OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Clinical Isolates
2.2. Antimicrobial Susceptibility of the Collected Isolates
2.3. Phenotypic Detection of CPs
2.3.1. Blue-Carba Test
2.3.2. Modified Carbapenem Inactivation Method (mCIM)
2.3.3. EDTA-Modified Carbapenem Inactivation Method (eCIM)
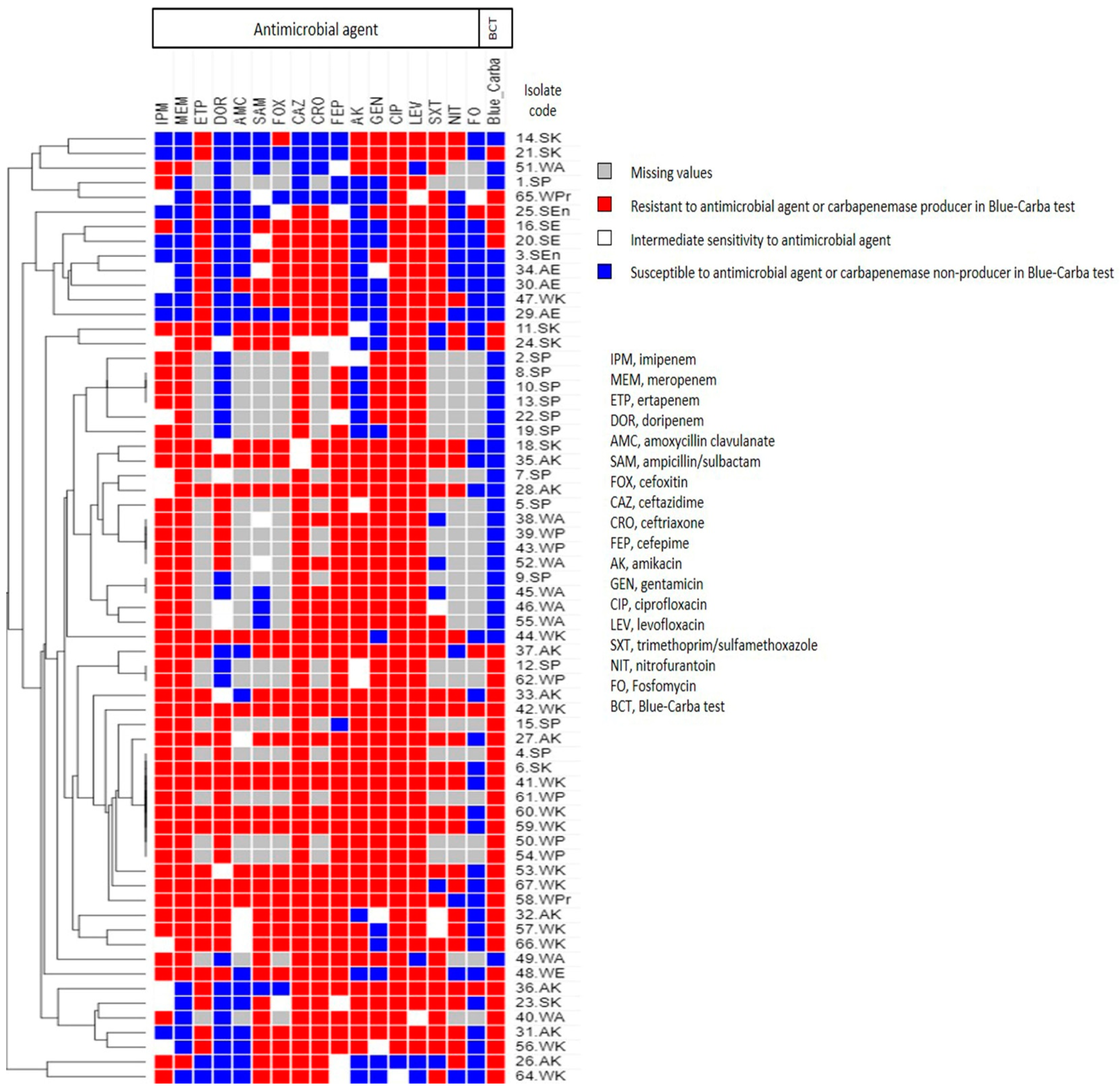
2.4. Phenotypic Analysis Using Heatmap Signature
2.5. Molecular Detection of Plasmid-Borne Carbapenemase Genes
2.5.1. Extraction of Plasmid DNA from Carbapenem-Resistant Isolates
2.5.2. Amplification of Some Plasmid-Encoded Carbapenemase Genes
2.5.3. Sequencing of PCR Products
2.5.4. Transformation
2.6. Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR) for Isolates Containing Plasmids
2.7. Statistical Analysis
3. Results
3.1. Collection of Clinical Isolates
3.2. Identification of the Collected GNB Clinical Isolates
3.3. Antimicrobial Susceptibility of the Collected Isolates
3.4. Phenotypic Detection of CPs
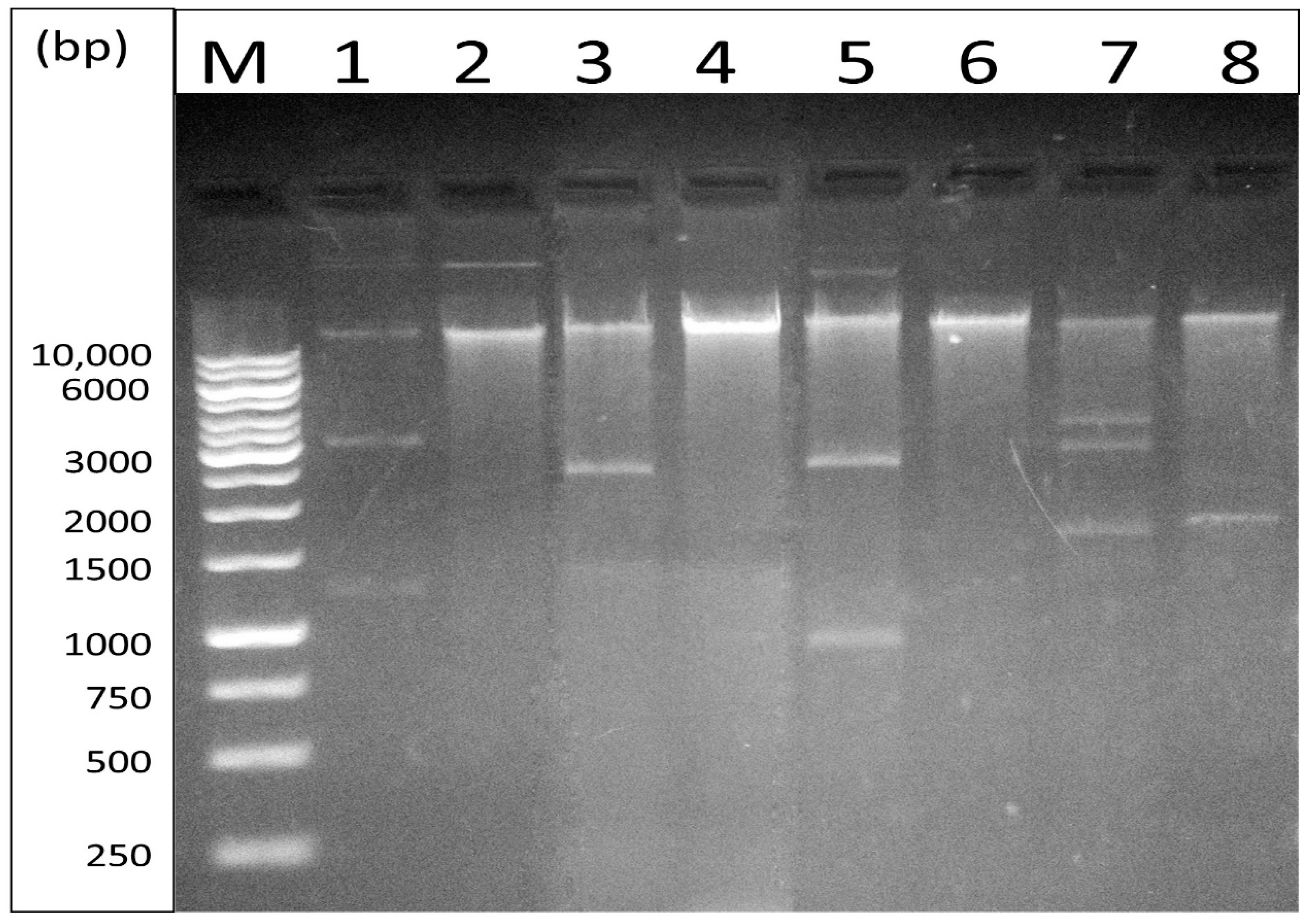
3.5. Extraction of Plasmid DNA and Amplification of Carbapenemase Genes
3.6. Transformation of Plasmids into Competent E. coli DH5α
3.7. Genotyping of CR-GNB Containing Plasmids
3.8. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McLellan, L.; Hunstad, D. Urinary tract infection: Pathogenesis and complications. Trends Mol. Med. 2016, 22, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Doesschate, T.T.; Van Der Vaart, T.; Damen, J.; Bonten, M.; Van Werkhoven, C. Carbapenem-alternative strategies for complicated urinary tract infections: A systematic review of randomized controlled trials. J. Infect. 2020, 81, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Barber, A.E.; Norton, J.P.; Spivak, A.M.; Mulvey, M.A. Urinary tract infections: Current and emerging management strategies. Clin. Infect. Dis. 2013, 57, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberger, P.; Hooton, T.M. Complicated urinary tract infections. Curr. Infect. Dis. Rep. 2008, 10, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Neal, D.E. Complicated Urinary Tract Infections. Urol. Clin. N. Am. 2008, 35, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pallett, A.; Hand, K. Complicated urinary tract infections: Practical solutions for the treatment of multiresistant Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, iii25–iii33. [Google Scholar] [CrossRef]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M.; Yassien, M.A.; Hassouna, N.A. Prevalence of plasmid-mediated resistance genes among multidrug-resistant uropathogens in Egypt. Afr. Health. Sci. 2020, 20, 190–198. [Google Scholar] [CrossRef]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2015, 3, 15–21. [Google Scholar] [CrossRef]
- Diene, S.M.; Rolain, J.-M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.N.A.; Tambyah, P.A.; Paterson, D.L. β-lactam and β-lactamase inhibitor combinations in the treatment of extended-spectrum β-lactamase producing Enterobacteriaceae: Time for a reappraisal in the era of few antibiotic options? Lancet Infect. Dis. 2015, 15, 475–485. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: Epidemiology, detection and treatment options. Futur. Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Munoz-Price, L.S. Mechanisms of disease: The New β-Lactamases. N. Engl. J. Med. 2005, 352, 380–391. [Google Scholar] [CrossRef]
- Gotte, M.; Berghuis, A.; Matlashewski, G.; Wainberg, M.A.; Sheppard, D. Handbook of Antimicrobial Resistance; Berghuis, A., Matlashewski, G., Wainberg, M.A., Sheppard, D., Eds.; Springer: New York, NY, USA, 2017; ISBN 978-1-4939-0693-2. [Google Scholar]
- Cantón, R.; Ruiz-Garbajosa, P. Co-resistance: An opportunity for the bacteria and resistance genes. Curr. Opin. Pharmacol. 2011, 11, 477–485. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)–structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef]
- Findlay, J.; Hopkins, K.; Meunier, D.; Woodford, N. Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J. Antimicrob. Chemother. 2015, 70, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Bergey, D.H.; Holt, J.G. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams & Wilkins: Baltimore, MD, USA, 1994; ISBN 978-0-683-00603-2. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests. CLSI Standard M02, 13th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1562388347.
- CLSI Performance Standards for Antimicrobial Susceptibility Testing. Informational Supplement. CLSI Document M100, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; ISBN 978-1-68440-067-6.
- Pires, J.; Novais, A.; Peixe, L. Blue-Carba, an Easy Biochemical Test for Detection of Diverse Carbapenemase Producers Directly from Bacterial Cultures. J. Clin. Microbiol. 2013, 51, 4281–4283. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.J.; Russell, D.D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 3, ISBN 0-87969-577-3. [Google Scholar]
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D.D. Laboratory Detection of Enterobacteriaceae That Produce Carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.; Walsh, T.R. How To Detect NDM-1 Producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-producingEnterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Poirel, L.; Naas, T.; Nicolas, D.; Collet, L.; Bellais, S.; Cavallo, J.-D.; Nordmann, P. Characterization of VIM-2, a Carbapenem-Hydrolyzing Metallo-β-Lactamase and Its Plasmid- and Integron-Borne Gene from a Pseudomonas aeruginosa Clinical Isolate in France. Antimicrob. Agents Chemother. 2000, 44, 891–897. [Google Scholar] [CrossRef]
- Woodford, N.; Tierno, P.M.; Young, K.; Tysall, L.; Palepou, M.-F.I.; Ward, E.; Painter, R.E.; Suber, D.F.; Shungu, D.; Silver, L.L.; et al. Outbreak of Klebsiella pneumoniae Producing a New Carbapenem-Hydrolyzing Class A β-Lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004, 48, 4793–4799. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.; Aboshanab, K.M.A.; Elkhatib, W.F.; Ashour, M.S. Aminoglycoside Resistance Patterns of Certain Gram Negative Uropathogens Recovered from Hospitalized Egyptian Patients. Br. Microbiol. Res. J. 2013, 3, 678–691. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Jay, C.; Metchock, B.; Berkowitz, F.; Weigel, L.; Crellin, J.; Steward, C.; Hill, B.; Medeiros, A.; Tenover, F.C. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 1997, 41, 647–653. [Google Scholar] [CrossRef]
- Bonnet, R.; Dutour, C.; Sampaio, J.L.M.; Chanal, C.; Sirot, D.; Labia, R.; De Champs, C.; Sirot, J. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240 Gly. Antimicrob. Agents Chemother. 2001, 45, 2269–2275. [Google Scholar] [CrossRef][Green Version]
- Codjoe, F.S.; Brown, C.A.; Smith, T.J.; Miller, K.; Donkor, E.S. Genetic relatedness in carbapenem-resistant isolates from clinical specimens in Ghana using ERIC-PCR technique. PLoS ONE 2019, 14, e0222168. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ishikawa, J. FramePlot: A new implementation of the Frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 1999, 174, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Hunter, P.R. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 1990, 28, 1903–1905. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, L.A.; Feeney, B.C. A Simple Guide to IBM SPSS Statistics for Version 20.0, 12th ed.; Wadsworth Cengage Learning: Belmont, CA, USA, 2013; ISBN 9781285086019. [Google Scholar]
- Tandogdu, Z.; Wagenlehner, F.M. Global epidemiology of urinary tract infections. Curr. Opin. Infect. Dis. 2016, 29, 73–79. [Google Scholar] [CrossRef]
- Hirsch, E.B.; Zucchi, P.C.; Chen, A.; Raux, B.R.; Kirby, J.E.; McCoy, C.; Eliopoulos, G.M. Susceptibility of Multidrug-Resistant Gram-Negative Urine Isolates to Oral Antibiotics. Antimicrob. Agents Chemother. 2016, 60, 3138–3140. [Google Scholar] [CrossRef]
- Bader, M.S.; Loeb, M.; Brooks, A. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad. Med. 2016, 129, 242–258. [Google Scholar] [CrossRef]
- Kotb, S.; Lyman, M.; Ismail, G.; El Fattah, M.A.; Girgis, S.A.; Etman, A.; Hafez, S.; El-Kholy, J.; Zaki, M.E.S.; Rashed, H.-A.G.; et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare–associated Infections Surveillance Data, 2011–2017. Antimicrob. Resist. Infect. Control. 2020, 9, 1–9. [Google Scholar] [CrossRef]
- Livermore, D.M.; Mushtaq, S.; Warner, M.; Zhang, J.C.; Maharjan, S.; Doumith, M.; Woodford, N. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J. Antimicrob. Chemother. 2011, 66, 48–53. [Google Scholar] [CrossRef]
- Livermore, D.M.; Warner, M.; Mushtaq, S.; Doumith, M.; Zhang, J.; Woodford, N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int. J. Antimicrob. Agents 2011, 37, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, S.S.; Abdellatif, G.R.; El-Ansary, M.R.; Aboshanab, K.M.; Ragab, Y.M. Carbapenemase Producers Among Extensive Drug-Resistant Gram-Negative Pathogens Recovered from Febrile Neutrophilic Patients in Egypt. Infect. Drug Resist. 2020, 13, 3113–3124. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Xu-McCrae, L.; Mushtaq, S.; Wu, H.H.T.; Ellington, M.J.; Lancaster, O.; Davies, F.; Donaldson, H.; Rao, G.G.; Verma, A.; et al. Prevalence of carbapenem resistance and carbapenemase production among Enterobacteriaceae isolated from urine in the UK: Results of the UK infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-UK). J. Antimicrob. Chemother. 2018, 73, 698–702. [Google Scholar] [CrossRef]
- Giani, T.; Antonelli, A.; Sennati, S.; Di Pilato, V.; Chiarelli, A.; Cannatelli, A.; Gatsch, C.; Luzzaro, F.; Spanu, T.; Stefani, S.; et al. Results of the Italian infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-IT): Activity of ceftazidime/avibactam against Enterobacterales isolated from urine. J. Antimicrob. Chemother. 2020, 75, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Eshetie, S.; Unakal, C.; Gelaw, A.; Ayelign, B.; Endris, M.; Moges, F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob. Resist. Infect. Control. 2015, 4, 12. [Google Scholar] [CrossRef]
- Kamel, N.A.; El-Tayeb, W.N.; El-Ansary, M.; Mansour, M.T.; Aboshanab, K.M. Phenotypic screening and molecular characterization of carbapenemase-producing Gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PLoS ONE 2018, 13, e0202119. [Google Scholar] [CrossRef]
- El-Domany, R.A.; Emara, M.; El-Magd, M.A.; Moustafa, W.H.; Abdeltwab, N.M. Emergence of Imipenem-ResistantPseudomonas aeruginosaClinical Isolates from Egypt CoharboringVIMandIMPCarbapenemases. Microb. Drug Resist. 2017, 23, 682–686. [Google Scholar] [CrossRef]
- Göttig, S.; Gruber, T.M.; Stecher, B.; Wichelhaus, T.A.; Kempf, V.A.J. In Vivo Horizontal Gene Transfer of the Carbapenemase OXA-48 During a Nosocomial Outbreak. Clin. Infect. Dis. 2015, 60, 1808–1815. [Google Scholar] [CrossRef] [PubMed]


| PCR Reaction | Gene | Primer | Primer Sequence (5′ → 3′) | Expected PCR Product Size (bp) | Ta (°C) | References |
|---|---|---|---|---|---|---|
| Multiplex | blaKPC | Pf | TGTCACTGTATCGCCGTC | 1011 | 50 | [28] |
| Pr | CTCAGTGCTCTACAGAAAACC | |||||
| blaNDM | Pf | GGTTTGGCGATCTGGTTTTC | 621 | [29,30] | ||
| Pr | CGGAATGGCTCATCACGAT | |||||
| Multiplex | blaVIM | Pf | TCTACATGACCGCGTCTGTC | 748 | 50 | [31] |
| Pr | TGTGCTTTGACAACGTTCGC | |||||
| blaOXA-48 | Pf | GCGTGGTTAAGGATGAACAC | 438 | [28,30] | ||
| Pr | CATCAAGTTCAACCCAACCG | |||||
| Monoplex | blaIMP | Pf | CTACCGCAGCAGAGTCTTTG | 587 | 50 | [32] |
| Pr | AACCAGTTTTGCCTTACCAT | |||||
| Multiplex | aac(6′)-Ib | Pf | TTGCGATGCTCTATGAGTGG | 358 | 49 | [33] |
| Pr | CGTTTGGATCTTGGTGACCT | |||||
| blaSHV | Pf | GGTTATGCGTTATATTCGCC | 867 | [34] | ||
| Pr | TTAGCGTTGCCAGTGCTC | |||||
| Multiplex | blaCTX-M | Pf | CGCTTTGCGATGTGCAG | 550 | 51 | [35] |
| Pr | ACCGCGATATCGTTGGT | |||||
| blaTEM | Pf | ATGAGTATTCAACATTTCCG | 867 | [34] | ||
| Pr | CTGACAGTTACCAATGCTTA | |||||
| ERIC-PCR | Pf | AAGTAAGTGACTGGGGTGAGCG | Variable | 45 | [36] | |
| Pr | ATGTAAGCTCCTGGGGATTCAC | |||||
| PCR Reaction | Primary Denaturation | Secondary Denaturation | Annealing | Extension | No. of Cycles | Final Extension |
|---|---|---|---|---|---|---|
| Multiplex blaKPC/blaNDM | 95 °C 4 min | 95 °C 30 s | 50 °C 45 s | 72 °C 1 min | 30 | 72 °C 10 min |
| Multiplex blaVIM/blaOXA-48 | 95 °C 4 min | 95 °C 30 s | 50 °C 45 s | 72 °C 1 min | 30 | 72 °C 10 min |
| Monoplex blaIMP | 95 °C 4 min | 95 °C 30 s | 50 °C 45 s | 72 °C 1 min | 30 | 72 °C 10 min |
| Multiplex aac(6′)-Ib/blaSHV | 95 °C 4 min | 95 °C 30 s | 49 °C 45 s | 72 °C 1 min | 30 | 72 °C 10 min |
| Multiplex blaCTX-M/blaTEM | 95 °C 4 min | 95 °C 30 s | 51 °C 45 s | 72 °C 1 min | 30 | 72 °C 10 min |
| ERIC-PCR | 95 °C 15 min | 94 °C 30 s | 45 °C 45 s | 72 °C 7 min | 45 | 72 °C 10 min |
| Antimicrobial Class | Antimicrobial Agent | Percentage of Resistance (%) | |||||
|---|---|---|---|---|---|---|---|
| Klebsiella spp. (n = 28) | P. aeruginosa (n = 19) | A. baumannii (n = 8) | E. coli (n = 6) | Proteus mirabilis (n = 2) | Enterobacter cloacae (n = 2) | ||
| Carbapenems | Imipenem (10 µg) | 64.3 | 89.5 | 100 | 33.3 | 50 | 0 |
| Meropenem (10 µg) | 71.4 | 94.7 | 87.5 | 16.7 | 50 | 0 | |
| Ertapenem (10 µg) | 92.9 | ND | ND | 100 | 100 | 100 | |
| Doripenem (10 µg) | 46.4 | 42.1 | 25 | 16.7 | 50 | 0 | |
| β-lactam combination agents | Amoxicillin/clavulanic acid (30 µg) | 46.4 | ND | ND | 16.7 | 50 | 0 |
| Ampicillin/sulbactam (20 µg) | 89.3 | ND | 25 | 50 | 50 | 50 | |
| Cephalosporins | Cefoxitin (30 µg) | 89.3 | ND | ND | 83.3 | 50 | 50 |
| Ceftazidime (30 µg) | 85.7 | 94.7 | 87.5 | 100 | 50 | 100 | |
| Ceftriaxone (30 µg) | 89.3 | ND | 87.5 | 100 | 50 | 100 | |
| Cefepime (30 µg) | 78.6 | 78.9 | 87.5 | 100 | 50 | 50 | |
| Aminoglycosides | Amikacin (30 µg) | 78.6 | 47.4 | 100 | 0 | 50 | 0 |
| Gentamicin (10 µg) | 64.3 | 89.5 | 100 | 0 | 50 | 100 | |
| Fluoroquinolones | Ciprofloxacin (5 µg) | 92.9 | 100 | 100 | 100 | 100 | 100 |
| Levofloxacin (5 µg) | 92.9 | 100 | 62.5 | 100 | 50 | 100 | |
| Folate pathway inhibitors | Trimethoprim/sulfamethoxazole (25 µg) | 78.6 | ND | 50 | 100 | 100 | 100 |
| Nitrofurans | Nitrofurantoin (300 µg) | 92.9 | ND | ND | 0 | 0 | 0 |
| Fosfomycins | Fosfomycin (200 µg) | 10.7 | ND | ND | 0 | 0 | 50 |
| Tested Isolates | Blue-Carba Test (n = 65) [26] | Modified Carbapenem Inactivation Method (mCIM) (n = 57) [25] | EDTA-Modified Carbapenem Inactivation Method (eCIM) (n = 29) [25] | |||
|---|---|---|---|---|---|---|
| No. of CPs/Total No. of Tested Isolates | % | No. of CPs/Total No. of Tested Isolates | % | No. of MBL Producers/Total No. of Tested Isolates | % | |
| Klebsiella spp. | 22/28 | 78.6 | 23/28 | 82.1 | 12/23 | 52.2 |
| P. aeruginosa | 7/19 | 36.8 | 2/19 | 10.5 | ND | ND |
| A. baumannii | 1/8 | 12.5 | ND | ND | ND | ND |
| E. coli | 3/6 | 50 | 4/6 | 66.7 | 3/4 | 75 |
| Proteus mirabilis | 2/2 | 100 | 1/2 | 50 | 0/1 | 0 |
| Enterobacter cloacae | 1/2 | 50 | 1/2 | 50 | 1/1 | 100 |
| TS24.SK | 24.SK | TS37.AK | 37.AK | TS59.WK | 59.WK | TS66.WK | 66.WK | E. coli DH5α | |
|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial susceptibility testing | |||||||||
| Imipenem (10 µg) | I | I | I | R | I | R | I | I | S |
| Meropenem (10 µg) | R | R | S | R | S | R | S | R | S |
| Ertapenem (10 µg) | R | R | S | R | S | R | S | R | S |
| Doripenem (10 µg) | S | I | S | S | S | R | S | R | S |
| Amoxicillin/clavulanic acid (30 µg) | S | R | S | S | S | R | S | I | S |
| Ampicillin/sulbactam (20 µg) | R | R | R | R | R | R | R | R | S |
| Cefoxitin (30 µg) | R | R | R | R | R | R | R | R | S |
| Ceftazidime (30 µg) | R | R | R | R | R | R | R | R | S |
| Ceftriaxone (30 µg) | I | I | I | R | I | R | R | R | S |
| Cefepime (30 µg) | SDD | SDD | S | R | S | R | SDD | R | S |
| Amikacin (30 µg) | S | S | S | R | S | R | S | R | S |
| Gentamicin (10 µg) | S | S | S | R | S | R | S | S | S |
| Ciprofloxacin (5 µg) | R | R | S | R | S | R | S | R | S |
| Levofloxacin (5 µg) | R | R | S | R | S | R | S | R | S |
| Trimethoprim/sulfamethoxazole (25 µg) | S | S | S | R | S | R | S | R | S |
| Nitrofurantoin (300 µg) | S | R | S | S | S | R | S | R | S |
| Fosfomycin (200 µg) | S | S | S | R | S | S | S | S | S |
| Blue-Carba test | |||||||||
| + | + | + | + | + | + | + | + | - | |
| Modified carbapenem inactivation method (mCIM) | |||||||||
| + | + | + | + | + | + | + | + | - | |
| Parental Strain | Transformant Code | Plasmid Code | Acquired Transformant Phenotype | Plasmid Genotype | Plasmid Size in Gel | |
|---|---|---|---|---|---|---|
| 24.SK | TS24.SK | pKPT24 | Imipenem (10 µg) | I | blaOXA-48/blaCTX-M | 2 plasmid bands: Large plasmid: >10 kb Small plasmid: ≈1700 bp |
| Meropenem (10 µg) | R | |||||
| Ertapenem (10 µg) | R | |||||
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| Cefepime (30 µg) | SDD | |||||
| Ciprofloxacin (5 µg) | R | |||||
| Levofloxacin (5 µg) | R | |||||
| 37.AK | TS37.AK | pKPT37 | Imipenem (10 µg) | I | blaOXA-48 | 2 plasmid bands: Large plasmid: >10 kb Small plasmid: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| 59.WK | TS59.WK | pKPT59 | Imipenem (10 µg) | I | blaOXA-48/blaTEM/blaaac(6′)-Ib | 1 plasmid band: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | I | |||||
| 66.WK | TS66.WK | pKPT66 | Imipenem (10 µg) | I | blaOXA-48/blaCTX-M | 1 plasmid band: >10 kb |
| Ampicillin/sulbactam (20 µg) | R | |||||
| Cefoxitin (30 µg) | R | |||||
| Ceftazidime (30 µg) | R | |||||
| Ceftriaxone (30 µg) | R | |||||
| Cefepime (30 µg) | SDD | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshamy, A.A.; Saleh, S.E.; Alshahrani, M.Y.; Aboshanab, K.M.; Aboulwafa, M.M.; Hassouna, N.A. OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections. Biology 2021, 10, 889. https://doi.org/10.3390/biology10090889
Elshamy AA, Saleh SE, Alshahrani MY, Aboshanab KM, Aboulwafa MM, Hassouna NA. OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections. Biology. 2021; 10(9):889. https://doi.org/10.3390/biology10090889
Chicago/Turabian StyleElshamy, Ann A., Sarra E. Saleh, Mohammad Y. Alshahrani, Khaled M. Aboshanab, Mohammad M. Aboulwafa, and Nadia A. Hassouna. 2021. "OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections" Biology 10, no. 9: 889. https://doi.org/10.3390/biology10090889
APA StyleElshamy, A. A., Saleh, S. E., Alshahrani, M. Y., Aboshanab, K. M., Aboulwafa, M. M., & Hassouna, N. A. (2021). OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections. Biology, 10(9), 889. https://doi.org/10.3390/biology10090889







