Phenotype Switching in Metal-Tolerant Bacteria Isolated from a Hyperaccumulator Plant
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
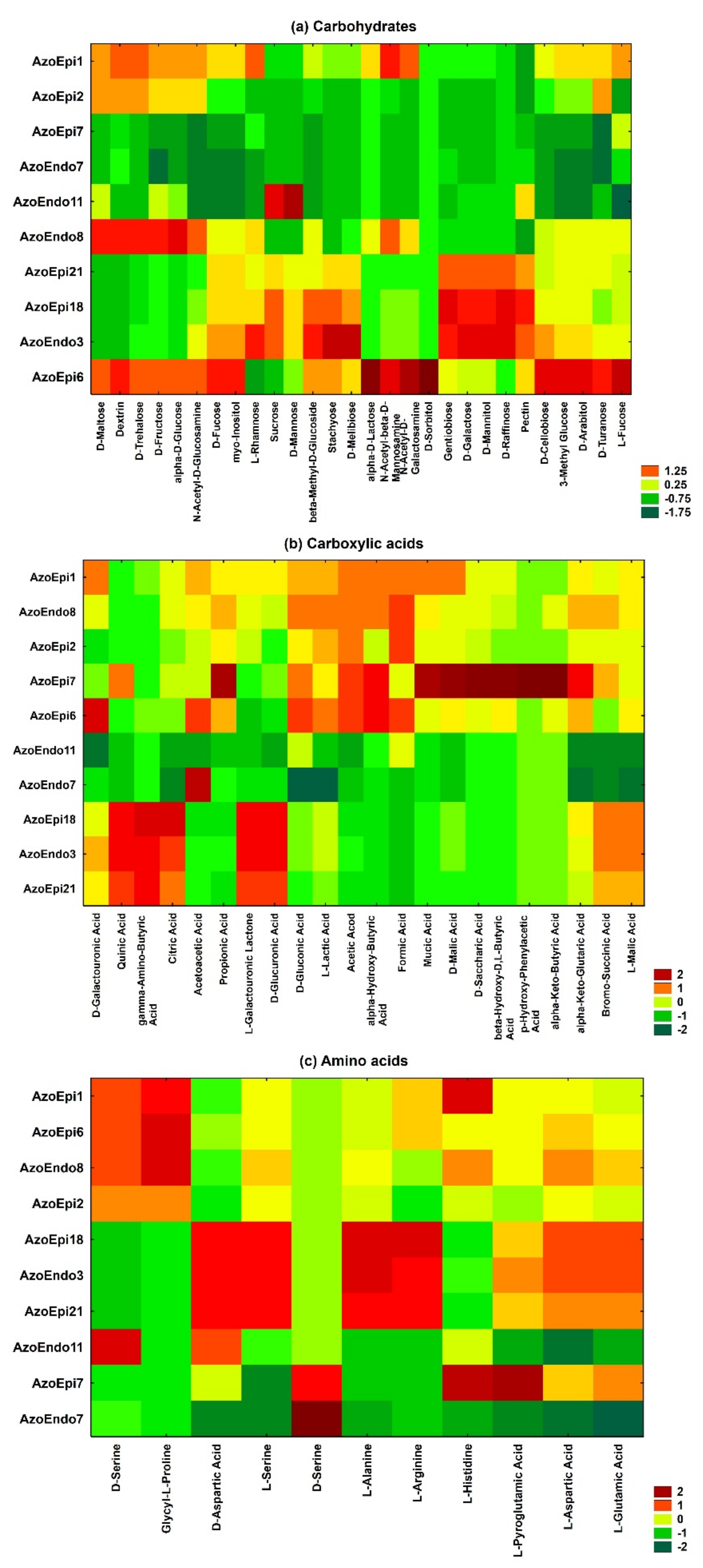
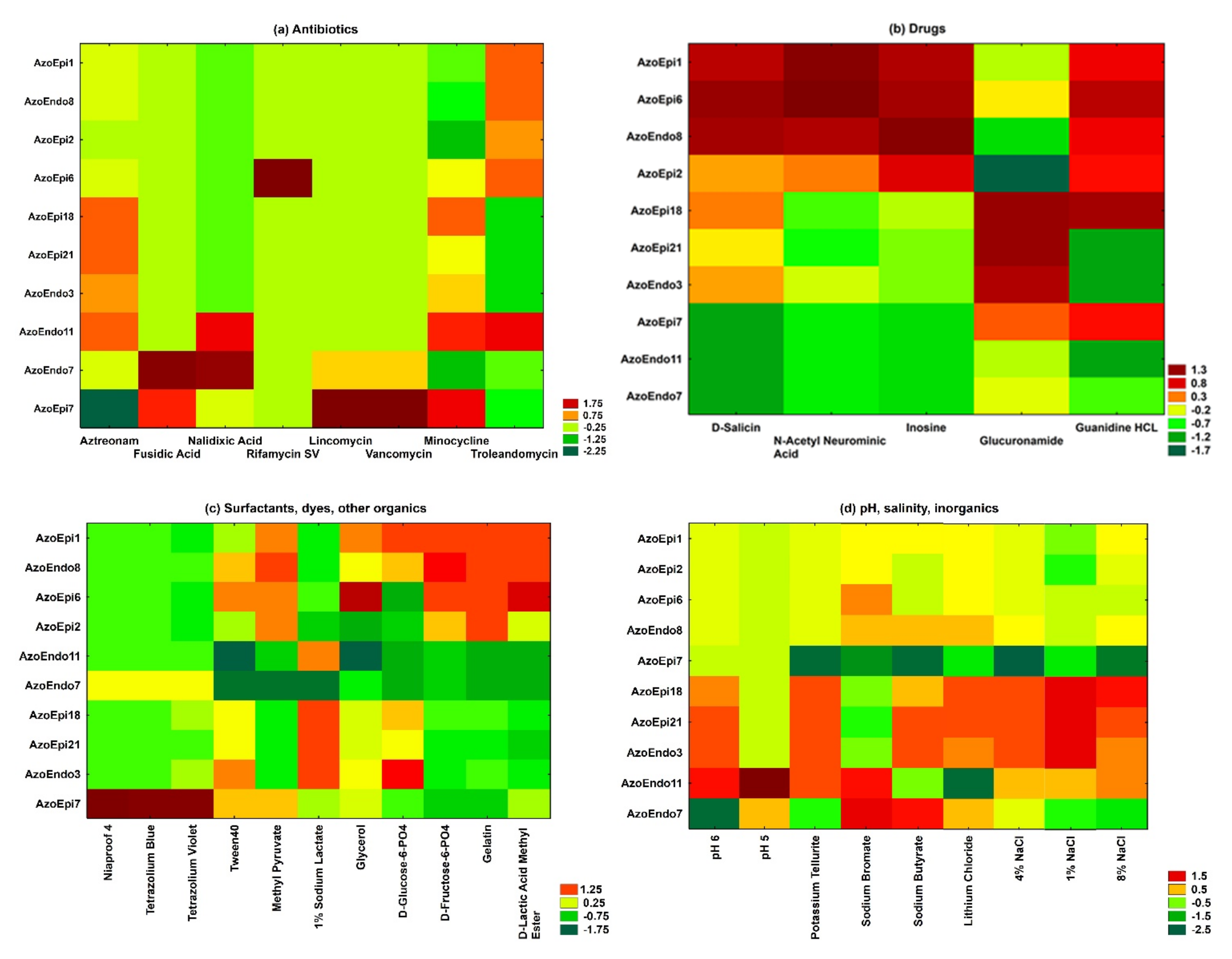
2.1. Phenotypic Profile Using BiologTM GEN III MicroPlates
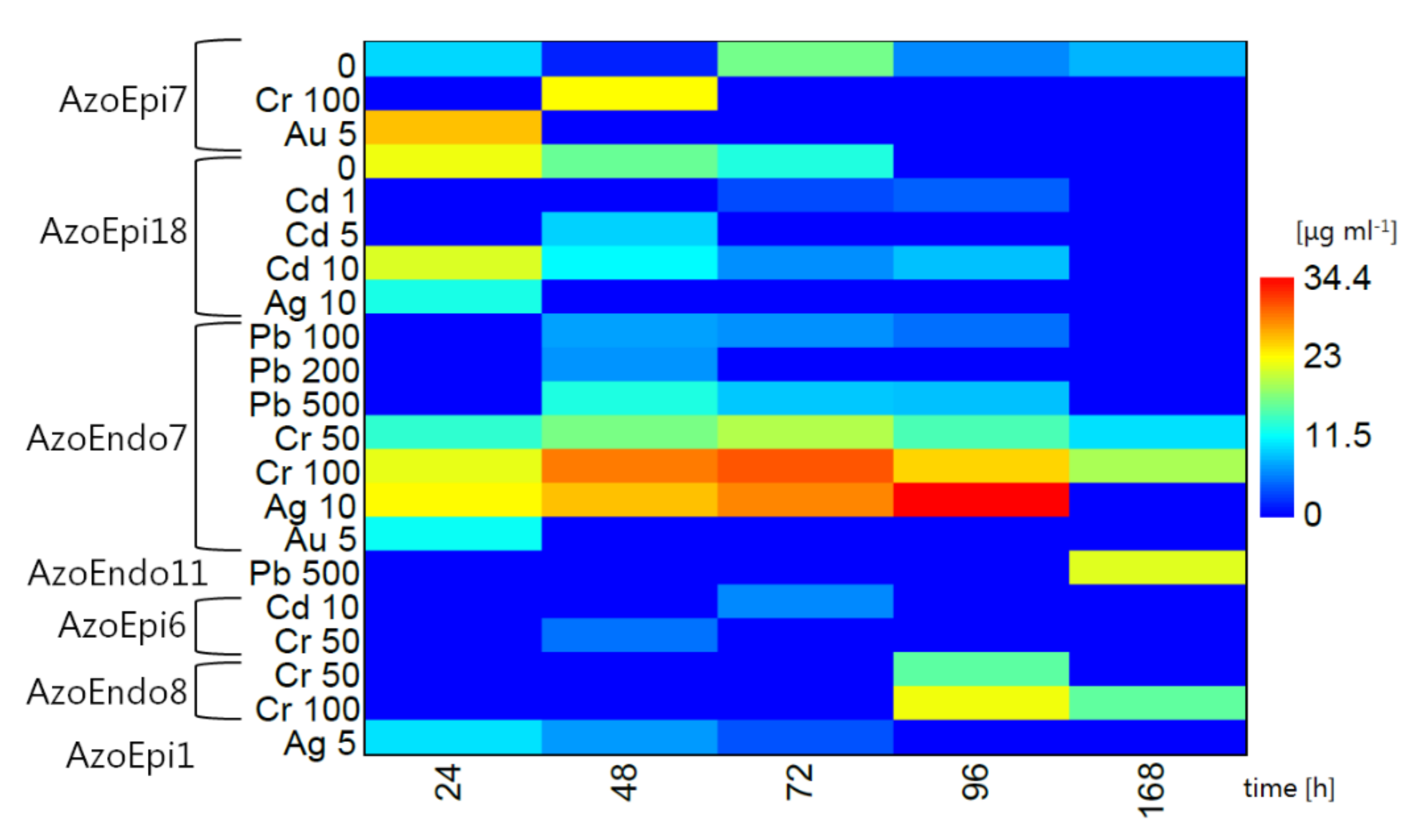
2.2. Metal Tolerance
2.3. Synthesis of Indole-3-Acetic Acid
2.4. Enzymatic Assays
2.4.1. Cellulolytic Activity
2.4.2. Xylanolytic Activity
2.4.3. Proteolytic Activity
2.5. Statistical Procedures
3. Results
3.1. Phenotypic Characterization of Bacteria Using BiologTM GEN III MicroPlates
3.2. Tolerance of Microorganisms to Heavy Metals
3.3. Synthesis of IAA under Heavy Metal Stress
3.4. Cellulolytic Activities in the Presence of Heavy Metals
3.5. Proteolytic Activities in the Presence of Heavy Metals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef] [Green Version]
- Vandana, U.K.; Rajkumari, J.; Singha, L.P.; Satish, L.; Alavilli, H.; Sudheer, P.D.V.N.; Chauhan, S.; Ratnala, R.; Satturu, V.; Mazumder, P.B.; et al. The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion. Biology 2021, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Megharaj, M.; Wu, C.-Y.; Subashchandrabose, S.R.; Dai, C.-C. Endophyte-assisted phytoremediation: Mechanisms and current application strategies for soil mixed pollutants. Crit. Rev. Biotechnol. 2020, 40, 31–45. [Google Scholar] [CrossRef]
- Acin-Albiac, M.; Filannino, P.; Gobbetti, M.; Di Cagno, R. Microbial high throughput phenomics: The potential of an irreplaceable omics. Comput. Struct. Biotechnol. J. 2020, 18, 2290–2299. [Google Scholar] [CrossRef]
- Madsen, E.L. Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Nazli, F.; Mustafa, A.; Ahmad, M.; Hussain, A.; Jamil, M.; Wang, X.; Shakeel, Q.; Imtiaz, M.; El-Esawi, M.A. A Review on Practical Application and Potentials of Phytohormone-Producing Plant Growth-Promoting Rhizobacteria for Inducing Heavy Metal Tolerance in Crops. Sustainability 2020, 12, 9056. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Sesin, V.; Kisiala, A.; Emery, R.J.N. Phytohormonal Roles in Plant Responses to Heavy Metal Stress: Implications for Using Macrophytes in Phytoremediation of Aquatic Ecosystems. Environ. Toxicol. Chem. 2021, 40, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Banach, A.M.; Banach, K.; Stępniewska, Z. Phytoremediation as a promising technology for water and soil purification: Azolla caroliniana Willd. As a case study. Acta Agrophysica 2012, 19, 241–252. [Google Scholar]
- Banach, A.; Kuźniar, A.; Mencfel, R.; Wolińska, A. The Study on the Cultivable Microbiome of the Aquatic Fern Azolla Filiculoides L. as New Source of Beneficial Microorganisms. Appl. Sci. 2019, 9, 2143. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.; Weber, R. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Karry, F.; Abdallah, M.B.; Kallel, N.; Hamza, M.; Fakhfakh, M.; Sayadi, S. Extracellular hydrolytic enzymes produced by halophilic bacteria and archaea isolated from hypersaline lake. Mol. Biol. Rep. 2018, 45, 1297–1309. [Google Scholar] [CrossRef]
- Menon, G.; Mody, K.; Keshri, J.; Jha, B. Isolation, purification, and characterization of haloalkaline xylanase from a marine Bacillus pumilus strain, GESF-1. Biotechnol. Bioproc. E 2010, 15, 998–1005. [Google Scholar] [CrossRef]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.M.; Seo, C.W.; Lee, I.J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flimban, S.; Oh, S.-E.; Joo, J.H.; Hussein, K.A. Characterization and Identification of Cellulose-degrading Bacteria Isolated from a Microbial Fuel Cell Reactor. Biotechnol. Bioproc. E 2019, 24, 622–631. [Google Scholar] [CrossRef]
- Knapik, K.; Becerra, M.; González-Siso, M.-I. Microbial diversity analysis and screening for novel xylanase enzymes from the sediment of the Lobios Hot Spring in Spain. Sci. Rep. 2019, 9, 11195. [Google Scholar] [CrossRef] [PubMed]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3 Biotech 2020, 10, 361. [Google Scholar] [CrossRef]
- Aslam, S.; Hussain, A.; Qazi, J.I. Production of Cellulase by Bacillus amyloliquefaciens-ASK11 Under High Chromium Stress. Waste Biomass Valor. 2019, 10, 53–61. [Google Scholar] [CrossRef]
- Bücker-Neto, L.; Paiva, A.L.S.; Machado, R.D.; Arenhart, R.A.; Margis-Pinheiro, M. Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 2017, 40, 373–386. [Google Scholar] [CrossRef]
- Wang, K.; Cao, R.; Wang, M.; Lin, Q.; Zhan, R.; Xu, H.; Wang, S. A novel thermostable GH10 xylanase with activities on a wide variety of cellulosic substrates from a xylanolytic Bacillus strain exhibiting significant synergy with commercial Celluclast 1.5 L in pretreated corn stover hydrolysis. Biotechnol. Biofuels 2019, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Woźniak, M.; Gałązka, A.; Tyśkiewicz, R.; Jaroszuk-Ściseł, J. Endophytic Bacteria Potentially Promote Plant Growth by Synthesizing Different Metabolites and their Phenotypic/Physiological Profiles in the Biolog GEN III MicroPlateTM Test. Int. J. Mol. Sci. 2019, 20, 5283. [Google Scholar] [CrossRef] [Green Version]
- Mashiguchi, K.; Hisano, H.; Takeda-Kamiya, N.; Takebayashi, Y.; Ariizumi, T.; Gao, Y.; Ezura, H.; Sato, K.; Zhao, Y.; Hayashi, K.; et al. Agrobacterium tumefaciens Enhances Biosynthesis of Two Distinct Auxins in the Formation of Crown Galls. Plant Cell Physiol. 2019, 60, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Liu, Z.; Nan, L.; Wang, E.; Chen, W.; Lin, Y.; Wei, G. Isolation, characterization, and selection of heavy metal-resistant and plant growth-promoting endophytic bacteria from root nodules of Robinia pseudoacacia in a Pb/Zn mining area. Microbiol. Res. 2018, 217, 51–59. [Google Scholar] [CrossRef]
- Corretto, E.; Antonielli, L.; Sessitsch, A.; Höfer, C.; Puschenreiter, M.; Widhalm, S.; Swarnalakshmi, K.; Brader, G. Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance. Front. Microbiol. 2020, 11, 1869. [Google Scholar] [CrossRef] [PubMed]
- Morel, M.A.; Ubalde, M.C.; Braña, V.; Castro-Sowinski, S. Delftia sp. JD2: A potential Cr(VI)-reducing agent with plant growth-promoting activity. Arch. Microbiol. 2011, 193, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Luo, S.; Wu, Y.; Ye, J.; Wang, Q.; Xu, X.; Pan, F.; Khan, K.Y.; Feng, Y.; Yang, X. The Effects of the Endophytic Bacterium Pseudomonas fluorescens Sasm05 and IAA on the Plant Growth and Cadmium Uptake of Sedum alfredii Hance. Front. Microbiol. 2017, 8, 2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatima, H.; Ahmed, A. Indole-3-acetic acid synthesizing chromium-resistant bacteria can mitigate chromium toxicity in Helianthus annuus L. Plant Soil Environ. 2020, 66, 216–221. [Google Scholar] [CrossRef]
- He, X.; Xu, M.; Wei, Q.; Tang, M.; Guan, L.; Lou, L.; Xu, X.; Hu, Z.; Chen, Y.; Shen, Z.; et al. Promotion of growth and phytoextraction of cadmium and lead in Solanum nigrum L. mediated by plant-growth-promoting rhizobacteria. Ecotoxicol. Environ. Saf. 2020, 205, 111333. [Google Scholar] [CrossRef]
- Yu, X.; Li, Y.; Zhang, C.; Liu, H.; Liu, J.; Zheng, W.; Kang, X.; Leng, X.; Zhao, K.; Gu, Y.; et al. Culturable Heavy Metal-Resistant and Plant Growth Promoting Bacteria in V-Ti Magnetite Mine Tailing Soil from Panzhihua, China. PLoS ONE 2014, 9, e106618. [Google Scholar] [CrossRef] [PubMed]
- Efe, D. Potential Plant Growth-Promoting Bacteria with Heavy Metal Resistance. Potential Plant Growth-Promoting Bacteria with Heavy Metal Resistance. Curr. Microbiol. 2020, 77, 3861–3868. [Google Scholar] [CrossRef]
- Panichikkal, J.; Roshmi, T.; Jimtha, J.J.; Radhakrishnan, E.K. Biogenic Gold Nanoparticle Supplementation to Plant Beneficial Pseudomonas monteilii was Found to Enhance its Plant Probiotic Effect. Curr. Microbiol. 2019, 76, 503–509. [Google Scholar] [CrossRef]
- Kumar, P.; Pahal, V.; Gupta, A.; Vadhan, R.; Chandra, H.; Dubey, R.C. Effect of silver nanoparticles and Bacillus cereus LPR2 on the growth of Zea mays. Sci. Rep. 2020, 10, 20409. [Google Scholar] [CrossRef] [PubMed]
- Agustrina, R.; Ekowati, C.N.; Pasaribu, Y.S. Characterization of protease from Bacillus sp. on medium containing FeCl3 exposed to magnetic field 0.2 mt. IOP Conf. Ser. Earth Environ. Sci. 2018, 130, 012046. [Google Scholar] [CrossRef]
- Ahmad, E.; Sharma, S.K.; Sharma, P.K. Deciphering operation of tryptophan-independent pathway in high indole-3-acetic acid (IAA) producing Micrococcus aloeverae DCB-20. FEMS Microbiol. Lett. 2020, 367, fnaa190. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Hernández, J.C.; Perea-Vélez, Y.S.; Arriola-Morales, J.; Martínez-Simón, S.M.; Pérez-Osorio, G. Assessing the effects of heavy metals in ACC deaminase and IAA production on plant growth-promoting bacteria. Microbiol. Res. 2016, 188-189, 53–61. [Google Scholar] [CrossRef]
- Strader, L.C.; Beisner, E.R.; Bartel, B. Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell 2009, 21, 3585–3590. [Google Scholar] [CrossRef] [Green Version]
- Haddad, S.A.; Lemanowicz, J.; Abd El-Azeim, M.M. Cellulose decomposition in clay and sandy soils contaminated with heavy metals. Int. J. Environ. Sci. Technol. 2019, 16, 3275–3290. [Google Scholar] [CrossRef]
- Kandeler, E.; Kampichler, C.; Horak, O. Influence of heavy metals on the functional diversity of soil microbial communities. Biol. Fertil. Soils. 1996, 23, 299–306. [Google Scholar] [CrossRef]
- Li, Q.; Hu, Q.; Zhang, C.; Jin, Z. Effects of Pb, Cd, Zn, and Cu on Soil Enzyme Activity and Soil Properties Related to Agricultural Land-Use Practices in Karst Area Contaminated by Pb-Zn Tailings. Pol. J. Environ. Stud. 2018, 27, 2623–2632. [Google Scholar] [CrossRef]
- Kheirabadi, M.; Mahmoodi, R.; Mollania, N.; Kheirabadi, M. Fast chromium removal by Shewanella sp.: An enzymatic mechanism depending on serine protease. Int. J. Environ. Sci. Technol. 2020, 17, 143–152. [Google Scholar] [CrossRef]

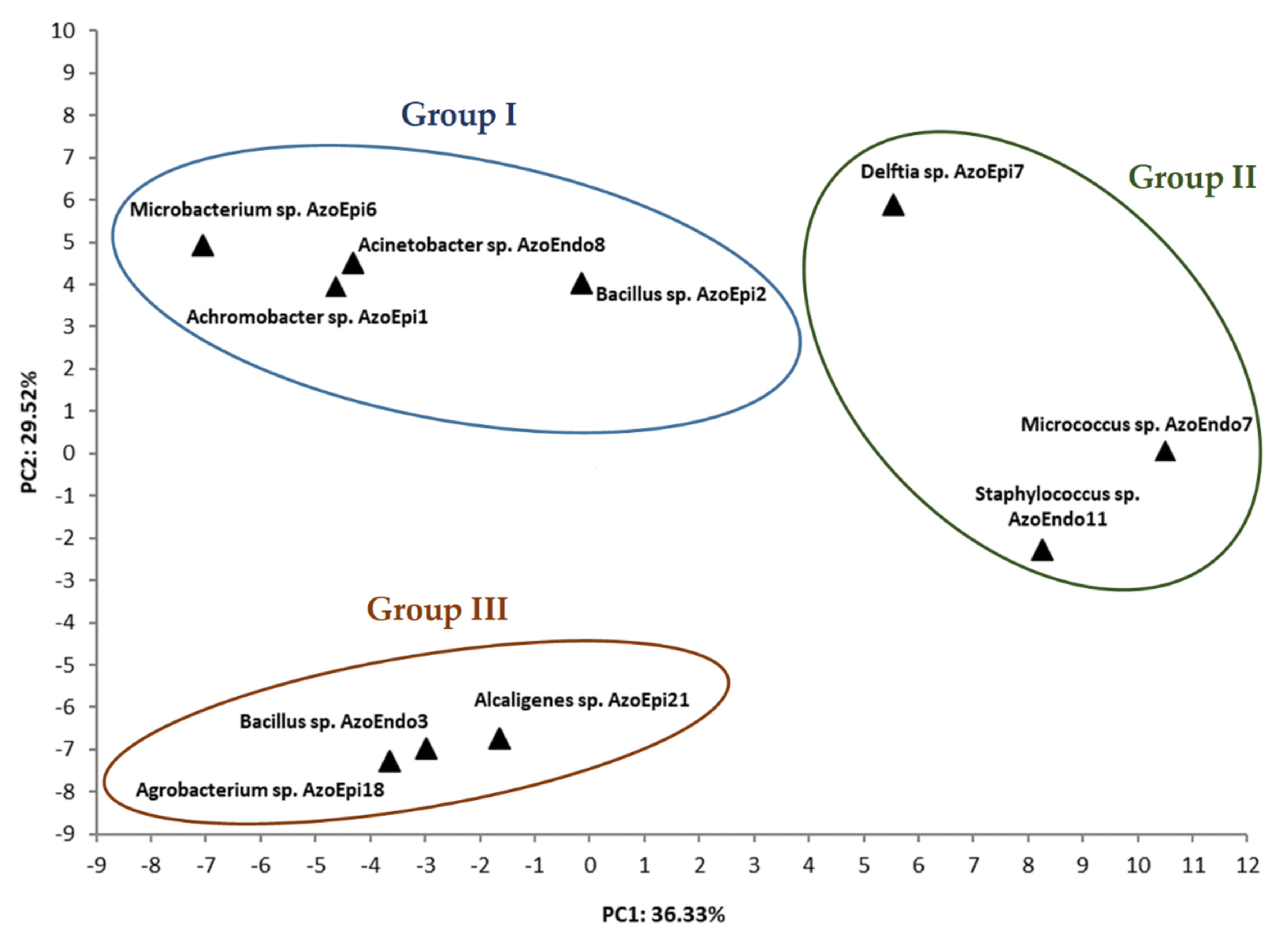
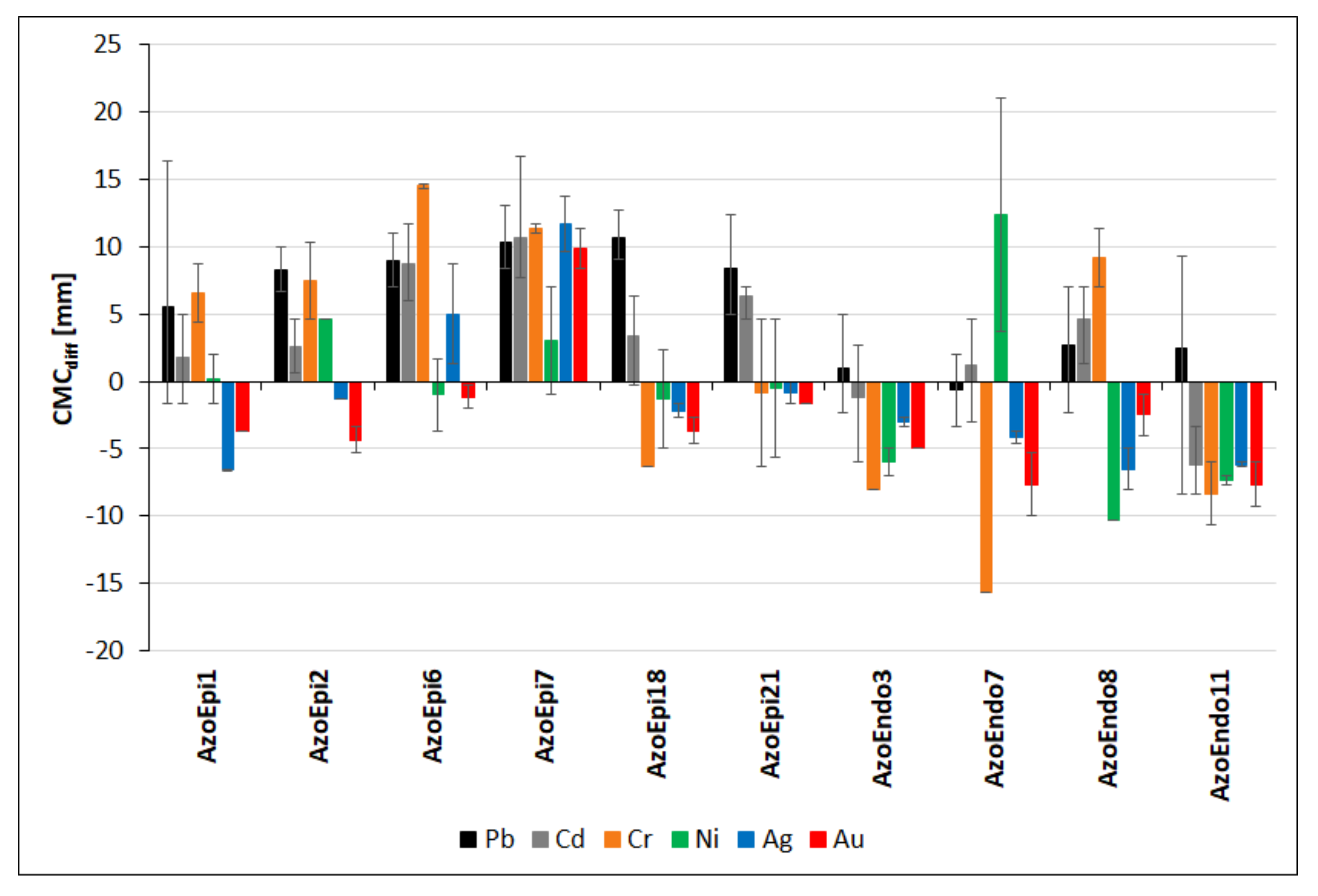
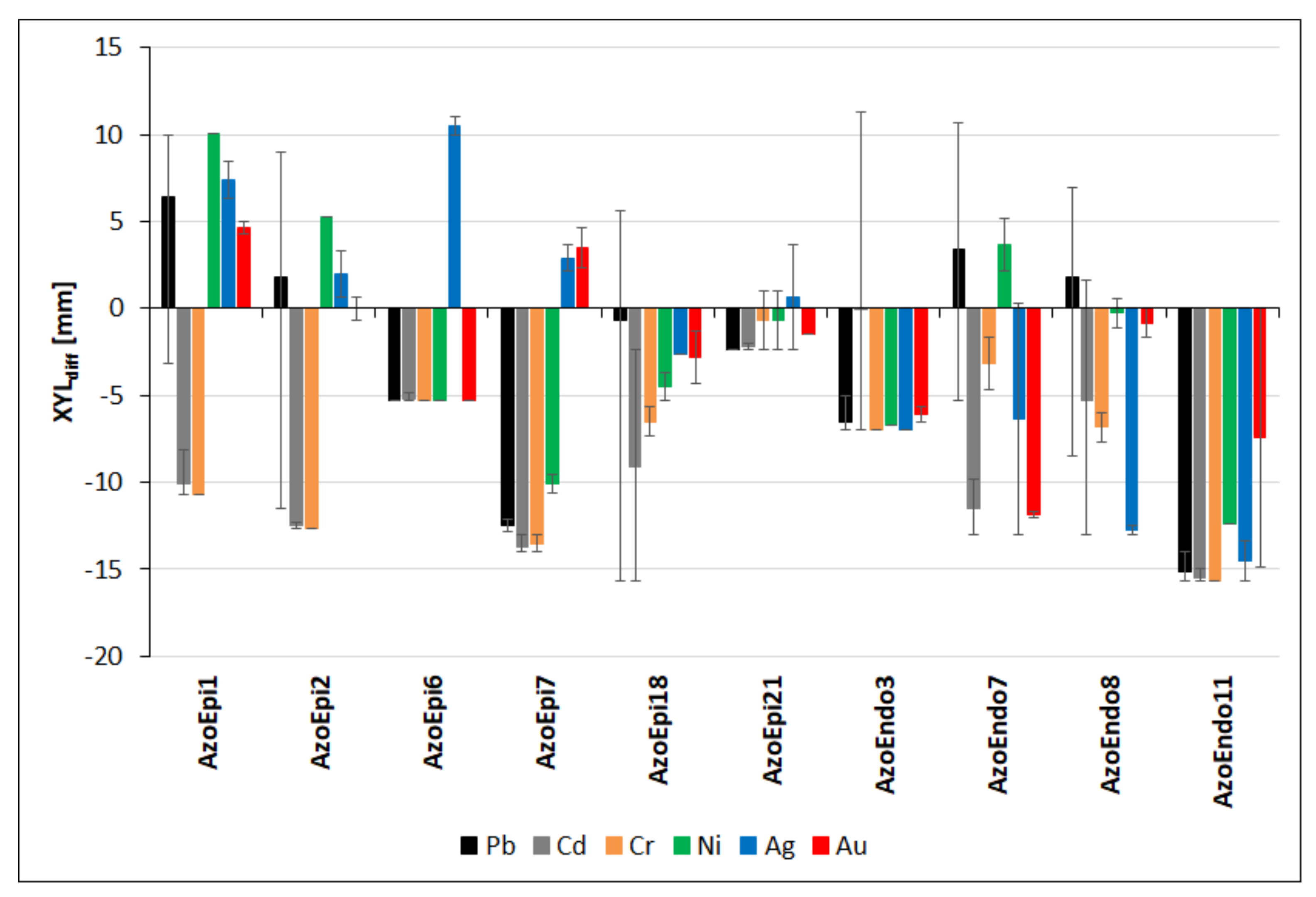
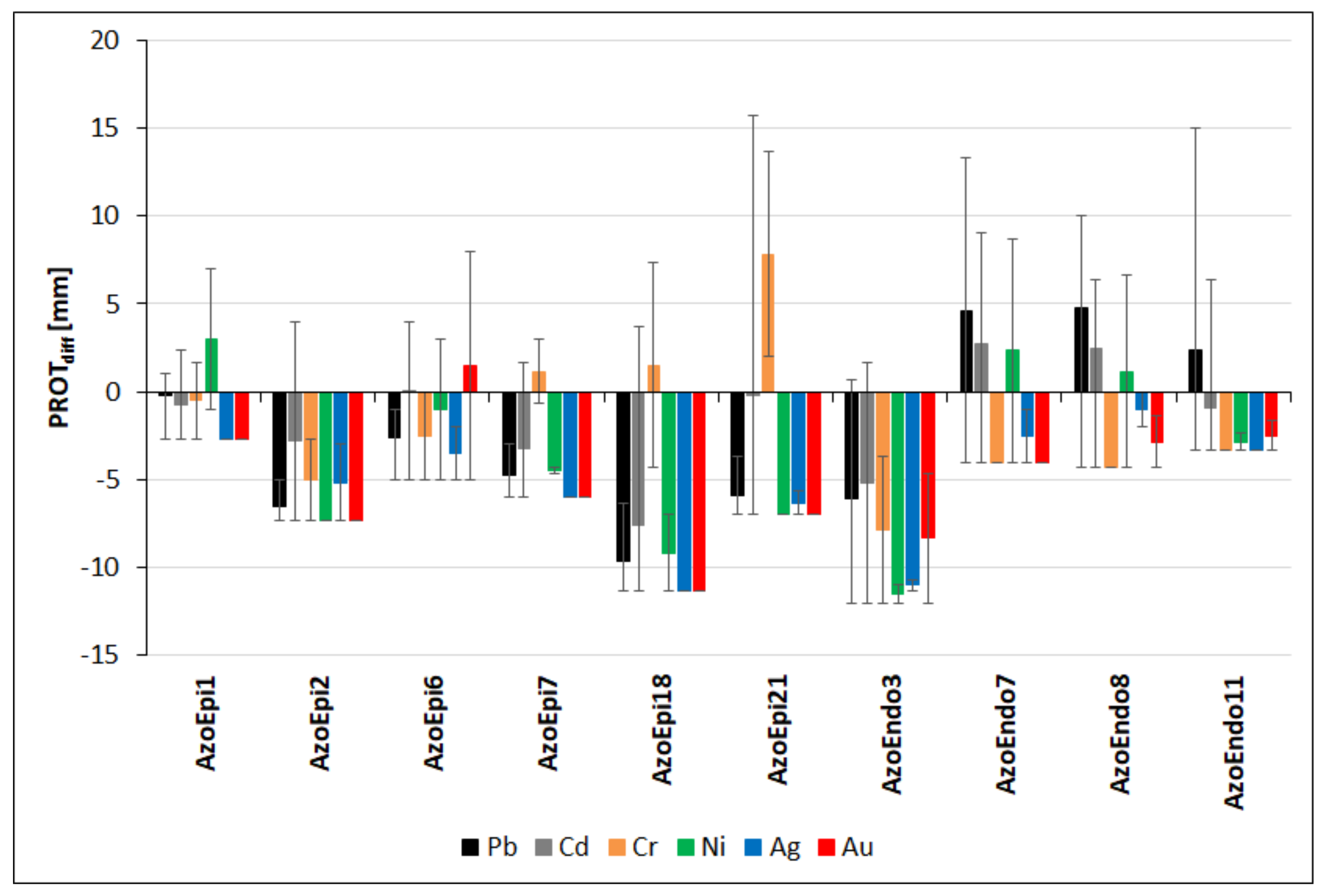
| Strain Name | Code | GenBank NO. |
|---|---|---|
| Achromobacter sp. AzoEpi1 | AzoEpi1 | MG881884 |
| Bacillus sp. AzoEpi2 | AzoEpi2 | MG881885 |
| Microbacterium sp. AzoEpi6 | AzoEpi6 | MG881889 |
| Delftia sp. AzoEpi7 | AzoEpi7 | MG881890 |
| Agrobacterium sp. AzoEpi18 | AzoEpi18 | MG881901 |
| Alcaligenes sp. AzoEpi21 | AzoEpi21 | MG881904 |
| Staphylococcus sp. AzoEndo11 | AzoEndo11 | MH605511 |
| Micrococcus sp. AzoEndo7 | AzoEndo7 | MG881917 |
| Bacillus sp. AzoEndo3 | AzoEndo3 | MG859254 |
| Acinetobacter sp. AzoEndo8 | AzoEndo8 | MG881918 |
| Strain | Pb | Cd | Cr | Ni | Ag | Au |
|---|---|---|---|---|---|---|
| Achromobacter sp. AzoEpi1 | 1000 | 10 | 100 | 100 | 10 | 5 |
| Bacillus sp. AzoEpi2 | 500 | 5 | 100 | 50 | 10 | 5 |
| Microbacterium sp. AzoEpi6 | 1000 | 10 | 100 | 50 | 10 | 5 |
| Delftia sp. AzoEpi7 | 200 | 0.5 | 100 | 100 | 10 | 10 |
| Agrobacterium sp. AzoEpi18 | 500 | 10 | 100 | 100 | 10 | 10 |
| Alcaligenes sp. AzoEpi21 | 200 | 10 | 100 | 100 | 10 | 5 |
| Staphylococcus sp. AzoEndo11 | 500 | 10 | 50 | 50 | 10 | 5 |
| Micrococcus sp. AzoEndo7 | 500 | 5 | 50 | 50 | 10 | 5 |
| Bacillus sp. AzoEndo3 | 1000 | 5 | 100 | 50 | 10 | 5 |
| Acinetobacter sp. AzoEndo8 | 1000 | 5 | 100 | 100 | 10 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banach, A.; Kuźniar, A.; Marzec-Grządziel, A.; Gałązka, A.; Wolińska, A. Phenotype Switching in Metal-Tolerant Bacteria Isolated from a Hyperaccumulator Plant. Biology 2021, 10, 879. https://doi.org/10.3390/biology10090879
Banach A, Kuźniar A, Marzec-Grządziel A, Gałązka A, Wolińska A. Phenotype Switching in Metal-Tolerant Bacteria Isolated from a Hyperaccumulator Plant. Biology. 2021; 10(9):879. https://doi.org/10.3390/biology10090879
Chicago/Turabian StyleBanach, Artur, Agnieszka Kuźniar, Anna Marzec-Grządziel, Anna Gałązka, and Agnieszka Wolińska. 2021. "Phenotype Switching in Metal-Tolerant Bacteria Isolated from a Hyperaccumulator Plant" Biology 10, no. 9: 879. https://doi.org/10.3390/biology10090879
APA StyleBanach, A., Kuźniar, A., Marzec-Grządziel, A., Gałązka, A., & Wolińska, A. (2021). Phenotype Switching in Metal-Tolerant Bacteria Isolated from a Hyperaccumulator Plant. Biology, 10(9), 879. https://doi.org/10.3390/biology10090879











