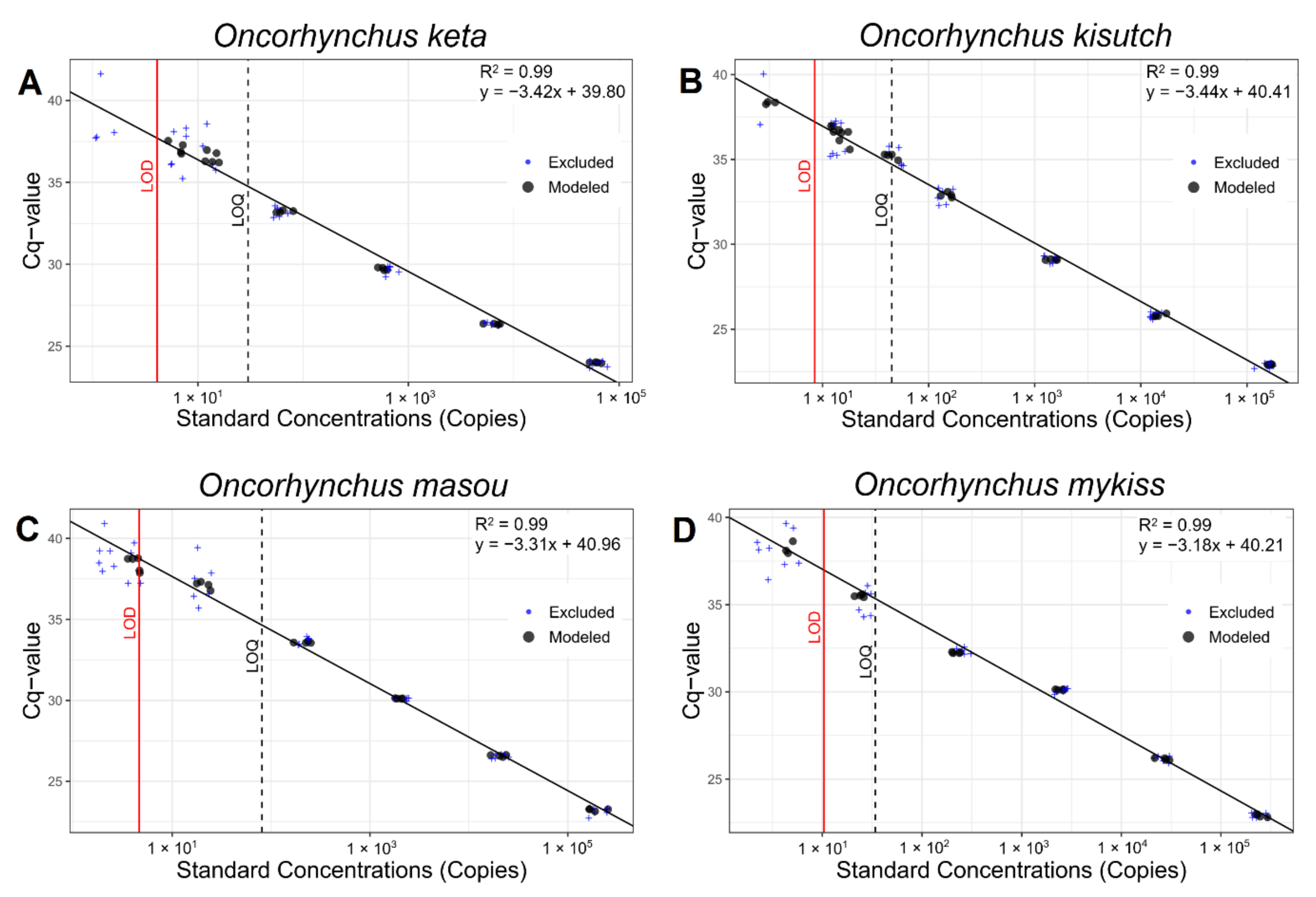
2.3. Establishment of Quantitative PCR Assay
2.4. Inhibition Test
2.5. Data Analysis
3. Results
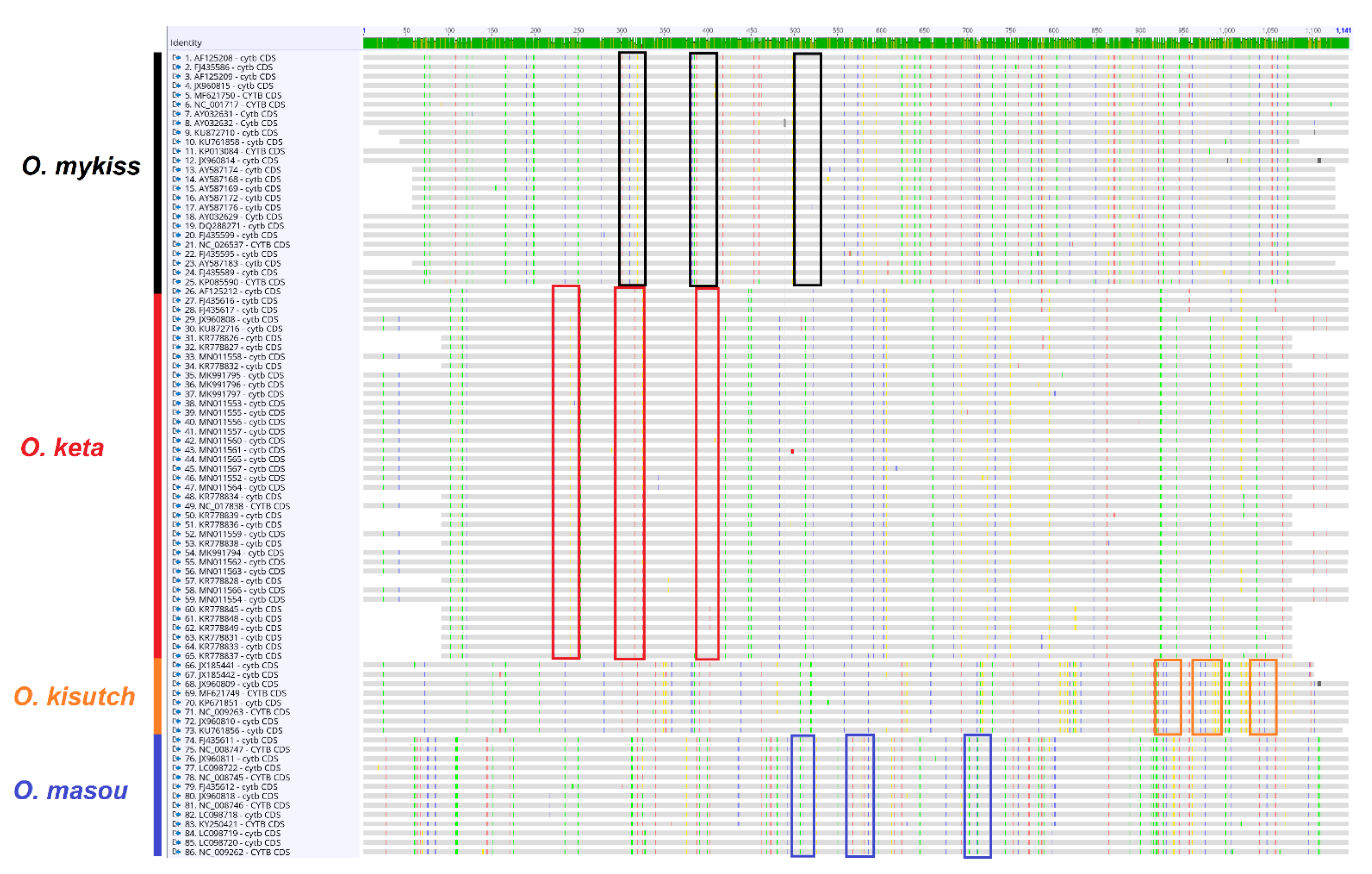
3.1. Profile and Performance of Primers and Probes
3.2. Inhibition in Environmental DNA Samples
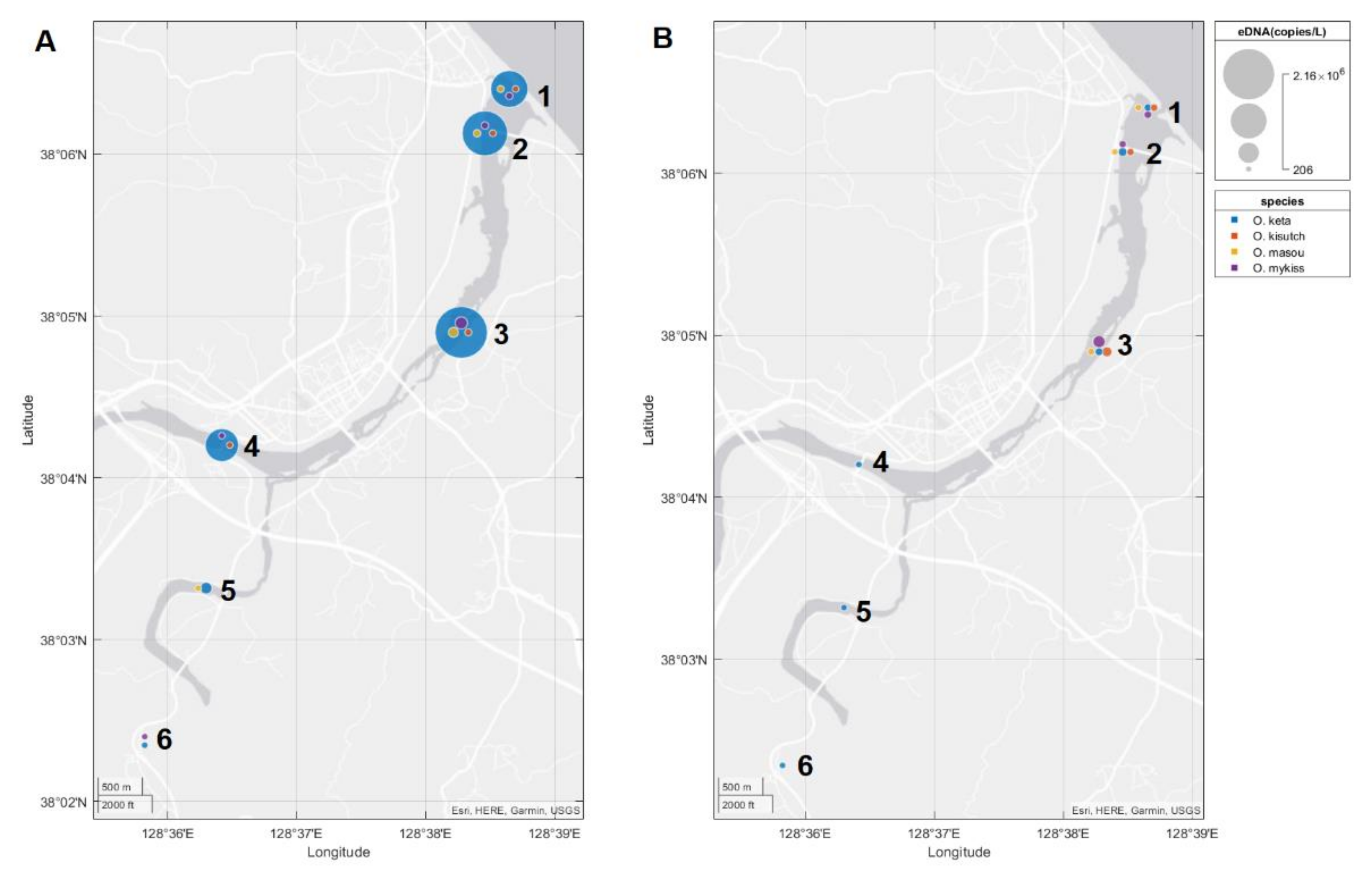
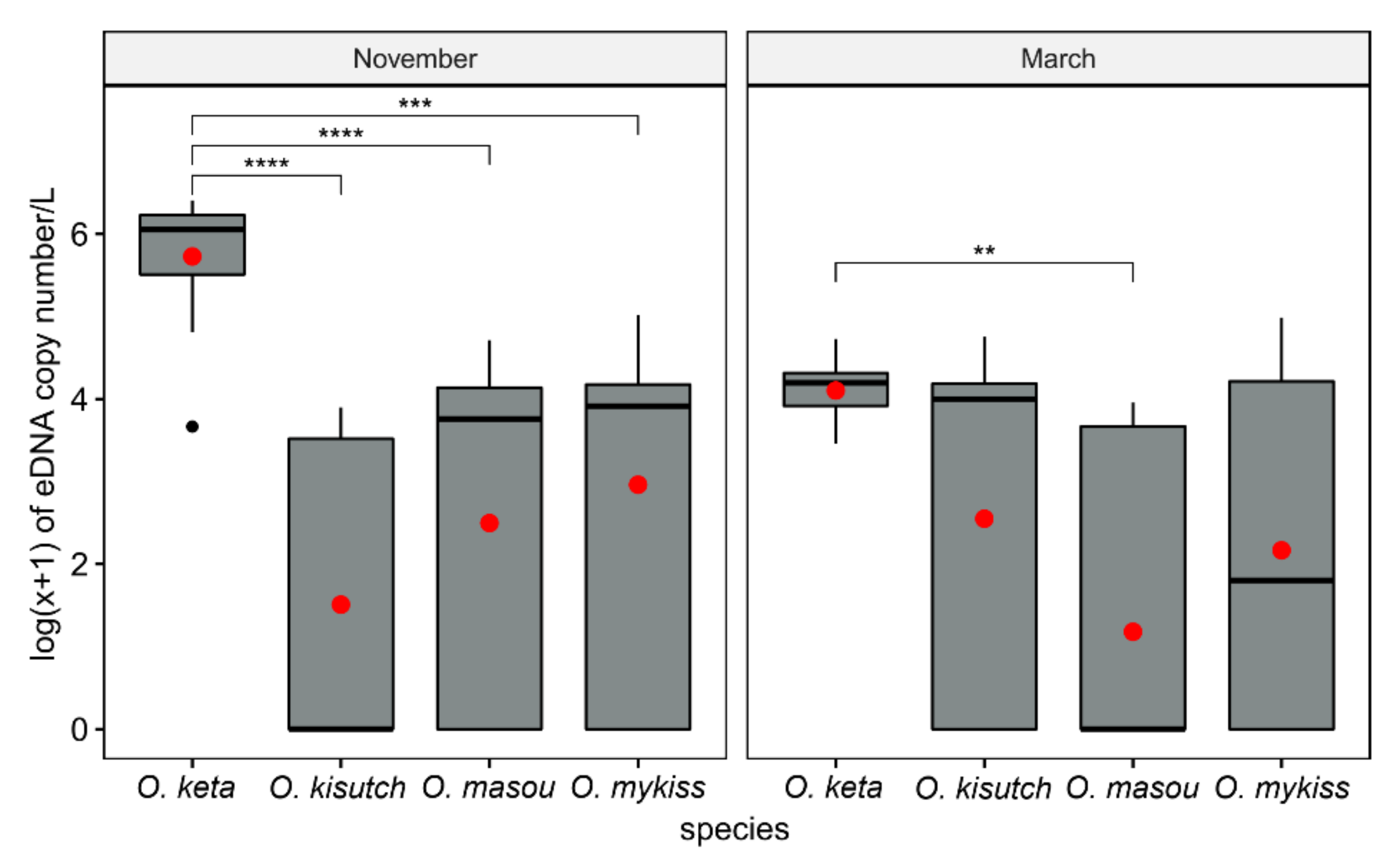
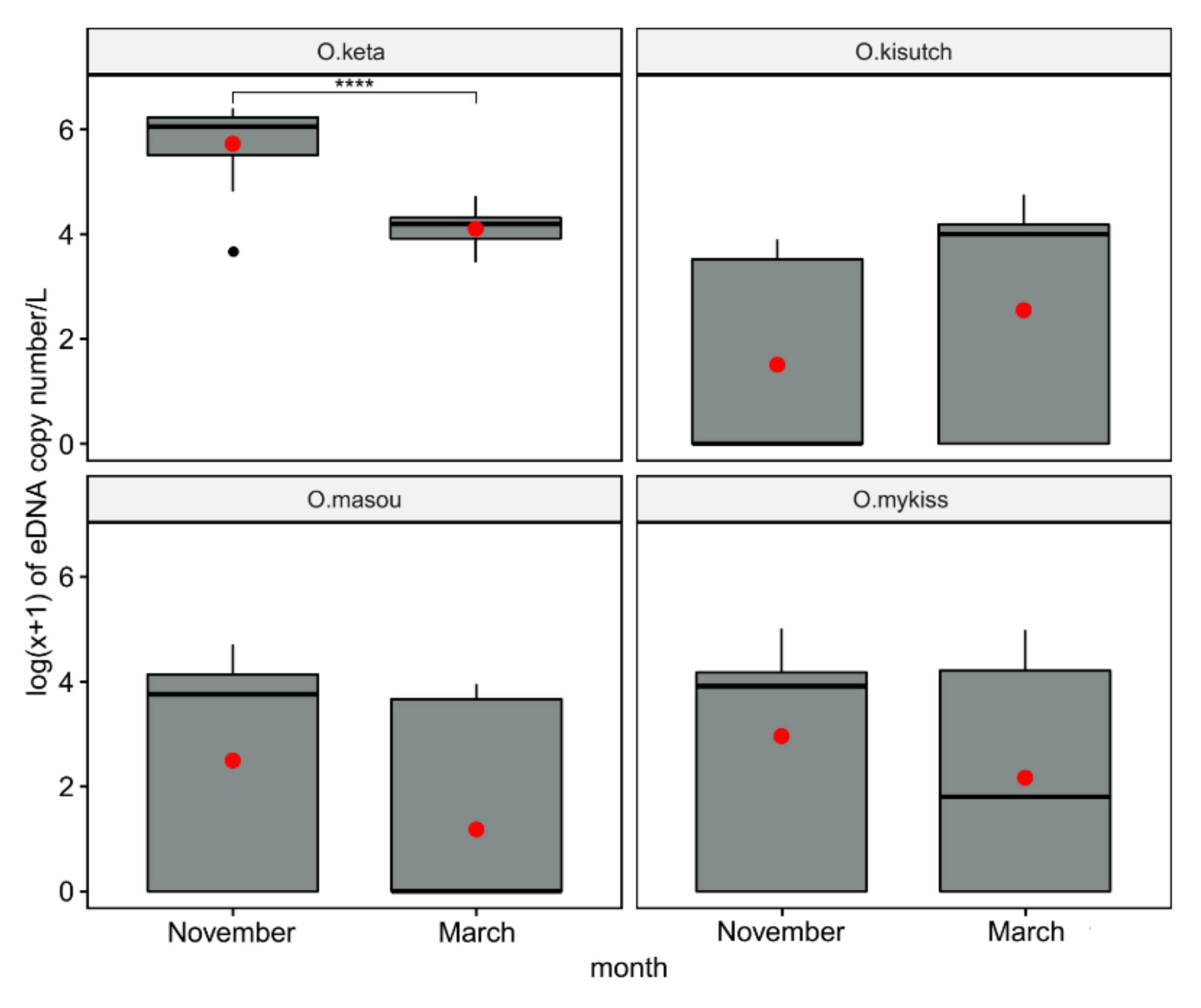
3.3. Environmental DNA Profiles in Field Testing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Species | GenBank Accession Number |
|---|---|
| Oncorhynchus keta | MN011557, MN011565, MN011555, MN011556, MN011562, MN011567, MK991795, MK991796, MK991797, MN011560, MN011558, JX960808, KU872716, MN011553, MN011561, MN011552, MN011564, KR778826, KR778827, KR778832, MN011563, MK991794, MN011554, MN011559, KR778834, NC_017838, KR778839, KR778838, KR778836, KR778845, KR778848, KR778849, KR778831, KR778833, KR778828, MN011566, KR778837, AF125212, FJ435616, FJ435617 |
| Oncorhynchus kisutch | KU761856, JX185441, JX185442, KU761856, NC_009263, JX960809, MF621749, KP671851, JX960810 |
| Oncorhynchus masou | JX960818, NC_008746, LC098718, FJ435612, NC_008747, FJ435611, NC_008745, JX960811, KY250421, LC098722, NC_009262, LC098720, LC098719 |
| Oncorhynchus mykiss | AF125208, FJ435586, JX960814, KP013084, KU872710, AY032629, KU761858, AY032632, AY587174, AY587168, AY587172, AY587176, AF125209, MF621750, JX960815, DQ288271, AY032631, AY587169, FJ435599, NC_026537, FJ435595, AY587183, FJ435589, NC_001717, KP085590 |
| O. keta | O. masou | O. mykiss | O. kisutch | ||
|---|---|---|---|---|---|
| qPCR | Luna® Universal Probe qPCR Master Mix (#M3004, New England Biolabs, Ipswich, MA, USA) | 10 µL | |||
| Forward primer | 0.4 µM | 0.4 µM | 0.4 µM | 0.5 µM | |
| Reverse primer | 0.4 µM | 0.4 µM | 0.4 µM | 0.5 µM | |
| Probe | 0.2 µM | 0.2 µM | 0.2 µM | 0.25 µM | |
| eDNA | 2 µL | ||||
| ddH2O | Up to 20 µL | ||||
| qPCR cycles | Initial denaturation | 95 °C, 5 min | |||
| Denaturation | 95 °C, 20 s | ||||
| Annealing | 63.6 °C, 20 s | 63.6 °C, 20 s | 64.9 °C, 20 s | 66.0 °C, 20 s | |
| Extension | 72 °C, 20 s | ||||
| Cycles | 40 | ||||
References
- Knudsen, E.E.; McDonald, D. Sustainable Fisheries Management: Pacific Salmon; CRC Press: New York, NY, USA, 2020. [Google Scholar]
- Schindler, D.E.; Scheuerell, M.D.; Moore, J.W.; Gende, S.M.; Francis, T.B.; Palen, W.J. Pacific salmon and the ecology of coastal ecosystems. Front. Ecol. Environ. 2003, 1, 31–37. [Google Scholar] [CrossRef]
- Sohn, D.; Kang, S.; Kim, S. Stock identification of chum salmon (Oncorhynchus keta) using trace elements in otoliths. J. Oceanogr. 2005, 61, 305–312. [Google Scholar] [CrossRef]
- Tsuboi, J.-I.; Endou, S.; Morita, K. Habitat fragmentation by damming threatens coexistence of stream-dwelling charr and salmon in the Fuji River, Japan. Hydrobiologia 2010, 650, 223–232. [Google Scholar] [CrossRef]
- Hilborn, R. Ocean and dam influences on salmon survival. Proc. Natl. Acad. Sci. USA 2013, 110, 6618–6619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, L.; Anulacion, B.; Arkoosh, M.; Olson, O.P.; Sloan, C.; Sol, S.Y.; Spromberg, J.; Teel, D.J.; Yanagida, G.; Ylitalo, G. Persistent organic pollutants in juvenile chinook salmon in the Columbia River Basin: Implications for stock recovery. Trans. Am. Fish. Soc. 2013, 142, 21–40. [Google Scholar] [CrossRef]
- Nehlsen, W.; Williams, J.E.; Lichatowich, J.A. Pacific Salmon at the Crossroads: Stocks at Risk from California, Oregon, Idaho, and Washington. Fisheries 1991, 16, 4–21. [Google Scholar] [CrossRef]
- Shanley, C.S.; Albert, D.M. Climate change sensitivity index for Pacific salmon habitat in Southeast Alaska. PLoS ONE 2014, 9, e104799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, A.W.; Bartz, K.K.; McClure, M.M. Interactive effects of water diversion and climate change for juvenile Chinook salmon in the Lemhi River Basin (USA). Conserv. Biol. 2013, 27, 1179–1189. [Google Scholar] [CrossRef]
- Vander Vorste, R.; Obedzinski, M.; Nossaman Pierce, S.; Carlson, S.M.; Grantham, T.E. Refuges and ecological traps: Extreme drought threatens persistence of an endangered fish in intermittent streams. Glob. Chang. Biol. 2020, 26, 3834–3845. [Google Scholar] [CrossRef]
- NPAFC. NPAFC Pacific Salmonid Hatchery Release Statistics (Updated 21 July 2020); North Pacific Anadromous Fish Commission: Vancouver, BC, Canada, 2020; Available online: https://npafc.org (accessed on 13 August 2020).
- NPAFC. NPAFC Pacific Salmonid Catch Statistics (Updated 21 July 2020); North Pacific Anadromous Fish Commission: Vancouver, BC, Canada, 2020; Available online: https://npafc.org (accessed on 13 August 2020).
- Kim, H.N.; Lee, S.W.; Lee, C.I.; Kim, N.R. Korean Salmon Catch Statistics and Hatchery Releases in 2019–2020; NPAFC Doc. 1897; North Pacific Anadromous Fish Commission: Vancouver, BC, Canada, 2020; p. 2. [Google Scholar]
- Greene, C.M.; Jensen, D.W.; Pess, G.R.; Steel, E.A.; Beamer, E. Effects of environmental conditions during stream, estuary, and ocean residency on chinook salmon return rates in the Skagit River, Washington. Trans. Am. Fish. Soc. 2005, 134, 1562–1581. [Google Scholar] [CrossRef]
- Holtby, L.B.; McMahon, T.E.; Scrivener, J.C. Stream temperatures and inter-annual variability in the emigration timing of coho salmon (Oncorhynchus kisutch) smolts and fry and chum salmon (O. keta) fry from Carnation Creek, British Columbia. Can. J. Fish. Aquat. Sci. 1989, 46, 1396–1405. [Google Scholar] [CrossRef]
- Yoon, H.-N.; Kim, K.-D.; Jeon, Y.-L.; Lee, J.-H.; Park, Y.-J. Stomach contents of the Manchurian trout (Brachymystax lenok tsinlingensis) and river salmon (Oncorhynchus masou masou) in the Odae mountain. Korean J. Ichthyol. 2013, 25, 90–105. [Google Scholar]
- Do, Y.; Ko, E.J.; Kim, Y.M.; Kim, H.G.; Joo, G.J.; Kim, J.Y.; Kim, H.W. Using text-mining method to identify research trends of freshwater exotic species in Korea. Korean J. Ecol. Environ. 2015, 48, 195–202. [Google Scholar] [CrossRef]
- Jensen, Ø.; Dempster, T.; Thorstad, E.; Uglem, I.; Fredheim, A. Escapes of fishes from Norwegian sea-cage aquaculture: Causes, consequences and prevention. Aquac. Environ. Interact. 2010, 1, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Sepúlveda, M.; Arismendi, I.; Soto, D.; Jara, F.; Farias, F. Escaped farmed salmon and trout in Chile: Incidence, impacts, and the need for an ecosystem view. Aquac. Environ. Interact. 2013, 4, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Levi, T.; Allen, J.M.; Bell, D.; Joyce, J.; Russell, J.R.; Tallmon, D.A.; Vulstek, S.C.; Yang, C.; Yu, D.W. Environmental DNA for the enumeration and management of Pacific salmon. Mol. Ecol. Resour. 2019, 19, 597–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonar, S.A.; Hubert, W.A.; Willis, D.W. Standard Methods for Sampling North American Freshwater Fishes; American Fisheries Society: Bethesda, MD, USA, 2009. [Google Scholar]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K.C. The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Douglas, W.Y.; De Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Tillotson, M.D.; Kelly, R.P.; Duda, J.J.; Hoy, M.; Kralj, J.; Quinn, T.P. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 2018, 220, 1–11. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigsgaard, E.E.; Carl, H.; Møller, P.R.; Thomsen, P.F. Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biol. Conserv. 2015, 183, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Piggott, M.P. Evaluating the effects of laboratory protocols on eDNA detection probability for an endangered freshwater fish. Ecol. Evol. 2016, 6, 2739–2750. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.V.; Garcia de Leaniz, C.; Rolla, M.; Consuegra, S. Monitoring the eradication of the highly invasive topmouth gudgeon (Pseudorasbora parva) using a novel eDNA assay. Environ. DNA 2019, 1, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Knudsen, S.W.; Ebert, R.B.; Hesselsøe, M.; Kuntke, F.; Hassingboe, J.; Mortensen, P.B.; Thomsen, P.F.; Sigsgaard, E.E.; Hansen, B.K.; Nielsen, E.E.; et al. Species-specific detection and quantification of environmental DNA from marine fishes in the Baltic Sea. J. Exp. Mar. Biol. Ecol. 2019, 510, 31–45. [Google Scholar] [CrossRef]
- Maruyama, A.; Sugatani, K.; Watanabe, K.; Yamanaka, H.; Imamura, A. Environmental DNA analysis as a non-invasive quantitative tool for reproductive migration of a threatened endemic fish in rivers. Ecol. Evol. 2018, 8, 11964–11974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, R.A.; Rees, C.B.; Coulter, A.A.; Merkes, C.M.; McCalla, S.G.; Touzinsky, K.F.; Walleser, L.; Goforth, R.R.; Amberg, J.J. Detecting the movement and spawning activity of bigheaded carps with environmental DNA. Mol. Ecol. Resour. 2016, 16, 957–965. [Google Scholar] [CrossRef]
- Yamanaka, H.; Minamoto, T. The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecol. Indic. 2016, 62, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Boothroyd, M.; Mandrak, N.E.; Fox, M.; Wilson, C.C. Environmental DNA (eDNA) detection and habitat occupancy of threatened spotted gar (Lepisosteus oculatus). Aquat. Conserv. 2016, 26, 1107–1119. [Google Scholar] [CrossRef]
- Bylemans, J.; Furlan, E.M.; Hardy, C.M.; McGuffie, P.; Lintermans, M.; Gleeson, D.M. An environmental DNA-based method for monitoring spawning activity: A case study, using the endangered Macquarie perch (Macquaria australasica). Methods Ecol. Evol. 2017, 8, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Hayer, C.-A.; Bayless, M.F.; George, A.; Thompson, N.; Richter, C.A.; Chapman, D.C. Use of environmental DNA to detect grass carp spawning events. Fishes 2020, 5, 27. [Google Scholar] [CrossRef]
- Tsuji, S.; Shibata, N. Identifying spawning events in fish by observing a spike in environmental DNA concentration after spawning. Environ. DNA 2021, 3, 190–199. [Google Scholar] [CrossRef]
- Pont, D.; Rocle, M.; Valentini, A.; Civade, R.; Jean, P.; Maire, A.; Roset, N.; Schabuss, M.; Zornig, H.; Dejean, T. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 2018, 8, 10361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, H.; Inui, R.; Akamatsu, Y.; Kanno, K.; Yamanaka, H.; Takahara, T.; Minamoto, T. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw. Biol. 2017, 62, 30–39. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z.I. Estimation of fish biomass using environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef] [Green Version]
- Itakura, H.; Wakiya, R.; Yamamoto, S.; Kaifu, K.; Sato, T.; Minamoto, T. Environmental DNA analysis reveals the spatial distribution, abundance, and biomass of Japanese eels at the river-basin scale. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Spear, M.J.; Embke, H.S.; Krysan, P.J.; Vander Zanden, M.J. Application of eDNA as a tool for assessing fish population abundance. Environ. DNA 2021, 3, 83–91. [Google Scholar] [CrossRef]
- Penaluna, B.E.; Allen, J.M.; Arismendi, I.; Levi, T.; Garcia, T.S.; Walter, J.K. Better boundaries: Identifying the upper extent of fish distributions in forested streams using eDNA and electrofishing. Ecosphere 2021, 12, e03332. [Google Scholar] [CrossRef]
- Evans, N.T.; Shirey, P.D.; Wieringa, J.G.; Mahon, A.R.; Lamberti, G.A. Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 2017, 42, 90–99. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Côté, G.; Leclerc, V.; Bernatchez, L. Quantifying relative fish abundance with eDNA: A promising tool for fisheries management. J. Appl. Ecol. 2016, 53, 1148–1157. [Google Scholar] [CrossRef]
- Hinlo, R.; Furlan, E.; Suitor, L.; Gleeson, D. Environmental DNA monitoring and management of invasive fish: Comparison of eDNA and fyke netting. Manag. Biol. Invasions 2017, 8, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Laramie, M.B.; Pilliod, D.S.; Goldberg, C.S. Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol. Conserv. 2015, 183, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Shelton, A.O.; Kelly, R.P.; O’Donnell, J.L.; Park, L.; Schwenke, P.; Greene, C.; Henderson, R.A.; Beamer, E.M. Environmental DNA provides quantitative estimates of a threatened salmon species. Biol. Conserv. 2019, 237, 383–391. [Google Scholar] [CrossRef]
- Minegishi, Y.; Wong, M.K.-S.; Kanbe, T.; Araki, H.; Kashiwabara, T.; Ijichi, M.; Kogure, K.; Hyodo, S. Spatiotemporal distribution of juvenile chum salmon in Otsuchi Bay, Iwate, Japan, inferred from environmental DNA. PLoS ONE 2019, 14, e0222052. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen Hellberg, R.S.; Morrissey, M.T.; Hanner, R.H. A multiplex PCR method for the identification of commercially important salmon and trout species (Oncorhynchus and Salmo) in North America. J. Food Sci. 2010, 75, C595–C606. [Google Scholar] [CrossRef]
- Kim, S.A.; Lee, C.S.; Kang, S.K. Present status and future prospect in salmon research in Korea. Sea 2007, 12, 57–60. [Google Scholar]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for methods utilizing environmental DNA for detection of fish species. Genes 2020, 11, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Jang, J.; Eom, J.; Cheong, D.; Lee, C. Monitoring of the estuary sand bar related with tidal inlet in Namdaecheon Stream using landsat imagery. Korean J. Remote Sens. 2017, 33, 481–493. [Google Scholar]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. BioTechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkes, C.; Klymus, K.; Allison, M.; Goldberg, C.; Helbing, C.; Hunter, M.; Jackson, C.; Lance, R.; Mangan, A.; Monroe, E.; et al. Generic qPCR Limit of Detection (LOD)/Limit of Quantification (LOQ) Calculator: R Script. Available online: https://github.com/cmerkes/qPCR_LOD_Calc (accessed on 8 January 2021).
- Forootan, A.; Sjöback, R.; Björkman, J.; Sjögreen, B.; Linz, L.; Kubista, M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantif. 2017, 12, 1–6. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klymus, K.E.; Merkes, C.M.; Allison, M.J.; Goldberg, C.S.; Helbing, C.C.; Hunter, M.E.; Jackson, C.A.; Lance, R.F.; Mangan, A.M.; Monroe, E.M.; et al. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2020, 2, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Brys, R.; Halfmaerten, D.; Neyrinck, S.; Mauvisseau, Q.; Auwerx, J.; Sweet, M.; Mergeay, J. Reliable eDNA detection and quantification of the European weather loach (Misgurnus fossilis). J. Fish Biol. 2021, 98, 399–414. [Google Scholar] [CrossRef]
- Thomas, A.C.; Howard, J.; Nguyen, P.L.; Seimon, T.A.; Goldberg, C.S. eDNA Sampler: A fully integrated environmental DNA sampling system. Methods Ecol. Evol. 2018, 9, 1379–1385. [Google Scholar] [CrossRef] [Green Version]
- Iwai, N.; Yasumiba, K.; Takahara, T. Efficacy of environmental DNA to detect and quantify stream tadpoles of Odorrana splendida. R. Soc. Open Sci. 2019, 6, 181798. [Google Scholar] [CrossRef] [Green Version]
- Baldigo, B.P.; Sporn, L.A.; George, S.D.; Ball, J.A. Efficacy of environmental DNA to detect and quantify brook trout populations in headwater streams of the Adirondack Mountains, New York. Trans. Am. Fish. Soc. 2017, 146, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Doi, H.; Katano, I.; Sakata, Y.; Souma, R.; Kosuge, T.; Nagano, M.; Ikeda, K.; Yano, K.; Tojo, K. Detection of an endangered aquatic heteropteran using environmental DNA in a wetland ecosystem. R. Soc. Open Sci. 2017, 4, 170568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, L.J.; Coyne, S.R.; Norwood, D.A. Development of a novel internal positive control for Taqman® based assays. Mol. Cell. Probes 2005, 19, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Schabacker, J.; Smith, S.; Al-Chokhachy, R.; Luikart, G.; Amish, S.J. Improved detection of rare, endangered and invasive trout in using a new large-volume sampling method for eDNA capture. Environ. DNA 2019, 1, 227–237. [Google Scholar] [CrossRef]
- Duda, J.J.; Hoy, M.S.; Chase, D.M.; Pess, G.R.; Brenkman, S.J.; McHenry, M.M.; Ostberg, C.O. Environmental DNA is an effective tool to track recolonizing migratory fish following large-scale dam removal. Environ. DNA 2021, 3, 121–141. [Google Scholar] [CrossRef]
- Xia, Z.; Zhan, A.; Johansson, M.L.; DeRoy, E.; Haffner, G.D.; MacIsaac, H.J. Screening marker sensitivity: Optimizing eDNA-based rare species detection. Divers. Distrib. 2021. [Google Scholar] [CrossRef]
- Hunter, M.E.; Dorazio, R.M.; Butterfield, J.S.S.; Meigs-Friend, G.; Nico, L.G.; Ferrante, J.A. Detection limits of quantitative and digital PCR assays and their influence in presence–absence surveys of environmental DNA. Mol. Ecol. Resour. 2017, 17, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Ellison, S.L.R.; English, C.A.; Burns, M.J.; Keer, J.T. Routes to improving the reliability of low level DNA analysis using real-time PCR. BMC Biotechnol. 2006, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Thalinger, B.; Deiner, K.; Harper, L.R.; Rees, H.C.; Blackman, R.C.; Sint, D.; Traugott, M.; Goldberg, C.S.; Bruce, K. A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environ. DNA 2021, 3, 823–836. [Google Scholar] [CrossRef]
- Lance, R.F.; Guan, X. Variation in inhibitor effects on qPCR assays and implications for eDNA surveys. Can. J. Fish. Aquat. Sci. 2020, 77, 23–33. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Miller, L.M.; Sorensen, P.W. Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Mol. Ecol. Resour. 2016, 16, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Foote, A.D.; Thomsen, P.F.; Sveegaard, S.; Wahlberg, M.; Kielgast, J.; Kyhn, L.A.; Salling, A.B.; Galatius, A.; Orlando, L.; Gilbert, M.T.P. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 2012, 7, e41781. [Google Scholar] [CrossRef] [PubMed]
- Jane, S.F.; Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Wood, Z.T.; Erdman, B.F.; York, G.; Trial, J.G.; Kinnison, M.T. Experimental assessment of optimal lotic eDNA sampling and assay multiplexing for a critically endangered fish. Environ. DNA 2020, 2, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Lacoursière-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.A.; Olds, B.; Jerde, C.; Bolster, D. Modelling the transport of environmental DNA through a porous substrate using continuous flow-through column experiments. J. R. Soc. Interface 2016, 13, 20160290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.; Olds, B.; Mahon, A.R.; Jerde, C.L.; Bolster, D. Controls on eDNA movement in streams: Transport, Retention, and Resuspension. Sci. Rep. 2017, 7, 5065. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.S.; Muir, A.M. Global introductions of salmon and trout in the genus Oncorhynchus: 1870–2007. Rev. Fish Biol. Fish. 2008, 18, 313–344. [Google Scholar] [CrossRef]
- Lee, W.O.; Ko, M.H.; Bak, J.M.; Kim, D.H.; Jeon, H.J.; Kim, K.H. Characteristics of fish fauna and community structure in Buk Stream of Goseong, Korea. Korean J. Ichthyol. 2010, 22, 238–248. [Google Scholar]
- Jeon, C.H.; Kim, S.R.; Kim, W.S.; Lee, C.H.; Seong, K.B.; Lee, C.S.; Oh, M.J.; Kim, J.H. Monitoring of viruses in chum salmon (Oncorhynchus keta) migrating to Korea. Arch. Virol. 2011, 156, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Lee, W.O.; Hong, K.E.; Lee, C.H.; Kim, J.H. Ichthyofauna and fish community structure in Namdae Stream, Yangyang, Korea. Korean J. Ichthyol. 2006, 18, 112–118. [Google Scholar]
- Lee, K.Y.; Lee, H.K.; Oh, Y.K.; Kim, J.C.; Choi, J.S. Disturbance of fish community and discontinuity of Inspection of river continuity in Yangyangnamdae stream, Korea. J. Environ. 2018, 13, 17–30. (In Korean) [Google Scholar]
- Havn, T.B.; Thorstad, E.B.; Borcherding, J.; Heermann, L.; Teichert, M.A.K.; Ingendahl, D.; Tambets, M.; Sæther, S.A.; Økland, F. Impacts of a weir and power station on downstream migrating Atlantic salmon smolts in a German river. River Res. Appl. 2020, 36, 784–796. [Google Scholar] [CrossRef] [Green Version]
- Aarestrup, K.; Koed, A. Survival of migrating sea trout (Salmo trutta) and Atlantic salmon (Salmo salar) smolts negotiating weirs in small Danish rivers. Ecol. Freshw. Fish 2003, 12, 169–176. [Google Scholar] [CrossRef]
- Crozier, L.G.; Hutchings, J.A. Plastic and evolutionary responses to climate change in fish. Evol. Appl. 2014, 7, 68–87. [Google Scholar] [CrossRef]
- Yu, J.N.; Choi, S.H.; Kweon, S.M.; Kim, S.K.; Jin, D.H.; Kim, G.S.; Yeo, J.H.; Kim, S. New polymorphic microsatellite markers for the masu salmon (Oncorhynchus masou masou) from Korea and their application to wild and hatchery populations. Biochem. Syst. Ecol. 2015, 61, 260–267. [Google Scholar] [CrossRef]
- Heu, M.-S.; Kim, H.-J.; Ham, J.-S.; Park, S.-H.; Kim, H.-S.; Kang, K.-T.; Jee, S.-J.; Lee, J.-H.; Kim, J.-S. Preparation and quality characteristics of seasoned and dried fish slice products using rainbow trout (Onchorhynchus mykiss). J. Korean Soc. Food Sci. Nutr. 2008, 37, 348–356. [Google Scholar] [CrossRef]
- Sandercock, F. Life history of coho salmon (Oncorhynchus kisutch). In Pacific Salmon Life Histories; Groot, C., Margolis, L., Eds.; UBC Press: Vancouver, BC, Canada, 1991; pp. 395–445. [Google Scholar]
- Lim, J.; Hong, S. Characterization of Aeromonas salmonicida and A. sobria isolated from cultured salmonid fish in Korea and development of a vaccine against furunculosis. J. Fish Dis. 2020, 43, 609–620. [Google Scholar] [CrossRef]
- Hong, Y.K.; Kim, K.H.; Kim, K.M.; Lim, G.H.; Song, M.Y.; Lee, W.O. Characteristics of fish fauna and community structure in Wangpicheon. Korean J. Environ. Ecol. 2016, 30, 874–887. [Google Scholar] [CrossRef]
- Ko, M.H.; Myung, R.Y.; Kim, H.S. Fish community characteristics and habitat aspects of endangered species, Rhodeus pseudosericeus in Heuk stream, a tributary of the Han river drainage system. Korean J. Environ. Ecol. 2019, 33, 266–279. [Google Scholar] [CrossRef]


| Species | Forward Primer (5′–3′) Reverse Primer (5′–3′) Probe (5′–3′) | Size (bp) |
|---|---|---|
| Oncorhynchus keta | CTACGGCTGACTAATTCGGAACATCCAC TCCTCACGGGAGGACGTAGCCC FAM-CGCCCGGGGACTTTATTACGGATCCTACCT-BHQ | 187 |
| Oncorhynchus kisutch | TTACACACCTCCAAACAACGAGGACTG TTGGCCGATAATGATGAATGGGTGTTCC FAM-CCCAATTCCTATTCTGGGCCTTGGTGGCG-BHQ | 138 |
| Oncorhynchus masou | GGGTTCTCTGTCGACAACGCCAC CTAAGGATGTTAGACAGAGAAGTATAGCTG FAM-CGTCATTACAGCTGCTGCAATCCTCCACCT-BHQ | 229 |
| Oncorhynchus mykiss | GAGGACTTTACTACGGCTCGTACCTC GTTAGAGTGGCGTTGTCAACGGAGAAG FAM-CTGCCTTTGTAGGCTACGTCCTCCCGTGAG-BHQ | 229 |
| Zenarchopterus dispar1 | CAGCAGCTATAAACGCATGAATTACAGG TTTTTGTCAGGTTGAGAGAATGAGTCCG | 188 |
| Species | LOD (Copy Numbers) | LOQ (Copy Numbers) | Efficiency (%) |
|---|---|---|---|
| Oncorhynchus keta | 4.11 | 30 | 96.26 |
| Oncorhynchus kisutch | 8.44 | 45 | 95.15 |
| Oncorhynchus masou | 4.65 | 81 | 100.67 |
| Oncorhynchus mykiss | 10.38 | 34 | 106.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu'adil Amin, M.H.; Lee, J.-H.; Kim, A.R.; Kim, J.-K.; Lee, C.-I.; Kim, H.-W. Development of a Quantitative PCR Assay for Four Salmon Species Inhabiting the Yangyangnamdae River Using Environmental DNA. Biology 2021, 10, 899. https://doi.org/10.3390/biology10090899
Fu'adil Amin MH, Lee J-H, Kim AR, Kim J-K, Lee C-I, Kim H-W. Development of a Quantitative PCR Assay for Four Salmon Species Inhabiting the Yangyangnamdae River Using Environmental DNA. Biology. 2021; 10(9):899. https://doi.org/10.3390/biology10090899
Chicago/Turabian StyleFu'adil Amin, Muhammad Hilman, Ji-Hyun Lee, Ah Ran Kim, Ju-Kyoung Kim, Chung-Il Lee, and Hyun-Woo Kim. 2021. "Development of a Quantitative PCR Assay for Four Salmon Species Inhabiting the Yangyangnamdae River Using Environmental DNA" Biology 10, no. 9: 899. https://doi.org/10.3390/biology10090899
APA StyleFu'adil Amin, M. H., Lee, J.-H., Kim, A. R., Kim, J.-K., Lee, C.-I., & Kim, H.-W. (2021). Development of a Quantitative PCR Assay for Four Salmon Species Inhabiting the Yangyangnamdae River Using Environmental DNA. Biology, 10(9), 899. https://doi.org/10.3390/biology10090899








